Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
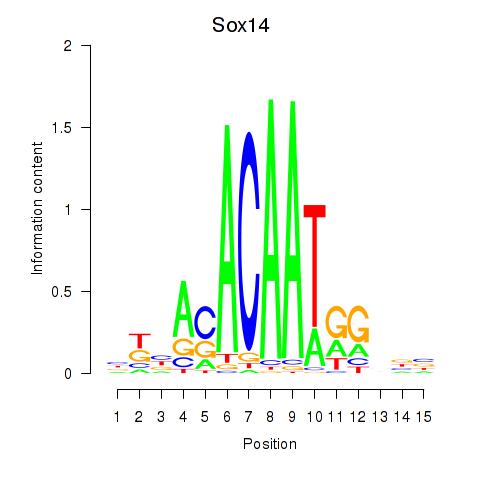
Results for Sox14
Z-value: 0.96
Transcription factors associated with Sox14
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Sox14
|
ENSRNOG00000022084 | SRY box 14 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Sox14 | rn6_v1_chr8_-_108109801_108109801 | 0.17 | 2.7e-03 | Click! |
Activity profile of Sox14 motif
Sorted Z-values of Sox14 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_+_30562178 | 28.25 |
ENSRNOT00000040998
|
LOC108348771
|
protocadherin beta-16-like |
| chr20_+_3422461 | 23.77 |
ENSRNOT00000084917
ENSRNOT00000079854 |
Tubb5
|
tubulin, beta 5 class I |
| chrX_+_39711201 | 22.76 |
ENSRNOT00000080512
ENSRNOT00000009802 |
Cnksr2
|
connector enhancer of kinase suppressor of Ras 2 |
| chr10_-_85974644 | 20.31 |
ENSRNOT00000006098
ENSRNOT00000082974 |
Cacnb1
|
calcium voltage-gated channel auxiliary subunit beta 1 |
| chr10_-_90393317 | 18.78 |
ENSRNOT00000028563
|
Fam171a2
|
family with sequence similarity 171, member A2 |
| chr2_-_96668222 | 18.13 |
ENSRNOT00000016567
|
Pkia
|
cAMP-dependent protein kinase inhibitor alpha |
| chr1_-_170404056 | 17.74 |
ENSRNOT00000024402
|
Apbb1
|
amyloid beta precursor protein binding family B member 1 |
| chr4_+_70572942 | 17.68 |
ENSRNOT00000051964
|
AC142181.1
|
|
| chr20_+_5049496 | 17.41 |
ENSRNOT00000088251
ENSRNOT00000001118 |
Ddah2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr1_-_236900904 | 16.93 |
ENSRNOT00000066846
|
AABR07006480.1
|
|
| chr5_+_152681101 | 16.44 |
ENSRNOT00000076052
ENSRNOT00000022574 |
Stmn1
|
stathmin 1 |
| chr4_+_64088900 | 16.04 |
ENSRNOT00000075341
|
Chrm2
|
cholinergic receptor, muscarinic 2 |
| chr15_+_52220578 | 15.61 |
ENSRNOT00000015104
|
Lgi3
|
leucine-rich repeat LGI family, member 3 |
| chr7_+_116632506 | 15.51 |
ENSRNOT00000009811
|
Gpihbp1
|
glycosylphosphatidylinositol anchored high density lipoprotein binding protein 1 |
| chr3_-_104502471 | 15.49 |
ENSRNOT00000040306
|
Ryr3
|
ryanodine receptor 3 |
| chr5_-_48165317 | 15.27 |
ENSRNOT00000088179
|
Ankrd6
|
ankyrin repeat domain 6 |
| chrX_+_28593405 | 14.99 |
ENSRNOT00000071708
|
Tmsb4x
|
thymosin beta 4, X-linked |
| chr1_+_264059374 | 14.79 |
ENSRNOT00000075397
|
Scd2
|
stearoyl-Coenzyme A desaturase 2 |
| chr12_+_48577905 | 14.45 |
ENSRNOT00000000888
|
Selplg
|
selectin P ligand |
| chr1_-_225283326 | 14.13 |
ENSRNOT00000027342
|
Scgb1a1
|
secretoglobin family 1A member 1 |
| chr10_+_55626741 | 14.11 |
ENSRNOT00000008492
|
Aurkb
|
aurora kinase B |
| chr7_-_136853957 | 14.08 |
ENSRNOT00000008985
|
Nell2
|
neural EGFL like 2 |
| chr12_-_5822874 | 14.01 |
ENSRNOT00000075920
|
Fry
|
FRY microtubule binding protein |
| chr8_-_65587427 | 13.96 |
ENSRNOT00000016491
|
Lrrc49
|
leucine rich repeat containing 49 |
| chrX_+_62727755 | 13.50 |
ENSRNOT00000077055
|
Pcyt1b
|
phosphate cytidylyltransferase 1, choline, beta |
| chr6_-_142903440 | 13.35 |
ENSRNOT00000075707
|
AABR07065823.2
|
|
| chr13_+_49850189 | 13.24 |
ENSRNOT00000042125
|
Pik3c2b
|
phosphatidylinositol-4-phosphate 3-kinase, catalytic subunit type 2 beta |
| chr9_-_61810417 | 13.23 |
ENSRNOT00000020910
|
Rftn2
|
raftlin family member 2 |
| chr2_-_111065413 | 13.00 |
ENSRNOT00000041111
|
Nlgn1
|
neuroligin 1 |
| chr1_+_192379543 | 12.98 |
ENSRNOT00000078705
|
Prkcb
|
protein kinase C, beta |
| chr6_-_41039437 | 12.95 |
ENSRNOT00000005774
|
Trib2
|
tribbles pseudokinase 2 |
| chr8_+_97291580 | 12.39 |
ENSRNOT00000018794
|
Rasgrf1
|
RAS protein-specific guanine nucleotide-releasing factor 1 |
| chrX_+_106360393 | 12.31 |
ENSRNOT00000004260
|
Gprasp2
|
G protein-coupled receptor associated sorting protein 2 |
| chr1_+_59765835 | 12.20 |
ENSRNOT00000014842
|
Fpr1
|
formyl peptide receptor 1 |
| chr18_+_30515962 | 12.19 |
ENSRNOT00000027172
|
LOC108348233
|
protocadherin beta-6-like |
| chr19_+_24329544 | 12.15 |
ENSRNOT00000080934
|
Tbc1d9
|
TBC1 domain family member 9 |
| chr13_+_47602692 | 12.10 |
ENSRNOT00000038822
|
Fcmr
|
Fc fragment of IgM receptor |
| chr18_+_29993361 | 12.01 |
ENSRNOT00000075810
|
Pcdha4
|
protocadherin alpha 4 |
| chr13_-_72789841 | 11.96 |
ENSRNOT00000004694
|
Mr1
|
major histocompatibility complex, class I-related |
| chr13_-_45318822 | 11.89 |
ENSRNOT00000091514
ENSRNOT00000005143 |
Cxcr4
|
C-X-C motif chemokine receptor 4 |
| chr9_-_80167033 | 11.71 |
ENSRNOT00000023530
|
Igfbp5
|
insulin-like growth factor binding protein 5 |
| chr10_-_56167426 | 11.39 |
ENSRNOT00000013955
|
Efnb3
|
ephrin B3 |
| chr11_+_61083757 | 11.29 |
ENSRNOT00000002790
|
Boc
|
BOC cell adhesion associated, oncogene regulated |
| chr15_+_41448064 | 11.19 |
ENSRNOT00000019551
|
Sacs
|
sacsin molecular chaperone |
| chr5_-_20189721 | 11.17 |
ENSRNOT00000014661
|
Tox
|
thymocyte selection-associated high mobility group box |
| chr17_-_44840131 | 10.75 |
ENSRNOT00000083417
|
Hist1h3b
|
histone cluster 1, H3b |
| chr7_+_116355698 | 10.73 |
ENSRNOT00000076693
|
Ly6e
|
lymphocyte antigen 6 complex, locus E |
| chr8_+_23113048 | 10.60 |
ENSRNOT00000029577
|
Cnn1
|
calponin 1 |
| chr1_-_228753422 | 10.53 |
ENSRNOT00000028626
|
Dtx4
|
deltex E3 ubiquitin ligase 4 |
| chr14_-_2700977 | 10.46 |
ENSRNOT00000000086
|
Mtf2
|
metal response element binding transcription factor 2 |
| chr14_-_82048251 | 10.44 |
ENSRNOT00000074734
|
Nat8l
|
N-acetyltransferase 8-like |
| chr18_+_55466373 | 10.35 |
ENSRNOT00000074629
|
LOC102555392
|
interferon-inducible GTPase 1-like |
| chr6_+_132510757 | 10.35 |
ENSRNOT00000080230
|
Evl
|
Enah/Vasp-like |
| chr1_+_140601791 | 10.34 |
ENSRNOT00000091588
|
Isg20
|
interferon stimulated exonuclease gene 20 |
| chr14_+_77380262 | 10.31 |
ENSRNOT00000008030
|
Nsg1
|
neuron specific gene family member 1 |
| chr7_+_116356324 | 10.18 |
ENSRNOT00000076407
ENSRNOT00000077022 ENSRNOT00000075873 |
Ly6e
|
lymphocyte antigen 6 complex, locus E |
| chr5_+_71742911 | 10.14 |
ENSRNOT00000047225
|
Zfp462
|
zinc finger protein 462 |
| chr4_-_55011415 | 10.05 |
ENSRNOT00000056996
|
Grm8
|
glutamate metabotropic receptor 8 |
| chr1_-_190914610 | 10.03 |
ENSRNOT00000023189
|
Cdr2
|
cerebellar degeneration-related protein 2 |
| chr6_-_142676432 | 9.96 |
ENSRNOT00000074947
|
AABR07065815.2
|
|
| chr15_-_108120279 | 9.88 |
ENSRNOT00000090572
|
Dock9
|
dedicator of cytokinesis 9 |
| chr8_-_80631873 | 9.85 |
ENSRNOT00000091661
ENSRNOT00000080662 |
Unc13c
|
unc-13 homolog C |
| chr18_+_30581530 | 9.76 |
ENSRNOT00000048166
|
Pcdhb20
|
protocadherin beta 20 |
| chr20_+_6923489 | 9.74 |
ENSRNOT00000093373
ENSRNOT00000000632 |
Pi16
|
peptidase inhibitor 16 |
| chr18_+_55463308 | 9.72 |
ENSRNOT00000073388
|
LOC100910979
|
interferon-inducible GTPase 1-like |
| chr3_+_35014538 | 9.70 |
ENSRNOT00000006341
|
Kif5c
|
kinesin family member 5C |
| chr2_-_96520137 | 9.68 |
ENSRNOT00000066966
|
Zc2hc1a
|
zinc finger, C2HC-type containing 1A |
| chr5_+_172364421 | 9.59 |
ENSRNOT00000018769
|
Hes5
|
hes family bHLH transcription factor 5 |
| chr6_-_139142218 | 9.49 |
ENSRNOT00000006975
|
Ighg
|
Immunoglobulin heavy chain (gamma polypeptide) |
| chr14_+_75880410 | 9.47 |
ENSRNOT00000014078
|
Hs3st1
|
heparan sulfate-glucosamine 3-sulfotransferase 1 |
| chr16_-_3765917 | 9.41 |
ENSRNOT00000088284
|
Duxbl1
|
double homeobox B-like 1 |
| chr17_+_53229785 | 9.33 |
ENSRNOT00000082040
|
Hecw1
|
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 1 |
| chr1_-_67065797 | 9.17 |
ENSRNOT00000048152
|
Vom1r46
|
vomeronasal 1 receptor 46 |
| chr7_+_116355911 | 9.15 |
ENSRNOT00000076182
|
Ly6e
|
lymphocyte antigen 6 complex, locus E |
| chr6_-_54020406 | 9.11 |
ENSRNOT00000086038
|
Hdac9
|
histone deacetylase 9 |
| chr12_+_38160464 | 9.04 |
ENSRNOT00000032249
|
Hcar2
|
hydroxycarboxylic acid receptor 2 |
| chr17_+_70684340 | 9.03 |
ENSRNOT00000051067
|
Pfkfb3
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 |
| chr9_-_113504605 | 9.00 |
ENSRNOT00000067974
|
Rab31
|
RAB31, member RAS oncogene family |
| chr8_-_65587658 | 8.97 |
ENSRNOT00000091982
|
Lrrc49
|
leucine rich repeat containing 49 |
| chr2_-_30458542 | 8.95 |
ENSRNOT00000072926
|
Naip6
|
NLR family, apoptosis inhibitory protein 6 |
| chr6_+_137164535 | 8.93 |
ENSRNOT00000018225
|
Inf2
|
inverted formin, FH2 and WH2 domain containing |
| chr7_-_50638798 | 8.84 |
ENSRNOT00000048880
|
Syt1
|
synaptotagmin 1 |
| chr2_+_226900619 | 8.81 |
ENSRNOT00000019638
|
Pde5a
|
phosphodiesterase 5A |
| chr13_+_52854031 | 8.61 |
ENSRNOT00000013738
ENSRNOT00000086004 |
Tmem9
|
transmembrane protein 9 |
| chr7_+_116356163 | 8.52 |
ENSRNOT00000076412
ENSRNOT00000009333 |
Ly6e
|
lymphocyte antigen 6 complex, locus E |
| chr18_+_30527705 | 8.47 |
ENSRNOT00000027168
|
Pcdhb14
|
protocadherin beta 14 |
| chr1_-_219438779 | 8.45 |
ENSRNOT00000029237
|
Tbc1d10c
|
TBC1 domain family, member 10C |
| chr2_-_210282352 | 8.28 |
ENSRNOT00000075653
|
Slc6a17
|
solute carrier family 6 member 17 |
| chr18_+_30509393 | 8.26 |
ENSRNOT00000043846
|
Pcdhb12
|
protocadherin beta 12 |
| chr12_+_25173005 | 8.20 |
ENSRNOT00000033200
ENSRNOT00000091328 |
Clip2
|
CAP-GLY domain containing linker protein 2 |
| chr1_-_101236065 | 8.09 |
ENSRNOT00000066834
|
Cd37
|
CD37 molecule |
| chr1_+_81230612 | 8.08 |
ENSRNOT00000026489
|
Kcnn4
|
potassium calcium-activated channel subfamily N member 4 |
| chr4_+_78694447 | 8.07 |
ENSRNOT00000011945
|
Gpnmb
|
glycoprotein nmb |
| chr7_-_136853154 | 8.06 |
ENSRNOT00000087376
|
Nell2
|
neural EGFL like 2 |
| chr17_-_43627629 | 8.06 |
ENSRNOT00000022965
|
Hist1h2af
|
histone cluster 1, H2af |
| chr5_-_173233188 | 8.04 |
ENSRNOT00000055343
|
Tmem88b
|
transmembrane protein 88B |
| chr18_+_30435119 | 8.03 |
ENSRNOT00000027190
|
Pcdhb8
|
protocadherin beta 8 |
| chrX_+_15113878 | 8.00 |
ENSRNOT00000007464
|
Wdr13
|
WD repeat domain 13 |
| chr18_+_30381322 | 7.99 |
ENSRNOT00000027216
|
Pcdhb3
|
protocadherin beta 3 |
| chr16_-_6675746 | 7.98 |
ENSRNOT00000025858
|
Prkcd
|
protein kinase C, delta |
| chr3_+_16817051 | 7.96 |
ENSRNOT00000071666
|
AABR07051550.1
|
|
| chr2_+_72006099 | 7.93 |
ENSRNOT00000034044
|
Cdh12
|
cadherin 12 |
| chr18_+_15467870 | 7.89 |
ENSRNOT00000091696
|
B4galt6
|
beta-1,4-galactosyltransferase 6 |
| chr18_+_55391388 | 7.85 |
ENSRNOT00000071612
|
LOC100910934
|
interferon-inducible GTPase 1-like |
| chr7_-_63407241 | 7.85 |
ENSRNOT00000024679
|
Tbc1d30
|
TBC1 domain family, member 30 |
| chr7_+_141346398 | 7.84 |
ENSRNOT00000077175
|
Asic1
|
acid sensing ion channel subunit 1 |
| chr10_+_44271587 | 7.80 |
ENSRNOT00000047700
|
Trim58
|
tripartite motif-containing 58 |
| chr6_+_8669722 | 7.75 |
ENSRNOT00000048550
|
Camkmt
|
calmodulin-lysine N-methyltransferase |
| chr10_+_55627025 | 7.71 |
ENSRNOT00000091016
|
Aurkb
|
aurora kinase B |
| chr20_+_13670066 | 7.69 |
ENSRNOT00000031400
|
LOC103694874
|
stromelysin-3 |
| chr18_+_55505993 | 7.64 |
ENSRNOT00000043736
|
RGD1309362
|
similar to interferon-inducible GTPase |
| chr16_-_19399851 | 7.64 |
ENSRNOT00000089056
ENSRNOT00000021073 |
Tpm4
|
tropomyosin 4 |
| chr9_+_6970507 | 7.63 |
ENSRNOT00000079488
|
St6gal2
|
ST6 beta-galactoside alpha-2,6-sialyltransferase 2 |
| chr18_+_29966245 | 7.51 |
ENSRNOT00000074028
|
Pcdha4
|
protocadherin alpha 4 |
| chr7_-_12275609 | 7.45 |
ENSRNOT00000086061
|
Apc2
|
APC2, WNT signaling pathway regulator |
| chr1_-_82004538 | 7.39 |
ENSRNOT00000087572
|
Pou2f2
|
POU class 2 homeobox 2 |
| chr4_+_21317695 | 7.36 |
ENSRNOT00000007572
|
Grm3
|
glutamate metabotropic receptor 3 |
| chr2_-_23909016 | 7.34 |
ENSRNOT00000084044
|
Scamp1
|
secretory carrier membrane protein 1 |
| chr2_-_183213228 | 7.29 |
ENSRNOT00000067618
|
Trim2
|
tripartite motif-containing 2 |
| chr2_+_178679041 | 7.25 |
ENSRNOT00000013524
|
Fam198b
|
family with sequence similarity 198, member B |
| chr14_+_81837809 | 7.20 |
ENSRNOT00000020576
|
Haus3
|
HAUS augmin-like complex, subunit 3 |
| chr11_+_30550141 | 7.20 |
ENSRNOT00000002866
|
Hunk
|
hormonally upregulated Neu-associated kinase |
| chr6_+_100337226 | 7.10 |
ENSRNOT00000011220
|
Fut8
|
fucosyltransferase 8 |
| chr4_-_165541314 | 7.09 |
ENSRNOT00000013833
|
Styk1
|
serine/threonine/tyrosine kinase 1 |
| chr5_-_90282327 | 7.09 |
ENSRNOT00000065792
|
Frmd3
|
FERM domain containing 3 |
| chr4_+_179398621 | 7.04 |
ENSRNOT00000049474
ENSRNOT00000067506 |
Lrmp
|
lymphoid-restricted membrane protein |
| chr6_-_42473738 | 6.93 |
ENSRNOT00000033327
|
Kcnf1
|
potassium voltage-gated channel modifier subfamily F member 1 |
| chr1_+_81373340 | 6.90 |
ENSRNOT00000026814
|
Zfp428
|
zinc finger protein 428 |
| chr10_+_35870682 | 6.82 |
ENSRNOT00000067188
|
Hnrnph1
|
heterogeneous nuclear ribonucleoprotein H1 |
| chr14_+_7618022 | 6.73 |
ENSRNOT00000088508
ENSRNOT00000002819 |
Slc10a6
|
solute carrier family 10 member 6 |
| chr3_-_146690375 | 6.68 |
ENSRNOT00000010641
|
Abhd12
|
abhydrolase domain containing 12 |
| chr20_+_3553455 | 6.68 |
ENSRNOT00000090080
|
Ddr1
|
discoidin domain receptor tyrosine kinase 1 |
| chr2_-_198002625 | 6.68 |
ENSRNOT00000091888
|
BC028528
|
cDNA sequence BC028528 |
| chr3_-_148932878 | 6.66 |
ENSRNOT00000013881
|
Nol4l
|
nucleolar protein 4-like |
| chr16_-_74122889 | 6.58 |
ENSRNOT00000025763
|
Plat
|
plasminogen activator, tissue type |
| chr8_+_63379087 | 6.56 |
ENSRNOT00000012091
ENSRNOT00000012295 |
Nptn
|
neuroplastin |
| chr16_+_23553647 | 6.52 |
ENSRNOT00000041994
|
Psd3
|
pleckstrin and Sec7 domain containing 3 |
| chr1_+_64928503 | 6.41 |
ENSRNOT00000086274
|
Vom2r80
|
vomeronasal 2 receptor, 80 |
| chr12_-_22211071 | 6.40 |
ENSRNOT00000087817
ENSRNOT00000001910 |
Actl6b
|
actin-like 6B |
| chr4_-_77706994 | 6.32 |
ENSRNOT00000038517
|
Zfp777
|
zinc finger protein 777 |
| chrX_+_156909913 | 6.30 |
ENSRNOT00000084390
|
L1cam
|
L1 cell adhesion molecule |
| chr6_-_2316717 | 6.26 |
ENSRNOT00000082017
ENSRNOT00000066363 |
Cyp1b1
|
cytochrome P450, family 1, subfamily b, polypeptide 1 |
| chr15_-_28733513 | 6.25 |
ENSRNOT00000078180
|
Sall2
|
spalt-like transcription factor 2 |
| chr17_+_70684539 | 6.23 |
ENSRNOT00000025700
|
Pfkfb3
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 |
| chr18_+_29972808 | 6.18 |
ENSRNOT00000074051
|
Pcdha4
|
protocadherin alpha 4 |
| chr6_-_141008427 | 6.17 |
ENSRNOT00000074472
|
AABR07065778.2
|
|
| chr14_-_35149608 | 6.06 |
ENSRNOT00000003050
ENSRNOT00000090654 |
Kit
|
KIT proto-oncogene receptor tyrosine kinase |
| chr6_-_55647665 | 6.04 |
ENSRNOT00000007414
|
Bzw2
|
basic leucine zipper and W2 domains 2 |
| chr10_+_43542697 | 6.03 |
ENSRNOT00000003555
|
Cnot8
|
CCR4-NOT transcription complex, subunit 8 |
| chr4_+_69457472 | 6.00 |
ENSRNOT00000067597
|
Trbv19
|
T cell receptor beta, variable 19 |
| chr4_+_102262007 | 5.93 |
ENSRNOT00000079328
|
AABR07060992.2
|
|
| chr9_+_46657922 | 5.92 |
ENSRNOT00000019180
|
Map4k4
|
mitogen-activated protein kinase kinase kinase kinase 4 |
| chr18_+_30895831 | 5.92 |
ENSRNOT00000026985
|
Pcdhga10
|
protocadherin gamma subfamily A, 10 |
| chr13_+_60435946 | 5.91 |
ENSRNOT00000004342
|
B3galt2
|
Beta-1,3-galactosyltransferase 2 |
| chr6_-_61405195 | 5.88 |
ENSRNOT00000008655
|
Lrrn3
|
leucine rich repeat neuronal 3 |
| chr7_+_14643704 | 5.87 |
ENSRNOT00000044642
|
AABR07055875.1
|
|
| chr5_+_145188323 | 5.85 |
ENSRNOT00000038888
|
Tmem35b
|
transmembrane protein 35B |
| chr6_+_55648021 | 5.83 |
ENSRNOT00000064822
ENSRNOT00000091488 |
Ankmy2
|
ankyrin repeat and MYND domain containing 2 |
| chr10_-_34439470 | 5.66 |
ENSRNOT00000072081
|
Btnl9
|
butyrophilin-like 9 |
| chr4_-_164211819 | 5.59 |
ENSRNOT00000084796
|
LOC497796
|
hypothetical protein LOC497796 |
| chr16_-_81912738 | 5.55 |
ENSRNOT00000087095
|
Mcf2l
|
MCF.2 cell line derived transforming sequence-like |
| chrX_+_105402867 | 5.55 |
ENSRNOT00000057222
|
Rpl36a
|
ribosomal protein L36a |
| chr3_+_19366370 | 5.51 |
ENSRNOT00000086557
|
AABR07051689.1
|
|
| chr2_+_212435728 | 5.49 |
ENSRNOT00000046960
|
AABR07012868.1
|
|
| chr14_+_46001849 | 5.49 |
ENSRNOT00000076611
|
Rell1
|
RELT-like 1 |
| chr20_+_42966140 | 5.47 |
ENSRNOT00000000707
|
Marcks
|
myristoylated alanine rich protein kinase C substrate |
| chr11_+_34598492 | 5.47 |
ENSRNOT00000065600
|
Ttc3
|
tetratricopeptide repeat domain 3 |
| chr10_-_15125408 | 5.47 |
ENSRNOT00000026395
|
Msln
|
mesothelin |
| chr9_+_73334618 | 5.45 |
ENSRNOT00000092717
|
Map2
|
microtubule-associated protein 2 |
| chr5_-_158549496 | 5.45 |
ENSRNOT00000074336
|
Igsf21
|
immunoglobin superfamily, member 21 |
| chr3_+_100787449 | 5.40 |
ENSRNOT00000078543
|
Bdnf
|
brain-derived neurotrophic factor |
| chr1_+_101687855 | 5.36 |
ENSRNOT00000028546
ENSRNOT00000091597 |
Dbp
|
D-box binding PAR bZIP transcription factor |
| chr2_+_38230757 | 5.35 |
ENSRNOT00000048598
|
RGD1564606
|
similar to 60S ribosomal protein L23a |
| chr16_+_83206004 | 5.28 |
ENSRNOT00000018870
|
Ankrd10
|
ankyrin repeat domain 10 |
| chr6_+_55374984 | 5.28 |
ENSRNOT00000091682
|
Agr3
|
anterior gradient 3, protein disulphide isomerase family member |
| chr8_+_5606592 | 5.20 |
ENSRNOT00000011727
|
Mmp12
|
matrix metallopeptidase 12 |
| chr13_-_91776397 | 5.17 |
ENSRNOT00000073147
|
Mptx1
|
mucosal pentraxin 1 |
| chr19_+_38601786 | 5.10 |
ENSRNOT00000087655
|
Zfp90
|
zinc finger protein 90 |
| chr1_+_87655618 | 5.06 |
ENSRNOT00000035994
ENSRNOT00000073521 |
Zfp84
|
zinc finger protein 84 |
| chr1_+_177093387 | 5.04 |
ENSRNOT00000021858
|
Mical2
|
microtubule associated monooxygenase, calponin and LIM domain containing 2 |
| chr1_+_213040024 | 5.03 |
ENSRNOT00000050237
|
Olr300
|
olfactory receptor 300 |
| chr7_-_114380613 | 4.97 |
ENSRNOT00000011898
|
Ago2
|
argonaute 2, RISC catalytic component |
| chr13_+_49870976 | 4.95 |
ENSRNOT00000090170
|
Pik3c2b
|
phosphatidylinositol-4-phosphate 3-kinase, catalytic subunit type 2 beta |
| chr7_-_117880289 | 4.93 |
ENSRNOT00000052227
|
Arhgap39
|
Rho GTPase activating protein 39 |
| chr10_-_35187416 | 4.93 |
ENSRNOT00000084509
|
Cnot6
|
CCR4-NOT transcription complex, subunit 6 |
| chr3_-_152159749 | 4.92 |
ENSRNOT00000067490
|
Cpne1
|
copine 1 |
| chr7_-_120874308 | 4.90 |
ENSRNOT00000078320
|
Fam227a
|
family with sequence similarity 227, member A |
| chr6_-_138909105 | 4.87 |
ENSRNOT00000087855
|
AABR07065656.9
|
|
| chr10_-_66602987 | 4.85 |
ENSRNOT00000017949
|
Wsb1
|
WD repeat and SOCS box-containing 1 |
| chr10_-_110101872 | 4.83 |
ENSRNOT00000071893
|
Ccdc57
|
coiled-coil domain containing 57 |
| chr11_+_64761146 | 4.83 |
ENSRNOT00000051428
|
Poglut1
|
protein O-glucosyltransferase 1 |
| chr3_+_18787606 | 4.79 |
ENSRNOT00000090508
|
AABR07051658.1
|
|
| chr16_-_22561496 | 4.77 |
ENSRNOT00000016543
|
Lpl
|
lipoprotein lipase |
| chr11_+_45751812 | 4.72 |
ENSRNOT00000079336
|
RGD1310935
|
similar to Dermal papilla derived protein 7 |
| chr1_-_265792932 | 4.68 |
ENSRNOT00000040904
|
Ldb1
|
LIM domain binding 1 |
| chr4_+_176606649 | 4.68 |
ENSRNOT00000084700
|
Golt1b
|
golgi transport 1B |
| chr1_+_72230038 | 4.65 |
ENSRNOT00000058997
|
Vom1r38
|
vomeronasal 1 receptor 38 |
| chr20_+_6288267 | 4.55 |
ENSRNOT00000000627
|
Srsf3
|
serine and arginine rich splicing factor 3 |
| chr1_-_59802401 | 4.49 |
ENSRNOT00000064917
|
Fpr2
|
formyl peptide receptor 2 |
| chr3_-_23474170 | 4.48 |
ENSRNOT00000039410
|
Scai
|
suppressor of cancer cell invasion |
Network of associatons between targets according to the STRING database.
First level regulatory network of Sox14
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.5 | 21.8 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 5.2 | 15.5 | GO:0010986 | positive regulation of lipoprotein particle clearance(GO:0010986) |
| 4.3 | 13.0 | GO:0035408 | histone H3-T6 phosphorylation(GO:0035408) |
| 4.1 | 16.4 | GO:0070495 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 3.5 | 14.1 | GO:0032696 | negative regulation of interleukin-13 production(GO:0032696) |
| 3.5 | 14.0 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 3.4 | 10.3 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 3.2 | 13.0 | GO:0098923 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) trans-synaptic signaling by soluble gas(GO:0099543) trans-synaptic signaling by trans-synaptic complex(GO:0099545) |
| 3.2 | 12.9 | GO:0045074 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 3.2 | 16.0 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 3.2 | 9.6 | GO:2000974 | auditory receptor cell fate determination(GO:0042668) negative regulation of pro-B cell differentiation(GO:2000974) negative regulation of forebrain neuron differentiation(GO:2000978) |
| 3.2 | 9.5 | GO:0001788 | antibody-dependent cellular cytotoxicity(GO:0001788) |
| 3.0 | 11.9 | GO:1990478 | positive regulation of vascular wound healing(GO:0035470) response to ultrasound(GO:1990478) |
| 2.9 | 11.7 | GO:1904204 | regulation of skeletal muscle hypertrophy(GO:1904204) |
| 2.8 | 8.3 | GO:0015820 | leucine transport(GO:0015820) |
| 2.7 | 8.0 | GO:0060697 | positive regulation of phospholipid catabolic process(GO:0060697) |
| 2.6 | 10.5 | GO:1903966 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 2.6 | 10.4 | GO:0051582 | positive regulation of neurotransmitter uptake(GO:0051582) positive regulation of dopamine uptake involved in synaptic transmission(GO:0051586) positive regulation of catecholamine uptake involved in synaptic transmission(GO:0051944) |
| 2.5 | 7.4 | GO:0002380 | immunoglobulin secretion involved in immune response(GO:0002380) |
| 2.4 | 9.7 | GO:0098964 | dendritic transport of ribonucleoprotein complex(GO:0098961) dendritic transport of messenger ribonucleoprotein complex(GO:0098963) anterograde dendritic transport of messenger ribonucleoprotein complex(GO:0098964) |
| 2.3 | 38.6 | GO:0048242 | epinephrine secretion(GO:0048242) |
| 2.2 | 13.2 | GO:0033227 | dsRNA transport(GO:0033227) |
| 2.1 | 15.0 | GO:2000483 | negative regulation of interleukin-8 secretion(GO:2000483) |
| 2.1 | 21.4 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 2.1 | 6.3 | GO:0071387 | cellular response to cortisol stimulus(GO:0071387) |
| 2.0 | 8.1 | GO:0030886 | negative regulation of myeloid dendritic cell activation(GO:0030886) |
| 1.8 | 22.1 | GO:0046543 | development of secondary female sexual characteristics(GO:0046543) |
| 1.8 | 5.5 | GO:1901726 | negative regulation of histone deacetylase activity(GO:1901726) |
| 1.8 | 7.1 | GO:0046368 | GDP-L-fucose metabolic process(GO:0046368) |
| 1.8 | 8.8 | GO:0060282 | negative regulation of cardiac muscle contraction(GO:0055118) positive regulation of oocyte development(GO:0060282) |
| 1.7 | 10.4 | GO:1904387 | cellular response to L-phenylalanine derivative(GO:1904387) |
| 1.7 | 5.2 | GO:1902739 | type I interferon secretion(GO:0072641) interferon-alpha secretion(GO:0072642) regulation of interferon-alpha secretion(GO:1902739) positive regulation of interferon-alpha secretion(GO:1902741) |
| 1.7 | 10.3 | GO:0098887 | neurotransmitter receptor transport, endosome to postsynaptic membrane(GO:0098887) |
| 1.7 | 8.4 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 1.7 | 5.0 | GO:0019417 | sulfur oxidation(GO:0019417) |
| 1.7 | 6.7 | GO:0052651 | monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 1.6 | 6.6 | GO:0060279 | positive regulation of ovulation(GO:0060279) |
| 1.6 | 4.9 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 1.6 | 6.6 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 1.6 | 17.4 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 1.6 | 3.1 | GO:1904339 | negative regulation of dopaminergic neuron differentiation(GO:1904339) |
| 1.5 | 15.3 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 1.5 | 6.1 | GO:0097324 | Kit signaling pathway(GO:0038109) melanocyte migration(GO:0097324) positive regulation of vascular smooth muscle cell differentiation(GO:1905065) |
| 1.5 | 18.1 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 1.5 | 13.5 | GO:1904116 | response to vasopressin(GO:1904116) |
| 1.5 | 10.5 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 1.3 | 13.5 | GO:1990001 | inhibition of cysteine-type endopeptidase activity involved in apoptotic process(GO:1990001) |
| 1.3 | 10.6 | GO:1904706 | negative regulation of vascular smooth muscle cell proliferation(GO:1904706) |
| 1.3 | 9.1 | GO:0034983 | peptidyl-lysine deacetylation(GO:0034983) |
| 1.3 | 9.0 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 1.3 | 9.0 | GO:0070269 | pyroptosis(GO:0070269) |
| 1.3 | 15.3 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 1.3 | 3.8 | GO:0007161 | calcium-independent cell-matrix adhesion(GO:0007161) |
| 1.2 | 5.0 | GO:0035087 | siRNA loading onto RISC involved in RNA interference(GO:0035087) mRNA cleavage involved in gene silencing by siRNA(GO:0090625) |
| 1.2 | 9.7 | GO:0061052 | negative regulation of cell growth involved in cardiac muscle cell development(GO:0061052) |
| 1.2 | 4.7 | GO:0046985 | positive regulation of hemoglobin biosynthetic process(GO:0046985) |
| 1.1 | 11.4 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 1.1 | 18.2 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 1.1 | 7.8 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 1.1 | 17.4 | GO:0000052 | citrulline metabolic process(GO:0000052) |
| 1.1 | 8.6 | GO:0061193 | taste bud development(GO:0061193) |
| 1.1 | 5.3 | GO:2000791 | regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) negative regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000584) regulation of mesenchymal cell proliferation involved in lung development(GO:2000790) negative regulation of mesenchymal cell proliferation involved in lung development(GO:2000791) |
| 1.0 | 11.2 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 1.0 | 3.0 | GO:0045077 | negative regulation of interferon-gamma biosynthetic process(GO:0045077) |
| 1.0 | 4.0 | GO:0042339 | keratan sulfate metabolic process(GO:0042339) |
| 1.0 | 11.0 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 1.0 | 7.9 | GO:0022007 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 1.0 | 10.7 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 1.0 | 4.8 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 1.0 | 3.8 | GO:0002913 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 1.0 | 3.8 | GO:0060398 | regulation of growth hormone receptor signaling pathway(GO:0060398) |
| 0.9 | 2.8 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.9 | 3.7 | GO:0002322 | B cell proliferation involved in immune response(GO:0002322) |
| 0.9 | 5.5 | GO:1900019 | regulation of protein kinase C activity(GO:1900019) positive regulation of protein kinase C activity(GO:1900020) |
| 0.9 | 9.9 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.9 | 10.3 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.9 | 3.4 | GO:0030581 | intracellular transport of viral protein in host cell(GO:0019060) symbiont intracellular protein transport in host(GO:0030581) intracellular protein transport in other organism involved in symbiotic interaction(GO:0051708) |
| 0.8 | 3.3 | GO:0021747 | cochlear nucleus development(GO:0021747) |
| 0.8 | 3.3 | GO:0060244 | negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
| 0.8 | 2.4 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) nuclear retention of pre-mRNA at the site of transcription(GO:0071033) |
| 0.8 | 4.8 | GO:0055095 | lipoprotein particle mediated signaling(GO:0055095) low-density lipoprotein particle mediated signaling(GO:0055096) |
| 0.8 | 8.7 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.8 | 12.4 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.8 | 109.4 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.8 | 27.7 | GO:0035458 | cellular response to interferon-beta(GO:0035458) |
| 0.7 | 3.7 | GO:1902473 | regulation of protein localization to synapse(GO:1902473) |
| 0.7 | 8.1 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.7 | 4.3 | GO:2001025 | positive regulation of response to drug(GO:2001025) |
| 0.7 | 2.8 | GO:1904925 | positive regulation of macromitophagy(GO:1901526) positive regulation of mitophagy in response to mitochondrial depolarization(GO:1904925) |
| 0.7 | 2.8 | GO:0045829 | negative regulation of isotype switching(GO:0045829) |
| 0.7 | 6.7 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.7 | 7.9 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.7 | 3.3 | GO:1903142 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.6 | 8.8 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.6 | 8.0 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.6 | 6.1 | GO:0070317 | negative regulation of G0 to G1 transition(GO:0070317) |
| 0.6 | 1.7 | GO:0045401 | regulation of interleukin-3 production(GO:0032672) positive regulation of interleukin-3 production(GO:0032752) interleukin-3 biosynthetic process(GO:0042223) regulation of interleukin-3 biosynthetic process(GO:0045399) positive regulation of interleukin-3 biosynthetic process(GO:0045401) positive regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045425) |
| 0.6 | 3.9 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.6 | 2.2 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.6 | 4.4 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.5 | 20.3 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.5 | 7.9 | GO:0006688 | glycosphingolipid biosynthetic process(GO:0006688) |
| 0.5 | 14.4 | GO:0050901 | leukocyte tethering or rolling(GO:0050901) |
| 0.5 | 2.0 | GO:0061641 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.5 | 12.1 | GO:0051481 | negative regulation of cytosolic calcium ion concentration(GO:0051481) |
| 0.5 | 9.0 | GO:0090382 | phagosome maturation(GO:0090382) |
| 0.5 | 2.0 | GO:0033277 | abortive mitotic cell cycle(GO:0033277) |
| 0.5 | 1.0 | GO:2000298 | regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 0.5 | 2.4 | GO:0035822 | meiotic gene conversion(GO:0006311) gene conversion(GO:0035822) |
| 0.5 | 6.1 | GO:0033148 | positive regulation of intracellular estrogen receptor signaling pathway(GO:0033148) |
| 0.5 | 1.9 | GO:0009597 | detection of virus(GO:0009597) |
| 0.5 | 2.8 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.4 | 12.0 | GO:0032620 | interleukin-17 production(GO:0032620) |
| 0.4 | 3.3 | GO:0035635 | exocyst assembly(GO:0001927) entry of bacterium into host cell(GO:0035635) regulation of entry of bacterium into host cell(GO:2000535) |
| 0.4 | 15.8 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.4 | 11.3 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 0.4 | 4.4 | GO:0048935 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.4 | 6.3 | GO:0097503 | sialylation(GO:0097503) |
| 0.3 | 7.2 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.3 | 2.4 | GO:0000920 | cell separation after cytokinesis(GO:0000920) |
| 0.3 | 8.9 | GO:0090140 | regulation of mitochondrial fission(GO:0090140) |
| 0.3 | 16.0 | GO:0043392 | negative regulation of DNA binding(GO:0043392) |
| 0.3 | 28.9 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.3 | 17.7 | GO:0008542 | visual learning(GO:0008542) |
| 0.3 | 1.2 | GO:0019064 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 0.3 | 8.1 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.3 | 4.5 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.3 | 3.2 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.3 | 0.8 | GO:0042245 | RNA repair(GO:0042245) |
| 0.3 | 1.9 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) negative regulation of type B pancreatic cell apoptotic process(GO:2000675) |
| 0.3 | 6.5 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.3 | 0.8 | GO:0090669 | telomerase RNA stabilization(GO:0090669) |
| 0.2 | 1.7 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.2 | 3.1 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.2 | 2.2 | GO:0002087 | regulation of respiratory gaseous exchange by neurological system process(GO:0002087) |
| 0.2 | 0.7 | GO:0016094 | polyprenol biosynthetic process(GO:0016094) ketone body catabolic process(GO:0046952) |
| 0.2 | 3.5 | GO:0043584 | nose development(GO:0043584) |
| 0.2 | 2.8 | GO:0051895 | negative regulation of focal adhesion assembly(GO:0051895) |
| 0.2 | 0.7 | GO:0030327 | prenylated protein catabolic process(GO:0030327) |
| 0.2 | 7.1 | GO:0033006 | regulation of mast cell activation involved in immune response(GO:0033006) regulation of mast cell degranulation(GO:0043304) |
| 0.2 | 3.4 | GO:0045063 | T-helper 1 cell differentiation(GO:0045063) |
| 0.2 | 1.2 | GO:2000354 | regulation of ovarian follicle development(GO:2000354) |
| 0.2 | 2.0 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.2 | 5.3 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.2 | 2.4 | GO:0048199 | vesicle targeting, to, from or within Golgi(GO:0048199) |
| 0.2 | 1.2 | GO:0002943 | tRNA dihydrouridine synthesis(GO:0002943) |
| 0.2 | 2.7 | GO:0021819 | layer formation in cerebral cortex(GO:0021819) |
| 0.2 | 5.6 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.2 | 4.2 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.2 | 2.3 | GO:0035090 | maintenance of apical/basal cell polarity(GO:0035090) |
| 0.2 | 3.8 | GO:0008340 | determination of adult lifespan(GO:0008340) |
| 0.2 | 3.4 | GO:0061099 | negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.2 | 0.5 | GO:0021812 | neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 0.2 | 3.4 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.1 | 2.6 | GO:0097435 | fibril organization(GO:0097435) |
| 0.1 | 4.9 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.1 | 2.1 | GO:0032674 | regulation of interleukin-5 production(GO:0032674) |
| 0.1 | 2.3 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.1 | 0.9 | GO:0036123 | histone H3-K9 dimethylation(GO:0036123) |
| 0.1 | 2.9 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.1 | 2.2 | GO:0061003 | positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.1 | 3.7 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.1 | 1.7 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.1 | 0.6 | GO:0070102 | interleukin-6-mediated signaling pathway(GO:0070102) |
| 0.1 | 7.9 | GO:0034763 | negative regulation of transmembrane transport(GO:0034763) |
| 0.1 | 0.6 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.1 | 5.1 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.1 | 0.8 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.1 | 15.6 | GO:0017157 | regulation of exocytosis(GO:0017157) |
| 0.1 | 5.9 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.1 | 0.5 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.1 | 1.4 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.1 | 0.9 | GO:2001214 | positive regulation of vasculogenesis(GO:2001214) |
| 0.1 | 2.4 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.1 | 2.1 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.1 | 1.9 | GO:0051205 | protein insertion into membrane(GO:0051205) |
| 0.1 | 3.5 | GO:0001893 | maternal placenta development(GO:0001893) |
| 0.1 | 5.8 | GO:0008589 | regulation of smoothened signaling pathway(GO:0008589) |
| 0.1 | 0.5 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.1 | 0.1 | GO:1904637 | response to ionomycin(GO:1904636) cellular response to ionomycin(GO:1904637) |
| 0.1 | 0.8 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.1 | 7.8 | GO:0018022 | peptidyl-lysine methylation(GO:0018022) |
| 0.1 | 5.9 | GO:0010771 | negative regulation of cell morphogenesis involved in differentiation(GO:0010771) |
| 0.1 | 1.1 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.1 | 0.2 | GO:0002084 | protein depalmitoylation(GO:0002084) |
| 0.1 | 0.9 | GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.0 | 9.2 | GO:0043523 | regulation of neuron apoptotic process(GO:0043523) |
| 0.0 | 7.6 | GO:0001649 | osteoblast differentiation(GO:0001649) |
| 0.0 | 2.7 | GO:0046323 | glucose import(GO:0046323) |
| 0.0 | 0.7 | GO:0050691 | regulation of defense response to virus by host(GO:0050691) |
| 0.0 | 1.0 | GO:0002228 | natural killer cell mediated immunity(GO:0002228) |
| 0.0 | 1.3 | GO:0070613 | regulation of protein processing(GO:0070613) |
| 0.0 | 2.3 | GO:0000724 | double-strand break repair via homologous recombination(GO:0000724) |
| 0.0 | 1.1 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.0 | 2.3 | GO:0001764 | neuron migration(GO:0001764) |
| 0.0 | 1.2 | GO:0031016 | pancreas development(GO:0031016) |
| 0.0 | 3.8 | GO:0071805 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.0 | 1.0 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 0.9 | GO:0006338 | chromatin remodeling(GO:0006338) |
| 0.0 | 1.1 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.0 | 1.4 | GO:0002244 | hematopoietic progenitor cell differentiation(GO:0002244) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.4 | 17.7 | GO:1990812 | growth cone lamellipodium(GO:1990761) growth cone filopodium(GO:1990812) |
| 3.6 | 21.8 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 2.7 | 16.0 | GO:0032280 | symmetric synapse(GO:0032280) |
| 2.6 | 23.8 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 2.4 | 9.5 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 2.1 | 10.3 | GO:0098845 | postsynaptic endosome(GO:0098845) |
| 2.0 | 11.7 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 1.8 | 5.5 | GO:0042585 | germinal vesicle(GO:0042585) |
| 1.8 | 9.0 | GO:0072557 | IPAF inflammasome complex(GO:0072557) |
| 1.7 | 15.5 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 1.5 | 8.8 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 1.4 | 4.1 | GO:1990796 | photoreceptor cell terminal bouton(GO:1990796) |
| 1.3 | 3.8 | GO:0034666 | integrin alpha2-beta1 complex(GO:0034666) integrin alpha3-beta1 complex(GO:0034667) |
| 1.2 | 9.9 | GO:0044305 | calyx of Held(GO:0044305) |
| 1.0 | 14.4 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.9 | 2.8 | GO:0060187 | cell pole(GO:0060187) |
| 0.9 | 6.5 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.9 | 16.7 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.9 | 4.3 | GO:0043293 | apoptosome(GO:0043293) |
| 0.8 | 9.5 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.8 | 17.2 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.8 | 13.0 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.7 | 12.0 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.7 | 7.2 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.7 | 5.0 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 0.7 | 3.5 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.7 | 9.7 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.7 | 2.7 | GO:0097196 | Shu complex(GO:0097196) |
| 0.6 | 5.5 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.6 | 5.5 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.6 | 8.6 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.6 | 7.5 | GO:0016342 | catenin complex(GO:0016342) |
| 0.6 | 6.3 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.6 | 3.9 | GO:0001651 | dense fibrillar component(GO:0001651) |
| 0.6 | 2.8 | GO:0032059 | bleb(GO:0032059) |
| 0.5 | 2.1 | GO:0031592 | centrosomal corona(GO:0031592) |
| 0.5 | 8.4 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.5 | 10.5 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.5 | 16.7 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.5 | 8.4 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.5 | 10.4 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.5 | 8.9 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.5 | 7.4 | GO:0097449 | astrocyte projection(GO:0097449) |
| 0.5 | 6.4 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.4 | 4.3 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.4 | 11.3 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.4 | 2.8 | GO:0031415 | NatA complex(GO:0031415) |
| 0.4 | 8.2 | GO:0042599 | lamellar body(GO:0042599) |
| 0.4 | 4.8 | GO:0042627 | chylomicron(GO:0042627) |
| 0.4 | 10.4 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.3 | 20.3 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.3 | 3.3 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.3 | 3.2 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.3 | 20.5 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.3 | 2.4 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.2 | 2.0 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.2 | 2.6 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.2 | 6.1 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.2 | 8.1 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.2 | 3.4 | GO:0043657 | host(GO:0018995) host cell(GO:0043657) |
| 0.2 | 11.6 | GO:0015030 | Cajal body(GO:0015030) |
| 0.2 | 2.4 | GO:0000801 | central element(GO:0000801) |
| 0.2 | 0.4 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.2 | 5.9 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.2 | 3.3 | GO:0010369 | chromocenter(GO:0010369) |
| 0.2 | 13.7 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.2 | 16.6 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.2 | 3.3 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.2 | 29.0 | GO:0030427 | site of polarized growth(GO:0030427) |
| 0.1 | 2.4 | GO:0046540 | U4/U6 x U5 tri-snRNP complex(GO:0046540) |
| 0.1 | 22.0 | GO:0043204 | perikaryon(GO:0043204) |
| 0.1 | 2.1 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.1 | 10.1 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.1 | 7.4 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.1 | 0.5 | GO:0005607 | laminin-1 complex(GO:0005606) laminin-2 complex(GO:0005607) laminin-10 complex(GO:0043259) |
| 0.1 | 11.8 | GO:0000786 | nucleosome(GO:0000786) |
| 0.1 | 2.1 | GO:0008328 | ionotropic glutamate receptor complex(GO:0008328) AMPA glutamate receptor complex(GO:0032281) neurotransmitter receptor complex(GO:0098878) |
| 0.1 | 0.9 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.1 | 10.9 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 0.7 | GO:0034385 | very-low-density lipoprotein particle(GO:0034361) triglyceride-rich lipoprotein particle(GO:0034385) |
| 0.1 | 1.0 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.1 | 8.0 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.1 | 80.2 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.1 | 15.4 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.1 | 6.9 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.1 | 8.1 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.1 | 26.6 | GO:0098857 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.1 | 5.5 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 7.1 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.1 | 24.9 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.1 | 7.0 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.1 | 19.1 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.1 | 7.9 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.1 | 2.0 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.1 | 9.0 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 5.6 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 14.8 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 0.8 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 20.3 | GO:0005815 | microtubule organizing center(GO:0005815) |
| 0.0 | 0.9 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 1.9 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 12.4 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 0.9 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.0 | 5.5 | GO:0015630 | microtubule cytoskeleton(GO:0015630) |
| 0.0 | 4.5 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.0 | 0.5 | GO:0016592 | mediator complex(GO:0016592) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.8 | 17.4 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 5.3 | 16.0 | GO:1990763 | arrestin family protein binding(GO:1990763) |
| 4.3 | 13.0 | GO:0035403 | histone kinase activity (H3-T6 specific)(GO:0035403) |
| 3.9 | 15.5 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 3.6 | 21.4 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 2.8 | 14.1 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 2.8 | 38.6 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 2.7 | 13.5 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 2.7 | 8.0 | GO:0070976 | TIR domain binding(GO:0070976) |
| 2.6 | 10.5 | GO:0032896 | stearoyl-CoA 9-desaturase activity(GO:0004768) palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 2.6 | 7.8 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 2.6 | 15.5 | GO:0035473 | lipase binding(GO:0035473) |
| 2.6 | 10.2 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 2.5 | 10.1 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 2.5 | 7.4 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 2.3 | 18.2 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 2.1 | 10.3 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 2.0 | 11.9 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 2.0 | 7.9 | GO:0008489 | UDP-galactose:glucosylceramide beta-1,4-galactosyltransferase activity(GO:0008489) |
| 1.8 | 8.8 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 1.7 | 10.3 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 1.6 | 4.8 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 1.6 | 9.5 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 1.5 | 18.1 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 1.4 | 15.3 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 1.3 | 6.6 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 1.3 | 3.9 | GO:0034512 | box C/D snoRNA binding(GO:0034512) |
| 1.3 | 11.7 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 1.2 | 19.9 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 1.2 | 8.7 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 1.2 | 3.7 | GO:0052723 | inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 1.2 | 23.8 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 1.2 | 5.9 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 1.2 | 8.1 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 1.1 | 9.9 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 1.1 | 8.6 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 1.1 | 20.3 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 1.0 | 8.1 | GO:0045545 | syndecan binding(GO:0045545) |
| 1.0 | 13.0 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 1.0 | 9.7 | GO:0034190 | apolipoprotein receptor binding(GO:0034190) |
| 1.0 | 6.7 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.9 | 3.7 | GO:0099609 | microtubule lateral binding(GO:0099609) |
| 0.9 | 5.5 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.9 | 31.4 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.8 | 2.4 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.8 | 9.5 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.8 | 6.1 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.8 | 3.8 | GO:0098639 | C-X3-C chemokine binding(GO:0019960) collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.7 | 2.2 | GO:0008481 | sphinganine kinase activity(GO:0008481) |
| 0.7 | 9.1 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.7 | 2.8 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.7 | 12.9 | GO:0055106 | ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.7 | 4.0 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.6 | 10.9 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.6 | 8.8 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.6 | 5.0 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.6 | 10.3 | GO:0005522 | profilin binding(GO:0005522) |
| 0.6 | 2.4 | GO:0016426 | tRNA (adenine) methyltransferase activity(GO:0016426) |
| 0.6 | 7.1 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.6 | 4.7 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.6 | 12.8 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.5 | 6.3 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.5 | 3.1 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.5 | 4.1 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.5 | 6.7 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.5 | 8.2 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.5 | 4.6 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.5 | 15.0 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.5 | 27.6 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.5 | 2.8 | GO:0019158 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.4 | 3.1 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.4 | 1.7 | GO:0019863 | IgE binding(GO:0019863) |
| 0.4 | 2.1 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.4 | 9.0 | GO:0001614 | purinergic nucleotide receptor activity(GO:0001614) nucleotide receptor activity(GO:0016502) |
| 0.4 | 4.4 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) |
| 0.4 | 1.2 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 0.4 | 1.9 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.4 | 2.9 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.3 | 1.0 | GO:0023030 | MHC class Ib protein complex binding(GO:0023025) MHC class Ib protein binding, via antigen binding groove(GO:0023030) |
| 0.3 | 11.4 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.3 | 4.8 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.3 | 4.2 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.3 | 5.5 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.3 | 2.0 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.3 | 16.2 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.3 | 0.8 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.3 | 6.5 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.3 | 8.0 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.2 | 2.2 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.2 | 0.7 | GO:0016453 | C-acetyltransferase activity(GO:0016453) |
| 0.2 | 2.8 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.2 | 3.3 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.2 | 12.5 | GO:0016278 | lysine N-methyltransferase activity(GO:0016278) protein-lysine N-methyltransferase activity(GO:0016279) |
| 0.2 | 0.8 | GO:0035515 | DNA demethylase activity(GO:0035514) oxidative RNA demethylase activity(GO:0035515) |
| 0.2 | 9.2 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.2 | 5.0 | GO:0071949 | FAD binding(GO:0071949) |
| 0.2 | 2.8 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.2 | 1.2 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.2 | 19.5 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.2 | 4.1 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.2 | 6.5 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.2 | 2.7 | GO:0005351 | sugar:proton symporter activity(GO:0005351) |
| 0.2 | 4.0 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.2 | 6.8 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.2 | 4.9 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.2 | 1.9 | GO:0015194 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.2 | 1.1 | GO:0035375 | zymogen binding(GO:0035375) |
| 0.1 | 7.1 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.1 | 9.0 | GO:0019003 | GDP binding(GO:0019003) |
| 0.1 | 0.8 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.1 | 11.6 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.1 | 0.9 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.1 | 3.3 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.1 | 84.2 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.1 | 8.5 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.1 | 22.2 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.1 | 0.7 | GO:0016670 | oxidoreductase activity, acting on a sulfur group of donors, oxygen as acceptor(GO:0016670) |
| 0.1 | 8.0 | GO:0008080 | N-acetyltransferase activity(GO:0008080) |
| 0.1 | 0.5 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.1 | 3.3 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.1 | 1.9 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 2.2 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.1 | 3.3 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 10.9 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.1 | 17.5 | GO:0015631 | tubulin binding(GO:0015631) |
| 0.1 | 2.0 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 4.0 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.1 | 0.5 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.1 | 19.7 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.1 | 6.9 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.1 | 1.4 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.1 | 3.3 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 0.9 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.1 | 1.5 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.1 | 3.4 | GO:0003774 | motor activity(GO:0003774) |
| 0.1 | 2.4 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 12.8 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 2.6 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.0 | 1.1 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.0 | 3.4 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 16.0 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 0.6 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.5 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 2.7 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 0.2 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.7 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 1.9 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 4.1 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 5.3 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |
| 0.0 | 3.0 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.8 | GO:0001076 | transcription factor activity, RNA polymerase II transcription factor binding(GO:0001076) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 20.2 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.9 | 13.0 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.6 | 19.9 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.6 | 14.4 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.6 | 29.4 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.5 | 21.8 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.5 | 30.0 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.5 | 26.8 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.5 | 20.1 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.4 | 15.3 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.3 | 8.4 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.3 | 4.4 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.3 | 10.9 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.3 | 2.1 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.3 | 14.1 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.3 | 8.8 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.3 | 18.1 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.3 | 9.1 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.3 | 4.1 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.3 | 7.4 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.3 | 7.9 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.2 | 5.9 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.2 | 39.5 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.2 | 5.5 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.2 | 9.0 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.2 | 2.8 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.2 | 4.5 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.1 | 2.7 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.1 | 3.3 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.1 | 2.5 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.1 | 0.8 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.1 | 2.0 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 2.0 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.0 | 0.9 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.0 | 3.4 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.0 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.1 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.0 | 0.4 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 0.5 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 11.9 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.7 | 13.0 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.7 | 15.0 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.7 | 12.4 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.6 | 8.8 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.6 | 5.5 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.6 | 11.7 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.5 | 13.5 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.5 | 18.2 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.5 | 20.3 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.5 | 11.0 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.4 | 10.8 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.4 | 7.2 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.4 | 10.3 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.4 | 15.3 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.4 | 8.0 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.4 | 30.3 | REACTOME SIGNALING BY NOTCH1 | Genes involved in Signaling by NOTCH1 |
| 0.4 | 6.3 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.4 | 7.9 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.4 | 8.8 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.4 | 9.5 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.3 | 12.7 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.3 | 11.3 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.3 | 4.8 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.3 | 8.0 | REACTOME CA DEPENDENT EVENTS | Genes involved in Ca-dependent events |
| 0.3 | 19.8 | REACTOME APC C CDH1 MEDIATED DEGRADATION OF CDC20 AND OTHER APC C CDH1 TARGETED PROTEINS IN LATE MITOSIS EARLY G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |
| 0.3 | 6.3 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.3 | 6.5 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.3 | 4.1 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.3 | 6.0 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.3 | 3.1 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.2 | 7.0 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.2 | 2.8 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.2 | 2.0 | REACTOME NRIF SIGNALS CELL DEATH FROM THE NUCLEUS | Genes involved in NRIF signals cell death from the nucleus |
| 0.2 | 9.1 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.2 | 4.0 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.2 | 10.3 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.2 | 3.8 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.2 | 5.4 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.2 | 2.7 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.2 | 15.0 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.1 | 3.3 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.1 | 22.9 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.1 | 4.4 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.1 | 2.3 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.1 | 9.9 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.1 | 2.4 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.1 | 2.1 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.1 | 6.6 | REACTOME SIGNALING BY PDGF | Genes involved in Signaling by PDGF |
| 0.1 | 0.6 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.1 | 1.2 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.1 | 3.1 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 2.1 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.1 | 2.8 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.1 | 5.4 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.1 | 5.5 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.1 | 1.4 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.1 | 4.2 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.1 | 3.2 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.0 | 0.9 | REACTOME SIGNALING BY NOTCH | Genes involved in Signaling by NOTCH |
| 0.0 | 1.0 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 3.6 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |