Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
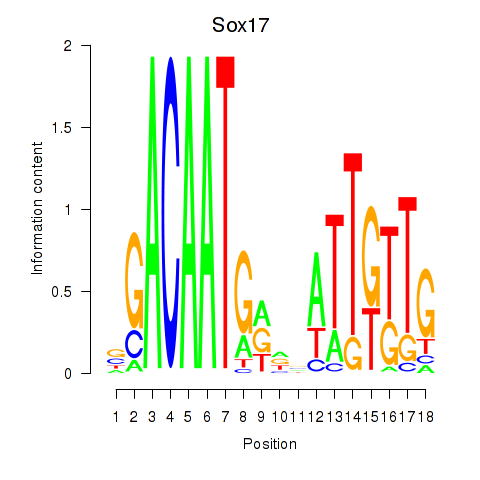
Results for Sox17
Z-value: 0.91
Transcription factors associated with Sox17
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Sox17
|
ENSRNOG00000027357 | SRY box 17 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Sox17 | rn6_v1_chr5_+_14890408_14890408 | -0.42 | 4.8e-15 | Click! |
Activity profile of Sox17 motif
Sorted Z-values of Sox17 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_161246351 | 37.25 |
ENSRNOT00000020348
|
Tnnc2
|
troponin C2, fast skeletal type |
| chr16_-_75581325 | 26.22 |
ENSRNOT00000058031
|
Defb33
|
defensin beta 33 |
| chr10_+_4953879 | 25.46 |
ENSRNOT00000003455
|
Tnp2
|
transition protein 2 |
| chr1_-_101210675 | 25.37 |
ENSRNOT00000028082
|
Dkkl1
|
dickkopf like acrosomal protein 1 |
| chr9_+_15582564 | 23.51 |
ENSRNOT00000020640
|
LOC100912849
|
uncharacterized LOC100912849 |
| chr1_+_215666628 | 20.53 |
ENSRNOT00000040598
ENSRNOT00000066135 ENSRNOT00000051425 ENSRNOT00000080339 ENSRNOT00000066896 ENSRNOT00000063918 |
Tnnt3
|
troponin T3, fast skeletal type |
| chr14_+_96300596 | 19.30 |
ENSRNOT00000071967
|
LOC302228
|
similar to Spindlin-like protein 2 (SPIN-2) |
| chr13_-_47331187 | 19.26 |
ENSRNOT00000035577
ENSRNOT00000049915 |
Zp3r
|
zona pellucida 3 receptor |
| chrX_+_43745604 | 19.01 |
ENSRNOT00000040825
|
LOC100362173
|
rCG36365-like |
| chr16_+_72086878 | 18.62 |
ENSRNOT00000023756
ENSRNOT00000078085 |
Adam3a
|
ADAM metallopeptidase domain 3A |
| chr1_-_67302751 | 17.62 |
ENSRNOT00000041518
|
Vom1r42
|
vomeronasal 1 receptor 42 |
| chrX_+_133227660 | 17.59 |
ENSRNOT00000004591
|
Actrt1
|
actin-related protein T1 |
| chr19_-_53477420 | 16.65 |
ENSRNOT00000071344
|
LOC687560
|
hypothetical protein LOC687560 |
| chr4_+_5644260 | 16.44 |
ENSRNOT00000041876
|
Actr3b
|
ARP3 actin related protein 3 homolog B |
| chr9_+_4731914 | 15.62 |
ENSRNOT00000061893
|
AABR07066201.1
|
|
| chr11_+_62584959 | 15.46 |
ENSRNOT00000071065
|
Gramd1c
|
GRAM domain containing 1C |
| chr10_-_70735742 | 15.32 |
ENSRNOT00000077035
|
Heatr9
|
HEAT repeat containing 9 |
| chr3_-_149356160 | 15.32 |
ENSRNOT00000037589
ENSRNOT00000081775 |
Sun5
|
Sad1 and UNC84 domain containing 5 |
| chr10_-_85717511 | 15.29 |
ENSRNOT00000005445
|
LOC691189
|
hypothetical protein LOC691189 |
| chr9_-_50820290 | 15.09 |
ENSRNOT00000050748
|
LOC102548013
|
uncharacterized LOC102548013 |
| chr13_-_47916185 | 14.97 |
ENSRNOT00000087793
|
Dyrk3
|
dual specificity tyrosine phosphorylation regulated kinase 3 |
| chr1_+_185673177 | 14.71 |
ENSRNOT00000048020
|
Sox6
|
SRY box 6 |
| chr5_-_77408323 | 14.71 |
ENSRNOT00000046857
ENSRNOT00000046760 |
LOC259245
Mup4
|
alpha-2u globulin PGCL5 major urinary protein 4 |
| chr3_+_15379109 | 14.68 |
ENSRNOT00000036495
|
Morn5
|
MORN repeat containing 5 |
| chr5_-_77492013 | 14.57 |
ENSRNOT00000012293
|
LOC259245
|
alpha-2u globulin PGCL5 |
| chr5_-_77342299 | 13.54 |
ENSRNOT00000075994
|
RGD1566134
|
similar to alpha-2u-globulin |
| chr11_+_88095170 | 13.33 |
ENSRNOT00000041557
|
Ccdc116
|
coiled-coil domain containing 116 |
| chr5_-_77433847 | 13.10 |
ENSRNOT00000076906
ENSRNOT00000043056 |
LOC500473
Mup4
|
similar to alpha-2u globulin PGCL2 major urinary protein 4 |
| chr8_-_84522588 | 12.92 |
ENSRNOT00000076213
|
Mlip
|
muscular LMNA-interacting protein |
| chr14_-_21944921 | 12.86 |
ENSRNOT00000060273
|
Csn1s2a
|
casein alpha s2-like A |
| chr5_-_77903062 | 12.80 |
ENSRNOT00000073954
ENSRNOT00000059320 ENSRNOT00000089382 ENSRNOT00000076074 ENSRNOT00000074349 |
LOC100912565
Rn50_5_0814.4
|
major urinary protein-like alpha2u globulin (LOC298111), mRNA |
| chr5_-_77316764 | 12.52 |
ENSRNOT00000071395
ENSRNOT00000076464 |
Mup4
|
major urinary protein 4 |
| chr13_+_83721300 | 12.49 |
ENSRNOT00000082677
|
Adcy10
|
adenylate cyclase 10 (soluble) |
| chr2_-_122756842 | 12.47 |
ENSRNOT00000080181
|
Ccdc144b
|
coiled-coil domain containing 144B |
| chr16_+_18648866 | 12.35 |
ENSRNOT00000014862
|
Dydc1
|
DPY30 domain containing 1 |
| chr8_+_18430302 | 12.12 |
ENSRNOT00000008341
|
Mbd3l1
|
methyl-CpG binding domain protein 3-like 1 |
| chr16_+_51748970 | 12.00 |
ENSRNOT00000059182
|
Adam26a
|
a disintegrin and metallopeptidase domain 26A |
| chrX_-_123092217 | 11.98 |
ENSRNOT00000039710
|
Gm14569
|
predicted gene 14569 |
| chr2_+_189423559 | 11.13 |
ENSRNOT00000029076
|
Tpm3
|
tropomyosin 3 |
| chr3_+_94035905 | 11.06 |
ENSRNOT00000014767
|
RGD1563222
|
similar to RIKEN cDNA A930018P22 |
| chr4_+_162077188 | 11.03 |
ENSRNOT00000090431
|
LOC100910725
|
C-type lectin domain family 2 member D-related protein-like |
| chrX_+_110233170 | 10.89 |
ENSRNOT00000036019
|
4930513O06Rik
|
RIKEN cDNA 4930513O06 gene |
| chrX_+_115721251 | 10.88 |
ENSRNOT00000060090
|
Trpc5os
|
TRPC5 opposite strand |
| chr3_-_14610546 | 10.74 |
ENSRNOT00000060011
|
LOC502618
|
ABO-family member 6 |
| chr7_+_66624512 | 10.42 |
ENSRNOT00000079410
|
Usp15
|
ubiquitin specific peptidase 15 |
| chr2_-_3124543 | 10.02 |
ENSRNOT00000036547
|
Fam81b
|
family with sequence similarity 81, member B |
| chr11_-_36533073 | 10.00 |
ENSRNOT00000033486
|
Lca5l
|
LCA5L, lebercilin like |
| chr13_+_95983809 | 9.92 |
ENSRNOT00000078258
|
AABR07021863.1
|
|
| chrX_+_158053164 | 9.65 |
ENSRNOT00000003235
|
Plac1
|
placenta-specific 1 |
| chr2_+_92549479 | 9.43 |
ENSRNOT00000082912
|
tGap1
|
GTPase activating protein testicular GAP1 |
| chr8_-_27907911 | 8.80 |
ENSRNOT00000010045
|
Glb1l3
|
galactosidase, beta 1-like 3 |
| chr1_+_190072420 | 8.63 |
ENSRNOT00000037026
|
Abca15
|
ATP-binding cassette, subfamily A (ABC1), member 15 |
| chr5_-_58484900 | 8.24 |
ENSRNOT00000012386
|
Fam214b
|
family with sequence similarity 214, member B |
| chr15_+_57709043 | 8.02 |
ENSRNOT00000030579
|
Erich6b
|
glutamate-rich 6B |
| chrX_-_60942845 | 7.85 |
ENSRNOT00000067446
|
Magebl1
|
melanoma antigen, family B-like 1 |
| chr7_-_130827152 | 7.82 |
ENSRNOT00000019406
|
Syt10
|
synaptotagmin 10 |
| chr4_+_166040503 | 7.56 |
ENSRNOT00000063869
|
Prb3
|
proline-rich protein BstNI subfamily 3 |
| chr6_-_141957537 | 7.55 |
ENSRNOT00000090358
|
Ighv13-2
|
immunoglobulin heavy variable 13-2 |
| chr19_+_6046665 | 7.51 |
ENSRNOT00000084126
|
Cdh8
|
cadherin 8 |
| chrX_+_124268129 | 7.13 |
ENSRNOT00000035489
|
Rhox13
|
Reproductive homeobox 13 |
| chr4_-_178441547 | 6.96 |
ENSRNOT00000087434
ENSRNOT00000055542 |
Sox5
|
SRY box 5 |
| chr8_+_53411316 | 6.95 |
ENSRNOT00000011107
ENSRNOT00000086957 |
Tmprss5
|
transmembrane protease, serine 5 |
| chr10_-_62664466 | 6.92 |
ENSRNOT00000078730
|
Git1
|
GIT ArfGAP 1 |
| chr3_+_3272193 | 6.78 |
ENSRNOT00000044284
|
Lcn9
|
lipocalin 9 |
| chr3_+_132052612 | 6.67 |
ENSRNOT00000030148
|
AABR07053879.1
|
|
| chr7_-_143956668 | 6.58 |
ENSRNOT00000018809
|
Aaas
|
aladin WD repeat nucleoporin |
| chr3_-_120404910 | 6.40 |
ENSRNOT00000051783
|
Bub1
|
BUB1 mitotic checkpoint serine/threonine kinase |
| chr20_+_27129291 | 6.37 |
ENSRNOT00000031053
|
AABR07044933.1
|
|
| chr2_+_194433808 | 6.23 |
ENSRNOT00000074310
|
RGD1563451
|
similar to TDPOZ2 |
| chr5_-_77294735 | 6.14 |
ENSRNOT00000018209
ENSRNOT00000056900 |
Zfp37
|
zinc finger protein 37 |
| chr3_-_45169118 | 6.06 |
ENSRNOT00000086371
|
Ccdc148
|
coiled-coil domain containing 148 |
| chr4_-_157486844 | 6.01 |
ENSRNOT00000038281
ENSRNOT00000022874 |
Cops7a
|
COP9 signalosome subunit 7A |
| chr10_+_89069256 | 5.72 |
ENSRNOT00000008576
|
Tubg2
|
tubulin, gamma 2 |
| chr2_-_187401786 | 5.70 |
ENSRNOT00000025624
|
Hapln2
|
hyaluronan and proteoglycan link protein 2 |
| chr1_+_83626639 | 5.65 |
ENSRNOT00000028261
|
Cyp2g1
|
cytochrome P450, family 2, subfamily g, polypeptide 1 |
| chr3_+_143109316 | 5.57 |
ENSRNOT00000006555
|
LOC257643
|
cystatin SC |
| chr7_+_71023976 | 5.35 |
ENSRNOT00000005679
|
Tac3
|
tachykinin 3 |
| chr18_-_37132223 | 5.24 |
ENSRNOT00000091634
|
Ppp2r2b
|
protein phosphatase 2, regulatory subunit B, beta |
| chr10_-_40381886 | 5.16 |
ENSRNOT00000050213
|
LOC100912427
|
nuclease-sensitive element-binding protein 1-like |
| chr2_-_149088787 | 4.85 |
ENSRNOT00000064833
|
Clrn1
|
clarin 1 |
| chr9_+_14779772 | 4.76 |
ENSRNOT00000040323
|
AABR07066753.1
|
|
| chr7_+_130357643 | 4.61 |
ENSRNOT00000040769
|
Klhdc7b
|
kelch domain containing 7B |
| chr5_+_162323373 | 4.13 |
ENSRNOT00000033150
|
Aadacl3
|
arylacetamide deacetylase-like 3 |
| chr1_-_228263198 | 4.06 |
ENSRNOT00000028572
|
Olr1874
|
olfactory receptor 1874 |
| chr6_+_111476768 | 4.02 |
ENSRNOT00000016479
|
Slirp
|
SRA stem-loop interacting RNA binding protein |
| chr11_-_71523267 | 4.01 |
ENSRNOT00000033026
|
Zdhhc19
|
zinc finger, DHHC-type containing 19 |
| chr3_-_22952063 | 4.00 |
ENSRNOT00000083850
|
Psmb7
|
proteasome subunit beta 7 |
| chr20_-_11556312 | 3.90 |
ENSRNOT00000091856
|
RGD1561557
|
similar to chromosome 21 open reading frame 29 |
| chrX_-_104726816 | 3.77 |
ENSRNOT00000005165
|
Tspan6
|
tetraspanin 6 |
| chr2_+_263212051 | 3.77 |
ENSRNOT00000089396
|
Negr1
|
neuronal growth regulator 1 |
| chr5_-_139505065 | 3.75 |
ENSRNOT00000013899
|
Ctps1
|
CTP synthase 1 |
| chr12_+_47103314 | 3.71 |
ENSRNOT00000064910
|
Rnf10
|
ring finger protein 10 |
| chr10_-_57606171 | 3.71 |
ENSRNOT00000009015
|
Nup88
|
nucleoporin 88 |
| chr9_+_6970507 | 3.67 |
ENSRNOT00000079488
|
St6gal2
|
ST6 beta-galactoside alpha-2,6-sialyltransferase 2 |
| chr14_-_21299068 | 3.62 |
ENSRNOT00000065778
|
Amtn
|
amelotin |
| chr3_-_143408604 | 3.52 |
ENSRNOT00000073262
|
AABR07054147.1
|
|
| chr3_+_167787111 | 3.45 |
ENSRNOT00000051082
|
AABR07054723.1
|
|
| chr1_-_169050963 | 3.43 |
ENSRNOT00000042637
|
Olr131
|
olfactory receptor 131 |
| chr1_+_171188133 | 3.40 |
ENSRNOT00000026511
|
Olr231
|
olfactory receptor 231 |
| chr16_+_2379480 | 3.07 |
ENSRNOT00000079215
|
Dnah12
|
dynein, axonemal, heavy chain 12 |
| chr8_-_104335729 | 3.07 |
ENSRNOT00000015821
|
Rnf7
|
ring finger protein 7 |
| chr2_+_195039054 | 3.00 |
ENSRNOT00000072634
ENSRNOT00000044412 |
LOC100911679
|
TD and POZ domain-containing protein 2-like |
| chr3_-_15379381 | 2.91 |
ENSRNOT00000084069
|
Ndufa8
|
NADH:ubiquinone oxidoreductase subunit A8 |
| chr4_-_165810121 | 2.82 |
ENSRNOT00000048210
|
Tas2r104
|
taste receptor, type 2, member 104 |
| chr12_+_18936594 | 2.82 |
ENSRNOT00000081587
|
Spry3
|
sprouty RTK signaling antagonist 3 |
| chr3_+_148035166 | 2.80 |
ENSRNOT00000055409
|
Defb19
|
defensin beta 19 |
| chr8_+_85605655 | 2.71 |
ENSRNOT00000086145
ENSRNOT00000085958 |
AABR07070816.1
|
|
| chr6_-_142635763 | 2.65 |
ENSRNOT00000048908
|
AABR07065815.2
|
|
| chr1_-_149914606 | 2.62 |
ENSRNOT00000044773
|
Olr19
|
olfactory receptor 19 |
| chr1_+_220423426 | 2.57 |
ENSRNOT00000072647
|
Brms1
|
breast cancer metastasis-suppressor 1 |
| chr1_-_61626454 | 2.53 |
ENSRNOT00000038190
|
Vom1r22
|
vomeronasal 1 receptor 22 |
| chr4_+_1658278 | 2.53 |
ENSRNOT00000073845
|
Olr1250
|
olfactory receptor 1250 |
| chr3_+_3000477 | 2.52 |
ENSRNOT00000064437
|
Obp2b
|
odorant binding protein 2B |
| chr19_-_17045349 | 2.50 |
ENSRNOT00000086829
|
Fto
|
FTO, alpha-ketoglutarate dependent dioxygenase |
| chrX_+_29504349 | 2.37 |
ENSRNOT00000005981
|
Tceanc
|
transcription elongation factor A N-terminal and central domain containing |
| chr19_-_29968424 | 2.37 |
ENSRNOT00000024981
|
Inpp4b
|
inositol polyphosphate-4-phosphatase type II B |
| chr9_+_88494676 | 2.36 |
ENSRNOT00000089451
|
LOC102556337
|
mitochondrial fission factor-like |
| chrX_-_106085878 | 2.29 |
ENSRNOT00000049735
|
Prame
|
preferentially expressed antigen in melanoma |
| chr5_-_165083487 | 2.28 |
ENSRNOT00000036647
|
Disp3
|
dispatched RND transporter family member 3 |
| chr10_-_75171774 | 2.19 |
ENSRNOT00000011735
|
Epx
|
eosinophil peroxidase |
| chr13_+_91711995 | 2.18 |
ENSRNOT00000040596
|
Olr1582
|
olfactory receptor 1582 |
| chr1_-_99135977 | 2.14 |
ENSRNOT00000056515
|
Vom2r38
|
vomeronasal 2 receptor, 38 |
| chr4_+_102876038 | 2.08 |
ENSRNOT00000085205
|
AABR07061022.3
|
|
| chrX_+_99250516 | 2.01 |
ENSRNOT00000039260
|
RGD1565129
|
similar to DNA segment, Chr 5, ERATO Doi 577, expressed |
| chr3_-_102484517 | 1.98 |
ENSRNOT00000006522
|
Olr750
|
olfactory receptor 750 |
| chr6_-_140572023 | 1.95 |
ENSRNOT00000072338
|
AABR07065772.1
|
|
| chr11_-_28979169 | 1.89 |
ENSRNOT00000061601
|
AABR07033580.1
|
|
| chr2_+_36093301 | 1.89 |
ENSRNOT00000074521
|
LOC100911241
|
serine protease inhibitor Kazal-type 12-like |
| chrX_-_43015896 | 1.89 |
ENSRNOT00000075045
|
LOC100911241
|
serine protease inhibitor Kazal-type 12-like |
| chr2_-_199529864 | 1.88 |
ENSRNOT00000023742
|
Olr390
|
olfactory receptor 390 |
| chr3_+_160660749 | 1.85 |
ENSRNOT00000018456
|
LOC103691965
|
seminal vesicle secretory protein 4 |
| chr10_+_65552897 | 1.79 |
ENSRNOT00000056217
|
Spag5
|
sperm associated antigen 5 |
| chr10_-_27427686 | 1.71 |
ENSRNOT00000087475
|
AABR07029410.1
|
|
| chr9_-_81880105 | 1.69 |
ENSRNOT00000022677
|
Rnf25
|
ring finger protein 25 |
| chr4_+_148623770 | 1.69 |
ENSRNOT00000051285
|
Olr828
|
olfactory receptor 828 |
| chr2_+_180936688 | 1.69 |
ENSRNOT00000016039
|
Asic5
|
acid sensing ion channel subunit family member 5 |
| chr3_+_79187055 | 1.68 |
ENSRNOT00000050785
|
Olr745
|
olfactory receptor 745 |
| chr1_-_216744066 | 1.68 |
ENSRNOT00000087961
|
Nap1l4
|
nucleosome assembly protein 1-like 4 |
| chr1_+_61095171 | 1.64 |
ENSRNOT00000042702
|
Vom1r19
|
vomeronasal 1 receptor 19 |
| chr9_+_52393779 | 1.58 |
ENSRNOT00000072809
|
RGD1564617
|
similar to large subunit ribosomal protein L36a |
| chr3_-_20479999 | 1.54 |
ENSRNOT00000050573
|
AABR07051733.2
|
|
| chr9_+_24170858 | 1.54 |
ENSRNOT00000081915
|
AC130064.1
|
|
| chr5_-_69578458 | 1.53 |
ENSRNOT00000082758
|
Olr851
|
olfactory receptor 851 |
| chr6_-_141321108 | 1.50 |
ENSRNOT00000040556
|
AABR07065789.3
|
|
| chr16_-_75262002 | 1.49 |
ENSRNOT00000046156
|
Defb50
|
defensin beta 50 |
| chr3_+_19141133 | 1.49 |
ENSRNOT00000058323
|
AABR07051675.1
|
|
| chr4_+_148567386 | 1.46 |
ENSRNOT00000047844
|
Olr826
|
olfactory receptor 826 |
| chr10_-_82374171 | 1.44 |
ENSRNOT00000032693
|
Eme1
|
essential meiotic structure-specific endonuclease 1 |
| chr15_-_27968358 | 1.44 |
ENSRNOT00000040616
|
Rnase9
|
ribonuclease A family member 9 |
| chr10_-_44082313 | 1.43 |
ENSRNOT00000051126
|
Olr1421
|
olfactory receptor 1421 |
| chr20_+_5008508 | 1.38 |
ENSRNOT00000001153
|
Vwa7
|
von Willebrand factor A domain containing 7 |
| chr15_+_18332910 | 1.28 |
ENSRNOT00000085822
|
Fam3d
|
family with sequence similarity 3, member D |
| chr10_-_56935141 | 1.25 |
ENSRNOT00000025910
|
Alox12e
|
arachidonate 12-lipoxygenase, epidermal |
| chr4_-_164015365 | 1.19 |
ENSRNOT00000078121
|
Ly49si2
|
immunoreceptor Ly49si2 |
| chr4_+_93888502 | 1.12 |
ENSRNOT00000090783
|
AABR07060792.1
|
|
| chr6_-_26135952 | 1.08 |
ENSRNOT00000084751
|
Mrpl33
|
mitochondrial ribosomal protein L33 |
| chr7_-_11018160 | 1.08 |
ENSRNOT00000092061
|
Aes
|
amino-terminal enhancer of split |
| chr4_+_157726941 | 1.07 |
ENSRNOT00000025081
|
Vamp1
|
vesicle-associated membrane protein 1 |
| chr1_+_213374895 | 1.05 |
ENSRNOT00000041052
|
Olr311
|
olfactory receptor 311 |
| chr15_-_29623597 | 1.05 |
ENSRNOT00000074483
|
AABR07017647.1
|
|
| chr4_-_166979391 | 1.05 |
ENSRNOT00000052034
|
Tas2r136
|
taste receptor, type 2, member 136 |
| chr6_-_123577695 | 1.04 |
ENSRNOT00000006604
|
Foxn3
|
forkhead box N3 |
| chr3_-_119075606 | 1.02 |
ENSRNOT00000041569
ENSRNOT00000086724 |
Hdc
|
histidine decarboxylase |
| chr1_+_89724303 | 1.01 |
ENSRNOT00000030687
|
Scgb2b2
|
secretoglobin, family 2B, member 2 |
| chr9_-_14654305 | 1.00 |
ENSRNOT00000065673
|
RGD1307182
|
similar to RIKEN cDNA B430306N03 gene |
| chr15_-_29360198 | 0.99 |
ENSRNOT00000072745
|
LOC688340
|
T-cell receptor alpha chain V region PHDS58-like |
| chr2_+_83393282 | 0.91 |
ENSRNOT00000044871
|
Ctnnd2
|
catenin delta 2 |
| chr3_-_103485806 | 0.88 |
ENSRNOT00000072524
|
Olr789
|
olfactory receptor 789 |
| chrX_+_135550834 | 0.81 |
ENSRNOT00000048922
|
Rbmx2
|
RNA binding motif protein, X-linked 2 |
| chr8_-_43024557 | 0.81 |
ENSRNOT00000073494
|
Olr1273
|
olfactory receptor 1273 |
| chr3_+_77152425 | 0.78 |
ENSRNOT00000092045
ENSRNOT00000082255 |
Olr653
|
olfactory receptor 653 |
| chr3_-_73653583 | 0.72 |
ENSRNOT00000073787
|
Olr493
|
olfactory receptor 493 |
| chr3_-_73801471 | 0.70 |
ENSRNOT00000090035
|
Olr507
|
olfactory receptor 507 |
| chr1_+_64849657 | 0.65 |
ENSRNOT00000081724
ENSRNOT00000086867 |
Vom2r12
|
vomeronasal 2 receptor, 12 |
| chr3_-_158936972 | 0.64 |
ENSRNOT00000074733
|
Olr1254
|
olfactory receptor 1254 |
| chr4_+_1584083 | 0.61 |
ENSRNOT00000087099
|
Olr1244
|
olfactory receptor 1244 |
| chrX_-_77559348 | 0.59 |
ENSRNOT00000047823
|
Fndc3c1
|
fibronectin type III domain containing 3C1 |
| chr2_+_159138758 | 0.59 |
ENSRNOT00000087942
|
AABR07011152.1
|
|
| chr15_+_32386816 | 0.58 |
ENSRNOT00000060322
|
AABR07017901.1
|
|
| chr1_+_66963035 | 0.56 |
ENSRNOT00000043603
|
LOC100911426
|
60S ribosomal protein L36a-like |
| chr3_+_19128400 | 0.52 |
ENSRNOT00000074272
|
AABR07051673.1
|
|
| chr1_-_200548317 | 0.48 |
ENSRNOT00000088610
|
AABR07005806.1
|
|
| chr10_-_43947494 | 0.46 |
ENSRNOT00000052223
|
Olr1416
|
olfactory receptor 1416 |
| chr15_+_31579478 | 0.44 |
ENSRNOT00000071642
|
AABR07017830.1
|
|
| chr6_+_128046780 | 0.40 |
ENSRNOT00000078064
|
LOC500712
|
Ab1-233 |
| chr13_-_91735361 | 0.40 |
ENSRNOT00000058090
|
Fcer1a
|
Fc fragment of IgE receptor Ia |
| chr10_+_34578356 | 0.38 |
ENSRNOT00000047454
|
Olr1389
|
olfactory receptor 1389 |
| chr6_-_142479939 | 0.36 |
ENSRNOT00000091268
|
AABR07065814.5
|
|
| chr8_+_20265891 | 0.36 |
ENSRNOT00000048326
|
Olr1166
|
olfactory receptor 1166 |
| chr5_+_169898302 | 0.28 |
ENSRNOT00000087911
|
AABR07050561.1
|
|
| chr9_+_84569597 | 0.23 |
ENSRNOT00000020161
|
Acsl3
|
acyl-CoA synthetase long-chain family member 3 |
| chr6_+_137997335 | 0.22 |
ENSRNOT00000006872
|
Tmem121
|
transmembrane protein 121 |
| chr8_-_39907478 | 0.21 |
ENSRNOT00000064141
|
Robo3
|
roundabout guidance receptor 3 |
| chr3_+_75322884 | 0.21 |
ENSRNOT00000066249
|
Olr556
|
olfactory receptor 556 |
| chr3_-_103203299 | 0.19 |
ENSRNOT00000047208
|
Olr783
|
olfactory receptor 783 |
| chr8_-_53758774 | 0.19 |
ENSRNOT00000068040
|
Ankk1
|
ankyrin repeat and kinase domain containing 1 |
| chr1_-_67134827 | 0.16 |
ENSRNOT00000045214
|
Vom1r45
|
vomeronasal 1 receptor 45 |
| chr3_+_77047466 | 0.16 |
ENSRNOT00000089424
|
Olr648
|
olfactory receptor 648 |
| chr7_-_114380613 | 0.14 |
ENSRNOT00000011898
|
Ago2
|
argonaute 2, RISC catalytic component |
| chr16_+_81089292 | 0.13 |
ENSRNOT00000087192
ENSRNOT00000026150 |
Tfdp1
|
transcription factor Dp-1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Sox17
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.6 | 25.5 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 3.6 | 21.7 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 3.0 | 15.0 | GO:0035617 | stress granule disassembly(GO:0035617) |
| 2.6 | 10.4 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 1.9 | 15.3 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 1.3 | 6.4 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 1.2 | 12.4 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 1.2 | 9.6 | GO:0090214 | spongiotrophoblast layer developmental growth(GO:0090214) |
| 1.2 | 3.6 | GO:0070175 | positive regulation of enamel mineralization(GO:0070175) |
| 1.2 | 37.9 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 1.1 | 5.7 | GO:0008065 | establishment of blood-nerve barrier(GO:0008065) |
| 1.1 | 57.8 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 1.1 | 11.0 | GO:0042270 | protection from natural killer cell mediated cytotoxicity(GO:0042270) |
| 1.0 | 10.7 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.8 | 4.0 | GO:0000957 | mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 0.7 | 3.7 | GO:0010626 | negative regulation of Schwann cell proliferation(GO:0010626) |
| 0.7 | 5.2 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.7 | 2.2 | GO:0002215 | defense response to nematode(GO:0002215) |
| 0.7 | 5.4 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.6 | 2.5 | GO:0035553 | oxidative RNA demethylation(GO:0035513) oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.6 | 12.1 | GO:0006346 | methylation-dependent chromatin silencing(GO:0006346) |
| 0.5 | 6.0 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.5 | 4.8 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.5 | 3.7 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.5 | 12.5 | GO:0003351 | epithelial cilium movement(GO:0003351) |
| 0.5 | 12.9 | GO:0010614 | negative regulation of cardiac muscle hypertrophy(GO:0010614) |
| 0.5 | 16.4 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.4 | 3.6 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.4 | 3.8 | GO:0039532 | negative regulation of viral-induced cytoplasmic pattern recognition receptor signaling pathway(GO:0039532) negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.4 | 6.9 | GO:0001771 | immunological synapse formation(GO:0001771) |
| 0.4 | 25.4 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.4 | 1.4 | GO:0072429 | response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 0.4 | 1.4 | GO:0060474 | positive regulation of sperm motility involved in capacitation(GO:0060474) |
| 0.3 | 5.2 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.3 | 3.1 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.3 | 2.4 | GO:1900244 | positive regulation of synaptic vesicle endocytosis(GO:1900244) |
| 0.3 | 3.8 | GO:0006241 | CTP biosynthetic process(GO:0006241) CTP metabolic process(GO:0046036) |
| 0.3 | 7.8 | GO:0045956 | positive regulation of calcium ion-dependent exocytosis(GO:0045956) |
| 0.3 | 1.0 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.3 | 2.3 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.2 | 5.7 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.2 | 5.7 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.2 | 3.7 | GO:0097503 | sialylation(GO:0097503) |
| 0.1 | 0.4 | GO:0045425 | regulation of interleukin-3 production(GO:0032672) positive regulation of interleukin-3 production(GO:0032752) interleukin-3 biosynthetic process(GO:0042223) regulation of interleukin-3 biosynthetic process(GO:0045399) positive regulation of interleukin-3 biosynthetic process(GO:0045401) positive regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045425) |
| 0.1 | 2.4 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.1 | 1.3 | GO:0019372 | lipoxygenase pathway(GO:0019372) |
| 0.1 | 1.8 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.1 | 25.5 | GO:0042742 | defense response to bacterium(GO:0042742) |
| 0.1 | 0.2 | GO:0061642 | chemoattraction of axon(GO:0061642) |
| 0.1 | 6.0 | GO:0009409 | response to cold(GO:0009409) |
| 0.1 | 7.1 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.1 | 0.2 | GO:2001245 | regulation of phosphatidylcholine biosynthetic process(GO:2001245) |
| 0.1 | 1.1 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.1 | 1.0 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.1 | 1.9 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.0 | 0.1 | GO:0035087 | siRNA loading onto RISC involved in RNA interference(GO:0035087) |
| 0.0 | 6.6 | GO:0009566 | fertilization(GO:0009566) |
| 0.0 | 3.8 | GO:0007631 | feeding behavior(GO:0007631) |
| 0.0 | 1.3 | GO:0046676 | negative regulation of insulin secretion(GO:0046676) |
| 0.0 | 1.7 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 3.6 | GO:0002244 | hematopoietic progenitor cell differentiation(GO:0002244) |
| 0.0 | 1.7 | GO:0051092 | positive regulation of NF-kappaB transcription factor activity(GO:0051092) |
| 0.0 | 0.1 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.0 | 0.9 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.0 | 3.1 | GO:0007018 | microtubule-based movement(GO:0007018) |
| 0.0 | 0.8 | GO:0071427 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.0 | 1.7 | GO:0035725 | sodium ion transmembrane transport(GO:0035725) |
| 0.0 | 2.5 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 2.3 | GO:0045665 | negative regulation of neuron differentiation(GO:0045665) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.8 | 19.3 | GO:0034774 | secretory granule lumen(GO:0034774) |
| 4.8 | 57.8 | GO:0005861 | troponin complex(GO:0005861) |
| 1.6 | 11.1 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 1.3 | 6.4 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 1.3 | 16.4 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 1.1 | 25.5 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.6 | 5.2 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.5 | 7.5 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.5 | 8.1 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.4 | 3.1 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.4 | 1.1 | GO:0035577 | azurophil granule membrane(GO:0035577) tertiary granule membrane(GO:0070821) |
| 0.3 | 15.0 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.3 | 1.4 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.3 | 5.7 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.3 | 15.3 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.2 | 5.2 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.2 | 17.3 | GO:0045178 | basal part of cell(GO:0045178) |
| 0.2 | 4.0 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.2 | 6.0 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.2 | 10.3 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.2 | 3.6 | GO:0005605 | basal lamina(GO:0005605) |
| 0.1 | 19.0 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.1 | 12.9 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 2.2 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.1 | 1.8 | GO:0035371 | centriolar satellite(GO:0034451) microtubule plus-end(GO:0035371) |
| 0.1 | 4.6 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 10.2 | GO:0030658 | transport vesicle membrane(GO:0030658) |
| 0.1 | 2.9 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.1 | 3.1 | GO:0030286 | dynein complex(GO:0030286) |
| 0.1 | 3.7 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 3.8 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 3.0 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.8 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 4.0 | GO:0036126 | sperm flagellum(GO:0036126) |
| 0.0 | 16.2 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 9.2 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 13.3 | GO:0005813 | centrosome(GO:0005813) |
| 0.0 | 38.0 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 10.2 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.0 | 0.1 | GO:0070578 | micro-ribonucleoprotein complex(GO:0035068) RISC-loading complex(GO:0070578) |
| 0.0 | 6.9 | GO:0045202 | synapse(GO:0045202) |
| 0.0 | 1.9 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 34.1 | GO:0005576 | extracellular region(GO:0005576) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 12.1 | 60.7 | GO:0005550 | pheromone binding(GO:0005550) |
| 5.1 | 20.5 | GO:0030172 | troponin C binding(GO:0030172) calcium-dependent ATPase activity(GO:0030899) |
| 1.8 | 12.9 | GO:0035375 | zymogen binding(GO:0035375) |
| 1.1 | 8.8 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 1.0 | 11.0 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.9 | 3.7 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.8 | 2.4 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.6 | 2.5 | GO:0035514 | DNA demethylase activity(GO:0035514) oxidative RNA demethylase activity(GO:0035515) |
| 0.6 | 12.1 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.5 | 3.1 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.5 | 37.3 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.5 | 12.5 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.3 | 14.6 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.3 | 1.7 | GO:0015252 | hydrogen ion channel activity(GO:0015252) |
| 0.3 | 8.1 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.3 | 1.3 | GO:0004052 | arachidonate 12-lipoxygenase activity(GO:0004052) |
| 0.2 | 11.1 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.2 | 5.7 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.2 | 10.4 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.2 | 15.2 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.2 | 18.6 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 4.0 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 7.0 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 7.8 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.1 | 5.7 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.1 | 1.4 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.1 | 4.0 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.1 | 4.3 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.1 | 12.9 | GO:0001191 | transcriptional repressor activity, RNA polymerase II transcription factor binding(GO:0001191) |
| 0.1 | 14.7 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.1 | 5.6 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.1 | 5.2 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.1 | 0.4 | GO:0019767 | IgE receptor activity(GO:0019767) |
| 0.1 | 3.8 | GO:0016879 | ligase activity, forming carbon-nitrogen bonds(GO:0016879) |
| 0.1 | 8.6 | GO:0042626 | ATPase activity, coupled to transmembrane movement of substances(GO:0042626) |
| 0.1 | 2.3 | GO:0042974 | retinoic acid receptor binding(GO:0042974) |
| 0.1 | 10.7 | GO:0016758 | transferase activity, transferring hexosyl groups(GO:0016758) |
| 0.0 | 2.2 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.0 | 5.2 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 3.1 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 3.8 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 9.6 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 1.1 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 1.9 | GO:0043621 | protein self-association(GO:0043621) |
| 0.0 | 1.0 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.1 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.0 | 0.2 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.0 | 7.4 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.9 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 4.6 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.2 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 1.4 | GO:0004540 | ribonuclease activity(GO:0004540) |
| 0.0 | 19.7 | GO:0003677 | DNA binding(GO:0003677) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 6.9 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.2 | 6.4 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 5.7 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 5.2 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 1.1 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 0.4 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 48.4 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.5 | 5.7 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.4 | 10.3 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.2 | 2.4 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.1 | 4.0 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.1 | 6.4 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.1 | 1.1 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 2.9 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 6.3 | REACTOME GENERIC TRANSCRIPTION PATHWAY | Genes involved in Generic Transcription Pathway |
| 0.0 | 5.4 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |