Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
Results for Runx2_Bcl11a
Z-value: 2.63
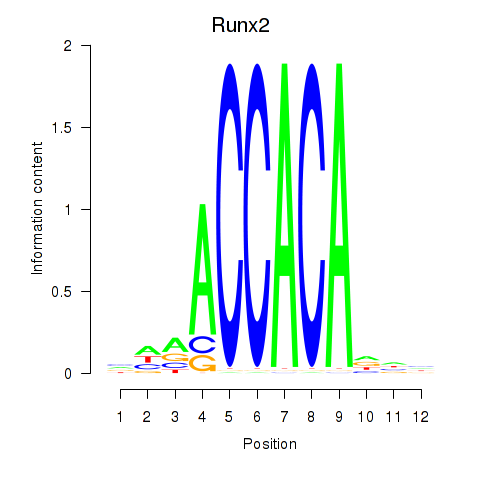
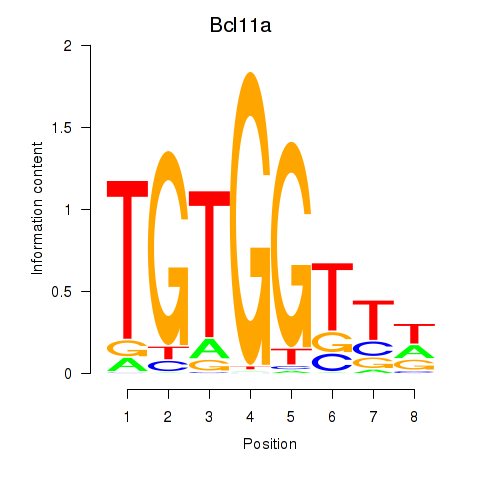
Transcription factors associated with Runx2_Bcl11a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Runx2
|
ENSRNOG00000020193 | runt-related transcription factor 2 |
|
Bcl11a
|
ENSRNOG00000007049 | B-cell CLL/lymphoma 11A |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Runx2 | rn6_v1_chr9_+_18564927_18564927 | 0.50 | 2.0e-21 | Click! |
| Bcl11a | rn6_v1_chr14_+_108826831_108826939 | 0.09 | 1.3e-01 | Click! |
Activity profile of Runx2_Bcl11a motif
Sorted Z-values of Runx2_Bcl11a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_206342066 | 127.91 |
ENSRNOT00000026556
|
Ptpn22
|
protein tyrosine phosphatase, non-receptor type 22 |
| chr15_+_32763067 | 117.83 |
ENSRNOT00000011998
|
AABR07017902.1
|
|
| chr3_+_19772056 | 107.96 |
ENSRNOT00000044455
|
AABR07051708.1
|
|
| chr10_-_70871066 | 107.30 |
ENSRNOT00000015139
|
Ccl3
|
C-C motif chemokine ligand 3 |
| chr15_-_35417273 | 102.44 |
ENSRNOT00000083961
ENSRNOT00000041430 |
Gzmb
|
granzyme B |
| chr3_-_91217491 | 101.04 |
ENSRNOT00000006115
|
Rag1
|
recombination activating 1 |
| chr11_+_86092468 | 99.49 |
ENSRNOT00000057971
|
LOC100361706
|
lambda-chain C1-region-like |
| chr4_+_70572942 | 99.37 |
ENSRNOT00000051964
|
AC142181.1
|
|
| chr3_+_18970574 | 90.25 |
ENSRNOT00000088394
|
AABR07051665.1
|
|
| chrX_-_71616997 | 87.74 |
ENSRNOT00000004406
|
Cxcr3
|
C-X-C motif chemokine receptor 3 |
| chr15_-_36472327 | 85.38 |
ENSRNOT00000032601
ENSRNOT00000059660 |
LOC102553861
|
granzyme-like protein 1-like |
| chr1_+_219173002 | 82.32 |
ENSRNOT00000024018
|
Unc93b1
|
unc-93 homolog B1 (C. elegans) |
| chr13_-_83202864 | 78.84 |
ENSRNOT00000003976
|
Xcl1
|
X-C motif chemokine ligand 1 |
| chr18_-_28438654 | 78.80 |
ENSRNOT00000036301
|
Mzb1
|
marginal zone B and B1 cell-specific protein |
| chr10_-_94500591 | 78.80 |
ENSRNOT00000015976
|
Cd79b
|
CD79b molecule |
| chr5_-_58198782 | 78.64 |
ENSRNOT00000023951
|
Ccl21
|
C-C motif chemokine ligand 21 |
| chr2_+_186703541 | 76.13 |
ENSRNOT00000093342
|
Fcrl1
|
Fc receptor-like 1 |
| chr15_-_36114752 | 75.85 |
ENSRNOT00000045209
|
Gzmbl3
|
Granzyme B-like 3 |
| chr3_-_44177689 | 75.59 |
ENSRNOT00000006387
|
Cytip
|
cytohesin 1 interacting protein |
| chr10_-_31493419 | 72.28 |
ENSRNOT00000009211
|
Itk
|
IL2-inducible T-cell kinase |
| chrX_+_15155230 | 70.14 |
ENSRNOT00000073289
ENSRNOT00000051439 |
Was
|
Wiskott-Aldrich syndrome |
| chr10_-_56270640 | 69.34 |
ENSRNOT00000056918
|
Cd68
|
Cd68 molecule |
| chr1_-_226887156 | 65.89 |
ENSRNOT00000054809
ENSRNOT00000028347 |
Cd6
|
Cd6 molecule |
| chr15_-_34693034 | 65.19 |
ENSRNOT00000083314
|
Mcpt8
|
mast cell protease 8 |
| chr10_+_104952458 | 63.29 |
ENSRNOT00000074082
|
RGD1559482
|
similar to immunoglobulin superfamily, member 7 |
| chr1_+_216191886 | 62.24 |
ENSRNOT00000054863
|
Tspan32
|
tetraspanin 32 |
| chr3_+_19045214 | 62.15 |
ENSRNOT00000070878
|
AABR07051670.1
|
|
| chr4_+_162541751 | 62.06 |
ENSRNOT00000010312
|
Klrb1c
|
killer cell lectin-like receptor subfamily B member 1C |
| chr11_-_33003021 | 61.63 |
ENSRNOT00000084134
|
Runx1
|
runt-related transcription factor 1 |
| chr19_-_43911057 | 59.88 |
ENSRNOT00000026017
|
Ctrb1
|
chymotrypsinogen B1 |
| chr4_+_14212925 | 58.80 |
ENSRNOT00000076946
|
LOC103690020
|
platelet glycoprotein 4-like |
| chr1_+_81230612 | 58.75 |
ENSRNOT00000026489
|
Kcnn4
|
potassium calcium-activated channel subfamily N member 4 |
| chr13_-_111917587 | 57.85 |
ENSRNOT00000007649
|
Traf3ip3
|
TRAF3 interacting protein 3 |
| chr10_+_104997205 | 56.80 |
ENSRNOT00000074003
|
RGD1561778
|
similar to dendritic cell-derived immunoglobulin(Ig)-like receptor 1, DIgR1 - mouse |
| chr4_+_78513459 | 56.48 |
ENSRNOT00000082905
|
Aoc1
|
amine oxidase, copper containing 1 |
| chr11_+_57108956 | 55.60 |
ENSRNOT00000035485
|
Cd96
|
CD96 molecule |
| chr4_-_155867708 | 55.51 |
ENSRNOT00000051525
|
Clec4a2
|
C-type lectin domain family 4, member A2 |
| chr3_-_6626284 | 53.26 |
ENSRNOT00000012494
|
Fcnb
|
ficolin B |
| chr20_-_5805627 | 52.95 |
ENSRNOT00000085996
|
Clps
|
colipase |
| chr12_-_20276121 | 52.85 |
ENSRNOT00000065873
|
LOC685157
|
similar to paired immunoglobin-like type 2 receptor beta |
| chr20_+_3156170 | 52.74 |
ENSRNOT00000082880
|
RT1-S2
|
RT1 class Ib, locus S2 |
| chr11_-_60547201 | 52.54 |
ENSRNOT00000093151
|
Btla
|
B and T lymphocyte associated |
| chr20_-_5806097 | 52.52 |
ENSRNOT00000000611
|
Clps
|
colipase |
| chr8_-_114617466 | 52.08 |
ENSRNOT00000082992
ENSRNOT00000038160 |
Col6a5
|
collagen type VI alpha 5 chain |
| chr15_-_34647421 | 51.14 |
ENSRNOT00000072426
|
Mcpt8
|
mast cell protease 8 |
| chr13_-_61591139 | 50.39 |
ENSRNOT00000005324
|
Rgs18
|
regulator of G-protein signaling 18 |
| chr9_-_14668297 | 50.26 |
ENSRNOT00000042404
|
Treml2
|
triggering receptor expressed on myeloid cells-like 2 |
| chr10_-_19574094 | 49.76 |
ENSRNOT00000059810
|
Dock2
|
dedicator of cytokinesis 2 |
| chr4_-_157154120 | 49.53 |
ENSRNOT00000056058
|
C1s
|
complement C1s |
| chr4_-_162025090 | 49.51 |
ENSRNOT00000085887
ENSRNOT00000009904 |
Klrb1a
|
killer cell lectin-like receptor subfamily B, member 1A |
| chr12_-_2245324 | 49.02 |
ENSRNOT00000001332
ENSRNOT00000001333 |
Fcer2
|
Fc fragment of IgE receptor II |
| chr6_-_138321002 | 47.79 |
ENSRNOT00000056845
|
Ighm
|
immunoglobulin heavy constant mu |
| chr3_-_7141522 | 47.75 |
ENSRNOT00000014572
|
Cel
|
carboxyl ester lipase |
| chr3_-_46457201 | 47.18 |
ENSRNOT00000010683
|
Ly75
|
lymphocyte antigen 75 |
| chr9_+_18564927 | 45.98 |
ENSRNOT00000061014
|
Runx2
|
runt-related transcription factor 2 |
| chr1_-_98493978 | 45.57 |
ENSRNOT00000023942
|
Nkg7
|
natural killer cell granule protein 7 |
| chr9_+_67546408 | 45.25 |
ENSRNOT00000013701
|
Cd28
|
Cd28 molecule |
| chr4_+_66670618 | 44.82 |
ENSRNOT00000010796
|
Tbxas1
|
thromboxane A synthase 1 |
| chr11_-_60546997 | 44.46 |
ENSRNOT00000083124
ENSRNOT00000050092 |
Btla
|
B and T lymphocyte associated |
| chr3_+_19141133 | 43.95 |
ENSRNOT00000058323
|
AABR07051675.1
|
|
| chr3_+_19366370 | 43.80 |
ENSRNOT00000086557
|
AABR07051689.1
|
|
| chr11_+_84972132 | 43.58 |
ENSRNOT00000034498
|
Lamp3
|
lysosomal-associated membrane protein 3 |
| chr10_+_104932616 | 43.20 |
ENSRNOT00000075706
|
RGD1562667
|
similar to leukocyte mono-Ig-like receptor2 |
| chr10_-_56506446 | 42.97 |
ENSRNOT00000021357
|
Acap1
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 1 |
| chr7_-_12741296 | 42.83 |
ENSRNOT00000060648
|
Arhgap45
|
Rho GTPase activating protein 45 |
| chr3_+_18787606 | 42.69 |
ENSRNOT00000090508
|
AABR07051658.1
|
|
| chr6_-_142635763 | 42.63 |
ENSRNOT00000048908
|
AABR07065815.2
|
|
| chr20_-_21689553 | 42.55 |
ENSRNOT00000038095
|
Tmem26
|
transmembrane protein 26 |
| chr6_-_139660666 | 42.45 |
ENSRNOT00000086098
|
AABR07065705.1
|
|
| chr1_+_88875375 | 42.23 |
ENSRNOT00000028284
|
Tyrobp
|
Tyro protein tyrosine kinase binding protein |
| chr10_-_34242985 | 42.08 |
ENSRNOT00000046438
|
RGD1559575
|
similar to novel protein |
| chr1_-_267463694 | 41.99 |
ENSRNOT00000084851
ENSRNOT00000016645 |
Col17a1
|
collagen type XVII alpha 1 chain |
| chr20_-_5076539 | 41.70 |
ENSRNOT00000076253
ENSRNOT00000037613 |
Ly6g6f
|
lymphocyte antigen 6 complex, locus G6F |
| chr6_-_143195445 | 40.72 |
ENSRNOT00000078672
|
AABR07065837.1
|
|
| chr4_+_119680112 | 40.60 |
ENSRNOT00000013282
|
Gp9
|
glycoprotein IX (platelet) |
| chr2_-_149325913 | 40.59 |
ENSRNOT00000036690
|
Gpr171
|
G protein-coupled receptor 171 |
| chr20_+_10438444 | 40.23 |
ENSRNOT00000071248
ENSRNOT00000075545 |
Cryaa
|
crystallin, alpha A |
| chr1_+_114046478 | 40.20 |
ENSRNOT00000032254
|
Siglech
|
sialic acid binding Ig-like lectin H |
| chr1_+_199555722 | 39.82 |
ENSRNOT00000054983
|
Itgax
|
integrin subunit alpha X |
| chr16_+_20110148 | 39.73 |
ENSRNOT00000080146
ENSRNOT00000025312 |
Jak3
|
Janus kinase 3 |
| chr16_+_20109200 | 39.00 |
ENSRNOT00000079784
|
Jak3
|
Janus kinase 3 |
| chr13_+_42008842 | 38.04 |
ENSRNOT00000038811
|
Gpr39
|
G protein-coupled receptor 39 |
| chrX_+_14578264 | 37.86 |
ENSRNOT00000038994
|
Cybb
|
cytochrome b-245 beta chain |
| chr10_+_15697227 | 37.56 |
ENSRNOT00000028008
|
Il9r
|
interleukin 9 receptor |
| chr2_-_44981458 | 37.47 |
ENSRNOT00000014134
|
Gzma
|
granzyme A |
| chr11_+_61970976 | 37.10 |
ENSRNOT00000078921
|
Tigit
|
T cell immunoreceptor with Ig and ITIM domains |
| chr15_-_34625121 | 37.09 |
ENSRNOT00000073555
|
Mcpt8l2
|
mast cell protease 8-like 2 |
| chr15_-_34694180 | 37.07 |
ENSRNOT00000079505
ENSRNOT00000027950 ENSRNOT00000079782 ENSRNOT00000080221 |
Mcpt8
|
mast cell protease 8 |
| chr16_-_9430743 | 36.88 |
ENSRNOT00000043811
|
Wdfy4
|
WDFY family member 4 |
| chr13_-_90022269 | 36.62 |
ENSRNOT00000035498
|
Ly9
|
lymphocyte antigen 9 |
| chr2_+_55835151 | 36.22 |
ENSRNOT00000018634
|
Fyb
|
FYN binding protein |
| chr7_-_12635943 | 36.21 |
ENSRNOT00000015029
|
Cfd
|
complement factor D |
| chr1_+_81230989 | 35.75 |
ENSRNOT00000077952
|
Kcnn4
|
potassium calcium-activated channel subfamily N member 4 |
| chr16_+_10417185 | 35.48 |
ENSRNOT00000082186
|
Anxa8
|
annexin A8 |
| chr15_-_28081465 | 34.91 |
ENSRNOT00000033739
|
Rnase6
|
ribonuclease, RNase A family, 6 |
| chrX_-_159841072 | 34.72 |
ENSRNOT00000001158
|
Arhgef6
|
Rac/Cdc42 guanine nucleotide exchange factor 6 |
| chr1_+_101214593 | 34.23 |
ENSRNOT00000028086
|
Tead2
|
TEA domain transcription factor 2 |
| chr1_-_227506822 | 33.86 |
ENSRNOT00000091506
|
Ms4a7
|
membrane spanning 4-domains A7 |
| chr5_-_155772040 | 33.55 |
ENSRNOT00000036788
|
Cela3b
|
chymotrypsin-like elastase family, member 3B |
| chr6_-_142676432 | 33.46 |
ENSRNOT00000074947
|
AABR07065815.2
|
|
| chr6_-_142585188 | 33.43 |
ENSRNOT00000067437
|
AABR07065815.1
|
|
| chr15_+_35032049 | 33.17 |
ENSRNOT00000091757
|
Mcpt2
|
mast cell protease 2 |
| chr3_+_19441604 | 33.11 |
ENSRNOT00000090581
|
AABR07051692.1
|
|
| chr9_+_47165342 | 32.80 |
ENSRNOT00000020144
|
Il1rl1
|
interleukin 1 receptor-like 1 |
| chr10_+_59652549 | 32.27 |
ENSRNOT00000073209
|
Itgae
|
integrin subunit alpha E |
| chr8_-_13251812 | 32.02 |
ENSRNOT00000049724
ENSRNOT00000078117 |
Piwil4
|
piwi-like RNA-mediated gene silencing 4 |
| chr1_-_237910012 | 31.40 |
ENSRNOT00000023664
|
Anxa1
|
annexin A1 |
| chr4_-_156276304 | 30.95 |
ENSRNOT00000078725
|
Clec4e
|
C-type lectin domain family 4, member E |
| chr13_+_51958834 | 30.78 |
ENSRNOT00000007833
|
Ptpn7
|
protein tyrosine phosphatase, non-receptor type 7 |
| chr1_-_82004538 | 30.65 |
ENSRNOT00000087572
|
Pou2f2
|
POU class 2 homeobox 2 |
| chr10_+_104952237 | 30.34 |
ENSRNOT00000085222
|
RGD1559482
|
similar to immunoglobulin superfamily, member 7 |
| chr4_-_119327822 | 30.30 |
ENSRNOT00000012645
|
Arhgap25
|
Rho GTPase activating protein 25 |
| chr4_+_119225040 | 30.12 |
ENSRNOT00000012365
|
Bmp10
|
bone morphogenetic protein 10 |
| chr14_+_10692764 | 29.74 |
ENSRNOT00000003012
|
LOC100910270
|
uncharacterized LOC100910270 |
| chr12_-_21923281 | 29.63 |
ENSRNOT00000075641
|
RGD1561143
|
similar to cell surface receptor FDFACT |
| chr15_-_51855325 | 29.37 |
ENSRNOT00000011663
|
Pdlim2
|
PDZ and LIM domain 2 |
| chr7_+_61337381 | 29.28 |
ENSRNOT00000009919
|
Ifng
|
interferon gamma |
| chr2_-_203680083 | 29.12 |
ENSRNOT00000021268
|
Cd2
|
Cd2 molecule |
| chr6_-_139911839 | 28.92 |
ENSRNOT00000077113
ENSRNOT00000084547 |
AABR07065714.1
|
|
| chr1_-_169456098 | 28.82 |
ENSRNOT00000030827
|
Trim30c
|
tripartite motif-containing 30C |
| chr8_+_2604962 | 28.66 |
ENSRNOT00000009993
|
Casp1
|
caspase 1 |
| chr13_+_88606894 | 28.64 |
ENSRNOT00000048692
|
Sh2d1b
|
SH2 domain containing 1B |
| chr4_-_70996395 | 28.24 |
ENSRNOT00000021485
|
Kel
|
Kell blood group, metallo-endopeptidase |
| chr2_-_198942732 | 27.97 |
ENSRNOT00000000109
ENSRNOT00000073913 |
Cd160
|
CD160 molecule |
| chr4_+_69386698 | 27.53 |
ENSRNOT00000091655
|
Trbv13-2
|
T cell receptor beta, variable 13-2 |
| chr12_-_21362205 | 27.46 |
ENSRNOT00000064787
|
LOC108348155
|
paired immunoglobulin-like type 2 receptor beta-2 |
| chr7_-_119797098 | 27.23 |
ENSRNOT00000009994
|
Rac2
|
ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2) |
| chr8_-_129371973 | 27.09 |
ENSRNOT00000042614
|
Snrpe
|
small nuclear ribonucleoprotein polypeptide E |
| chr15_-_34612432 | 27.02 |
ENSRNOT00000090206
|
Mcpt1l1
|
mast cell protease 1-like 1 |
| chr8_+_55603968 | 26.77 |
ENSRNOT00000066848
|
Pou2af1
|
POU class 2 associating factor 1 |
| chr1_-_167700332 | 26.73 |
ENSRNOT00000092890
|
Trim21
|
tripartite motif-containing 21 |
| chr11_+_88122271 | 26.58 |
ENSRNOT00000002540
|
Sdf2l1
|
stromal cell-derived factor 2-like 1 |
| chr3_+_20007192 | 26.56 |
ENSRNOT00000075229
|
AABR07051716.1
|
|
| chr14_-_18853315 | 26.40 |
ENSRNOT00000003794
|
Ppbp
|
pro-platelet basic protein |
| chr16_-_75340360 | 26.32 |
ENSRNOT00000018501
|
Defa5
|
defensin alpha 5 |
| chr14_+_13192347 | 26.23 |
ENSRNOT00000000092
|
Antxr2
|
anthrax toxin receptor 2 |
| chr20_+_30791422 | 26.22 |
ENSRNOT00000047394
ENSRNOT00000000683 |
Tbata
|
thymus, brain and testes associated |
| chr17_-_32783427 | 26.14 |
ENSRNOT00000059921
|
Serpinb6b
|
serine (or cysteine) peptidase inhibitor, clade B, member 6b |
| chr3_+_17180411 | 25.70 |
ENSRNOT00000058260
|
AABR07051565.1
|
|
| chr9_+_67763897 | 25.68 |
ENSRNOT00000071226
|
Icos
|
inducible T-cell co-stimulator |
| chr1_-_173528943 | 25.58 |
ENSRNOT00000020393
|
Nlrp10
|
NLR family, pyrin domain containing 10 |
| chr4_+_14213587 | 25.33 |
ENSRNOT00000067543
|
LOC103690020
|
platelet glycoprotein 4-like |
| chr18_-_28535828 | 25.29 |
ENSRNOT00000068386
|
Tmem173
|
transmembrane protein 173 |
| chr3_+_18805220 | 25.19 |
ENSRNOT00000071216
|
Igkv4-86
|
immunoglobulin kappa variable 4-86 |
| chr10_-_110701137 | 25.07 |
ENSRNOT00000074193
|
Znf750
|
zinc finger protein 750 |
| chr6_-_142933110 | 24.98 |
ENSRNOT00000046885
|
AABR07065823.1
|
|
| chr13_-_110784209 | 24.81 |
ENSRNOT00000071698
|
Traf5
|
TNF receptor-associated factor 5 |
| chr7_+_121841855 | 24.56 |
ENSRNOT00000024673
|
Grap2
|
GRB2-related adaptor protein 2 |
| chr1_+_63734135 | 24.48 |
ENSRNOT00000084439
|
Nilr1
|
neutrophil immunoglobulin-like receptor-1 |
| chr1_+_31835000 | 24.30 |
ENSRNOT00000020780
|
Cep72
|
centrosomal protein 72 |
| chr10_-_89900131 | 23.76 |
ENSRNOT00000028238
|
Sost
|
sclerostin |
| chr14_-_100184192 | 23.72 |
ENSRNOT00000007044
|
Plek
|
pleckstrin |
| chr20_-_4316715 | 23.65 |
ENSRNOT00000031704
|
C4b
|
complement C4B (Chido blood group) |
| chr3_+_19128400 | 23.54 |
ENSRNOT00000074272
|
AABR07051673.1
|
|
| chr20_-_4508197 | 23.44 |
ENSRNOT00000086027
ENSRNOT00000000514 |
C4a
|
complement component 4A (Rodgers blood group) |
| chr4_-_164049599 | 23.40 |
ENSRNOT00000078267
|
AABR07062183.1
|
|
| chr20_-_2678141 | 23.10 |
ENSRNOT00000072377
ENSRNOT00000083833 |
C4a
|
complement component 4A (Rodgers blood group) |
| chr1_+_165506361 | 23.02 |
ENSRNOT00000024156
|
Ucp2
|
uncoupling protein 2 |
| chr1_-_198662610 | 22.95 |
ENSRNOT00000055012
|
Sept1
|
septin 1 |
| chr14_-_5378726 | 22.92 |
ENSRNOT00000002896
|
Lrrc8c
|
leucine rich repeat containing 8 family, member C |
| chr6_-_107658741 | 22.74 |
ENSRNOT00000088712
|
Elmsan1
|
ELM2 and Myb/SANT domain containing 1 |
| chr4_-_165541314 | 22.42 |
ENSRNOT00000013833
|
Styk1
|
serine/threonine/tyrosine kinase 1 |
| chr1_-_169463760 | 22.41 |
ENSRNOT00000023100
|
Trim30c
|
tripartite motif-containing 30C |
| chr7_-_144880092 | 22.40 |
ENSRNOT00000055281
|
Nfe2
|
nuclear factor, erythroid 2 |
| chr14_-_22890958 | 22.22 |
ENSRNOT00000031916
|
LOC100911104
|
lymphocyte antigen 6B-like |
| chr1_+_197999037 | 22.13 |
ENSRNOT00000091065
|
Apobr
|
apolipoprotein B receptor |
| chr11_+_71211768 | 21.90 |
ENSRNOT00000079753
|
LOC100910650
|
uncharacterized LOC100910650 |
| chr10_-_57436368 | 21.64 |
ENSRNOT00000056608
|
Scimp
|
SLP adaptor and CSK interacting membrane protein |
| chr10_+_45258887 | 21.63 |
ENSRNOT00000048642
|
Btnl10
|
butyrophilin like 10 |
| chr10_+_1406486 | 21.58 |
ENSRNOT00000078773
|
LOC679822
|
similar to dynein, cytoplasmic, light peptide |
| chr3_+_20303979 | 21.57 |
ENSRNOT00000058331
|
AABR07051730.1
|
|
| chr3_-_92749121 | 21.49 |
ENSRNOT00000008760
|
Cd44
|
CD44 molecule (Indian blood group) |
| chr4_+_57855416 | 21.42 |
ENSRNOT00000029608
|
Cpa2
|
carboxypeptidase A2 |
| chr12_+_21678580 | 21.37 |
ENSRNOT00000039797
|
LOC100910669
|
paired immunoglobulin-like type 2 receptor alpha-like |
| chr3_+_20375699 | 21.31 |
ENSRNOT00000088492
|
AABR07051731.1
|
|
| chr20_-_4561062 | 21.20 |
ENSRNOT00000065044
ENSRNOT00000092698 ENSRNOT00000060607 |
Cfb
C2
|
complement factor B complement C2 |
| chr18_-_27520295 | 21.18 |
ENSRNOT00000027544
|
Gfra3
|
GDNF family receptor alpha 3 |
| chr11_+_84506703 | 21.13 |
ENSRNOT00000066953
|
Cyp2ab1
|
cytochrome P450, family 2, subfamily ab, polypeptide 1 |
| chr8_-_69121192 | 21.04 |
ENSRNOT00000012404
|
Zwilch
|
zwilch kinetochore protein |
| chrX_-_62661152 | 20.94 |
ENSRNOT00000075997
|
Pola1
|
DNA polymerase alpha 1, catalytic subunit |
| chr10_+_69423086 | 20.90 |
ENSRNOT00000000256
|
Ccl7
|
C-C motif chemokine ligand 7 |
| chr1_-_259357056 | 20.80 |
ENSRNOT00000022000
|
Pdlim1
|
PDZ and LIM domain 1 |
| chr10_-_86930947 | 20.74 |
ENSRNOT00000081440
|
Top2a
|
topoisomerase (DNA) II alpha |
| chr13_+_92249759 | 20.49 |
ENSRNOT00000089352
|
Olr1596
|
olfactory receptor 1596 |
| chr8_-_133002201 | 20.45 |
ENSRNOT00000008772
|
Ccr1
|
C-C motif chemokine receptor 1 |
| chr2_+_87418517 | 20.42 |
ENSRNOT00000048046
|
LOC100909879
|
tyrosine-protein phosphatase non-receptor type substrate 1-like |
| chr10_-_5253336 | 20.41 |
ENSRNOT00000085310
|
Ciita
|
class II, major histocompatibility complex, transactivator |
| chr5_-_160405050 | 20.40 |
ENSRNOT00000081899
|
Ctrc
|
chymotrypsin C |
| chr1_+_98398660 | 20.32 |
ENSRNOT00000047473
|
Cd33
|
CD33 molecule |
| chr13_-_61070599 | 20.30 |
ENSRNOT00000005251
|
Rgs1
|
regulator of G-protein signaling 1 |
| chr5_+_64566804 | 20.29 |
ENSRNOT00000073192
|
LOC103690035
|
TGF-beta receptor type-1-like |
| chr5_+_151181559 | 20.27 |
ENSRNOT00000085674
|
Fgr
|
FGR proto-oncogene, Src family tyrosine kinase |
| chr1_+_196996581 | 20.27 |
ENSRNOT00000021690
|
Il21r
|
interleukin 21 receptor |
| chr1_-_85220237 | 20.21 |
ENSRNOT00000026907
|
Sycn
|
syncollin |
| chr13_-_36101411 | 20.07 |
ENSRNOT00000074471
|
Tmem37
|
transmembrane protein 37 |
| chr15_+_31719866 | 20.03 |
ENSRNOT00000072127
|
AABR07017838.2
|
|
| chr13_+_27465930 | 19.90 |
ENSRNOT00000003314
|
Serpinb10
|
serpin family B member 10 |
| chr3_-_153250641 | 19.72 |
ENSRNOT00000008961
|
Samhd1
|
SAM and HD domain containing deoxynucleoside triphosphate triphosphohydrolase 1 |
| chr14_+_33010300 | 19.63 |
ENSRNOT00000002809
|
Igfbp7
|
insulin-like growth factor binding protein 7 |
| chrX_+_156873849 | 19.60 |
ENSRNOT00000085410
|
Arhgap4
|
Rho GTPase activating protein 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Runx2_Bcl11a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 68.9 | 206.7 | GO:0071663 | granzyme B production(GO:0071613) regulation of granzyme B production(GO:0071661) positive regulation of granzyme B production(GO:0071663) |
| 34.1 | 102.4 | GO:0060545 | positive regulation of necroptotic process(GO:0060545) |
| 26.2 | 78.7 | GO:0045210 | negative regulation of dendritic cell cytokine production(GO:0002731) FasL biosynthetic process(GO:0045210) |
| 26.2 | 78.6 | GO:0035759 | mesangial cell-matrix adhesion(GO:0035759) |
| 25.6 | 128.2 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 25.3 | 101.0 | GO:0002331 | pre-B cell allelic exclusion(GO:0002331) |
| 20.5 | 61.6 | GO:2000872 | positive regulation of progesterone secretion(GO:2000872) |
| 19.7 | 78.8 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) |
| 16.6 | 49.8 | GO:0002277 | myeloid dendritic cell activation involved in immune response(GO:0002277) |
| 16.5 | 82.3 | GO:0034154 | toll-like receptor 7 signaling pathway(GO:0034154) |
| 15.6 | 62.2 | GO:0030886 | negative regulation of myeloid dendritic cell activation(GO:0030886) |
| 15.2 | 45.6 | GO:0002380 | immunoglobulin secretion involved in immune response(GO:0002380) |
| 13.3 | 53.3 | GO:0002752 | cell surface pattern recognition receptor signaling pathway(GO:0002752) |
| 12.7 | 38.0 | GO:0035483 | gastric emptying(GO:0035483) |
| 12.6 | 37.9 | GO:1904844 | response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
| 12.3 | 49.0 | GO:0002925 | positive regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002925) |
| 11.8 | 118.5 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 11.7 | 70.1 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 11.7 | 46.7 | GO:0046061 | dATP catabolic process(GO:0046061) |
| 10.9 | 32.8 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 10.7 | 21.4 | GO:0002876 | regulation of chronic inflammatory response to antigenic stimulus(GO:0002874) positive regulation of chronic inflammatory response to antigenic stimulus(GO:0002876) |
| 9.8 | 29.3 | GO:0060559 | positive regulation of vitamin metabolic process(GO:0046136) positive regulation of vitamin D biosynthetic process(GO:0060557) positive regulation of calcidiol 1-monooxygenase activity(GO:0060559) |
| 9.6 | 95.7 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 9.6 | 66.9 | GO:0032429 | regulation of phospholipase A2 activity(GO:0032429) |
| 8.4 | 41.8 | GO:0071104 | response to interleukin-9(GO:0071104) |
| 8.1 | 24.3 | GO:0033566 | gamma-tubulin complex localization(GO:0033566) |
| 7.9 | 55.6 | GO:0002727 | natural killer cell cytokine production(GO:0002370) regulation of natural killer cell cytokine production(GO:0002727) |
| 7.9 | 23.8 | GO:2000054 | negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 7.7 | 46.0 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 7.7 | 23.0 | GO:0007056 | spindle assembly involved in female meiosis(GO:0007056) |
| 7.4 | 110.3 | GO:0001771 | immunological synapse formation(GO:0001771) |
| 7.2 | 28.7 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 6.9 | 20.7 | GO:0010752 | regulation of cGMP-mediated signaling(GO:0010752) |
| 6.9 | 13.8 | GO:2000422 | regulation of eosinophil chemotaxis(GO:2000422) positive regulation of eosinophil chemotaxis(GO:2000424) |
| 6.9 | 27.5 | GO:2000318 | positive regulation of T-helper 17 type immune response(GO:2000318) |
| 6.7 | 40.2 | GO:0070309 | lens fiber cell morphogenesis(GO:0070309) |
| 6.7 | 93.8 | GO:1900118 | negative regulation of execution phase of apoptosis(GO:1900118) |
| 6.6 | 72.3 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 6.2 | 37.5 | GO:0032074 | negative regulation of nuclease activity(GO:0032074) |
| 6.1 | 18.3 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 5.9 | 23.7 | GO:0070560 | protein secretion by platelet(GO:0070560) |
| 5.6 | 44.8 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 5.5 | 22.2 | GO:0060298 | positive regulation of sarcomere organization(GO:0060298) |
| 5.3 | 15.9 | GO:2001226 | negative regulation of inorganic anion transmembrane transport(GO:1903796) negative regulation of chloride transport(GO:2001226) |
| 5.2 | 20.9 | GO:0006272 | leading strand elongation(GO:0006272) |
| 5.2 | 36.6 | GO:0032740 | positive regulation of interleukin-17 production(GO:0032740) |
| 5.2 | 15.6 | GO:0019303 | D-ribose catabolic process(GO:0019303) |
| 5.2 | 15.6 | GO:1990868 | response to chemokine(GO:1990868) cellular response to chemokine(GO:1990869) |
| 5.0 | 55.5 | GO:0048304 | positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 5.0 | 15.0 | GO:0055073 | cadmium ion homeostasis(GO:0055073) |
| 5.0 | 44.8 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 4.7 | 28.2 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 4.7 | 42.2 | GO:2001204 | regulation of osteoclast development(GO:2001204) |
| 4.7 | 14.0 | GO:0042125 | protein glycosylation at cell surface(GO:0033575) protein galactosylation at cell surface(GO:0033580) protein galactosylation(GO:0042125) |
| 4.6 | 18.5 | GO:0010999 | regulation of eIF2 alpha phosphorylation by heme(GO:0010999) |
| 4.6 | 32.0 | GO:0010528 | regulation of transposition(GO:0010528) negative regulation of transposition(GO:0010529) |
| 4.4 | 13.3 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 4.4 | 13.3 | GO:0002414 | immunoglobulin transcytosis in epithelial cells(GO:0002414) |
| 4.3 | 8.6 | GO:0033668 | negative regulation by symbiont of host apoptotic process(GO:0033668) negative regulation by symbiont of host programmed cell death(GO:0052041) negative regulation by organism of programmed cell death in other organism involved in symbiotic interaction(GO:0052490) |
| 4.3 | 25.7 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 4.2 | 4.2 | GO:0039530 | MDA-5 signaling pathway(GO:0039530) |
| 4.2 | 29.1 | GO:2000484 | positive regulation of interleukin-8 secretion(GO:2000484) |
| 4.1 | 8.3 | GO:0034163 | regulation of toll-like receptor 9 signaling pathway(GO:0034163) |
| 4.1 | 12.3 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 4.1 | 16.4 | GO:0002884 | negative regulation of hypersensitivity(GO:0002884) |
| 4.0 | 11.9 | GO:0001788 | antibody-dependent cellular cytotoxicity(GO:0001788) |
| 3.9 | 47.2 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 3.9 | 11.8 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) |
| 3.9 | 27.2 | GO:0060753 | regulation of mast cell chemotaxis(GO:0060753) |
| 3.9 | 19.3 | GO:1990086 | lens fiber cell apoptotic process(GO:1990086) |
| 3.8 | 34.5 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 3.8 | 15.2 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 3.7 | 47.8 | GO:0006707 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 3.6 | 18.2 | GO:0072564 | blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 3.4 | 10.3 | GO:0042222 | interleukin-1 biosynthetic process(GO:0042222) |
| 3.4 | 20.4 | GO:0045341 | MHC class I biosynthetic process(GO:0045341) regulation of MHC class I biosynthetic process(GO:0045343) positive regulation of MHC class I biosynthetic process(GO:0045345) |
| 3.4 | 20.3 | GO:0061737 | leukotriene signaling pathway(GO:0061737) |
| 3.4 | 10.1 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 3.3 | 6.7 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 3.3 | 10.0 | GO:1901674 | histone H3-K27 acetylation(GO:0043974) regulation of histone H3-K27 acetylation(GO:1901674) |
| 3.3 | 9.8 | GO:0002148 | hypochlorous acid metabolic process(GO:0002148) hypochlorous acid biosynthetic process(GO:0002149) |
| 3.2 | 6.3 | GO:0098758 | response to interleukin-8(GO:0098758) cellular response to interleukin-8(GO:0098759) |
| 3.2 | 12.6 | GO:0046338 | phosphatidylethanolamine catabolic process(GO:0046338) |
| 3.2 | 37.9 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 3.2 | 12.6 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 3.1 | 12.4 | GO:0051106 | regulation of DNA ligation(GO:0051105) positive regulation of DNA ligation(GO:0051106) |
| 3.1 | 9.2 | GO:0006529 | asparagine biosynthetic process(GO:0006529) |
| 3.0 | 21.3 | GO:0003057 | regulation of the force of heart contraction by chemical signal(GO:0003057) |
| 3.0 | 39.5 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 3.0 | 27.1 | GO:0052697 | xenobiotic glucuronidation(GO:0052697) |
| 3.0 | 9.0 | GO:0060370 | susceptibility to T cell mediated cytotoxicity(GO:0060370) |
| 3.0 | 54.0 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 3.0 | 6.0 | GO:1904124 | microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 3.0 | 5.9 | GO:0071930 | negative regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071930) |
| 3.0 | 11.9 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 3.0 | 8.9 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 2.9 | 14.6 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 2.9 | 31.8 | GO:0046598 | positive regulation of viral entry into host cell(GO:0046598) |
| 2.8 | 56.5 | GO:0071280 | cellular response to copper ion(GO:0071280) |
| 2.8 | 8.4 | GO:0045077 | peripheral T cell tolerance induction(GO:0002458) peripheral tolerance induction(GO:0002465) negative regulation of interferon-gamma biosynthetic process(GO:0045077) |
| 2.8 | 8.4 | GO:0070638 | nicotinamide riboside catabolic process(GO:0006738) nicotinamide riboside metabolic process(GO:0046495) pyridine nucleoside metabolic process(GO:0070637) pyridine nucleoside catabolic process(GO:0070638) |
| 2.8 | 19.3 | GO:0021773 | striatal medium spiny neuron differentiation(GO:0021773) |
| 2.7 | 8.1 | GO:2000620 | positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 2.7 | 13.5 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 2.7 | 8.0 | GO:1903898 | cap-dependent translational initiation(GO:0002191) positive regulation of translation in response to endoplasmic reticulum stress(GO:0036493) negative regulation of PERK-mediated unfolded protein response(GO:1903898) |
| 2.7 | 26.6 | GO:0090070 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 2.7 | 13.3 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 2.6 | 37.1 | GO:0032695 | negative regulation of interleukin-12 production(GO:0032695) |
| 2.6 | 7.9 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 2.6 | 15.7 | GO:0060700 | regulation of ribonuclease activity(GO:0060700) |
| 2.6 | 10.5 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) positive regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900224) |
| 2.6 | 7.8 | GO:0060697 | positive regulation of phospholipid catabolic process(GO:0060697) |
| 2.6 | 10.4 | GO:0090290 | positive regulation of natural killer cell proliferation(GO:0032819) positive regulation of osteoclast proliferation(GO:0090290) |
| 2.6 | 7.8 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 2.6 | 5.1 | GO:0045626 | negative regulation of T-helper 1 cell differentiation(GO:0045626) |
| 2.6 | 5.1 | GO:0032752 | regulation of interleukin-3 production(GO:0032672) positive regulation of interleukin-3 production(GO:0032752) interleukin-3 biosynthetic process(GO:0042223) regulation of interleukin-3 biosynthetic process(GO:0045399) positive regulation of interleukin-3 biosynthetic process(GO:0045401) positive regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045425) |
| 2.5 | 17.7 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 2.5 | 25.3 | GO:0071360 | cellular response to exogenous dsRNA(GO:0071360) |
| 2.5 | 9.9 | GO:0055099 | regulation of Cdc42 protein signal transduction(GO:0032489) response to high density lipoprotein particle(GO:0055099) platelet dense granule organization(GO:0060155) |
| 2.4 | 26.6 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 2.4 | 48.2 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 2.4 | 33.6 | GO:1990845 | adaptive thermogenesis(GO:1990845) |
| 2.4 | 30.9 | GO:0038094 | Fc-gamma receptor signaling pathway(GO:0038094) |
| 2.4 | 4.7 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 2.4 | 18.9 | GO:0034214 | protein hexamerization(GO:0034214) |
| 2.3 | 9.4 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 2.3 | 11.5 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 2.3 | 29.7 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 2.3 | 20.3 | GO:0010759 | positive regulation of macrophage chemotaxis(GO:0010759) |
| 2.2 | 143.8 | GO:0032094 | response to food(GO:0032094) |
| 2.2 | 19.6 | GO:0051414 | response to cortisol(GO:0051414) |
| 2.2 | 8.7 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 2.1 | 6.4 | GO:0003221 | right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 2.1 | 19.0 | GO:0016446 | somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 2.1 | 8.4 | GO:0002536 | respiratory burst involved in inflammatory response(GO:0002536) |
| 2.1 | 39.9 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 2.1 | 21.0 | GO:0002517 | T cell tolerance induction(GO:0002517) |
| 2.1 | 10.5 | GO:0072366 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) regulation of cellular ketone metabolic process by positive regulation of transcription from RNA polymerase II promoter(GO:0072366) |
| 2.1 | 8.4 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 2.0 | 10.2 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 2.0 | 12.2 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 2.0 | 6.1 | GO:0048210 | Golgi vesicle fusion to target membrane(GO:0048210) |
| 2.0 | 32.4 | GO:0048368 | lateral mesoderm development(GO:0048368) |
| 2.0 | 9.9 | GO:0002786 | regulation of antimicrobial peptide production(GO:0002784) regulation of antibacterial peptide production(GO:0002786) |
| 2.0 | 11.9 | GO:0072615 | interleukin-17 secretion(GO:0072615) |
| 2.0 | 13.7 | GO:0010764 | negative regulation of fibroblast migration(GO:0010764) |
| 1.9 | 13.6 | GO:0002507 | tolerance induction(GO:0002507) |
| 1.9 | 5.7 | GO:0070495 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 1.9 | 3.8 | GO:0002589 | regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) |
| 1.9 | 7.5 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 1.8 | 5.5 | GO:1900111 | positive regulation of histone H3-K9 dimethylation(GO:1900111) positive regulation of histone H3-K9 trimethylation(GO:1900114) |
| 1.8 | 5.5 | GO:0071603 | endothelial cell-cell adhesion(GO:0071603) |
| 1.8 | 14.6 | GO:0002283 | neutrophil activation involved in immune response(GO:0002283) |
| 1.8 | 3.7 | GO:0032902 | nerve growth factor production(GO:0032902) |
| 1.8 | 14.6 | GO:0060481 | lobar bronchus epithelium development(GO:0060481) |
| 1.8 | 52.8 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 1.8 | 14.5 | GO:0042795 | snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 1.8 | 57.7 | GO:0035456 | response to interferon-beta(GO:0035456) |
| 1.8 | 5.3 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 1.8 | 19.3 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 1.8 | 5.3 | GO:0046333 | octopamine biosynthetic process(GO:0006589) octopamine metabolic process(GO:0046333) |
| 1.7 | 5.1 | GO:0009644 | response to high light intensity(GO:0009644) |
| 1.7 | 5.1 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 1.7 | 5.1 | GO:1904976 | cellular response to bleomycin(GO:1904976) |
| 1.7 | 73.6 | GO:0050853 | B cell receptor signaling pathway(GO:0050853) |
| 1.7 | 5.0 | GO:0032907 | transforming growth factor beta3 production(GO:0032907) regulation of transforming growth factor beta3 production(GO:0032910) positive regulation of transforming growth factor beta3 production(GO:0032916) |
| 1.6 | 26.3 | GO:0031640 | killing of cells of other organism(GO:0031640) disruption of cells of other organism(GO:0044364) |
| 1.6 | 6.5 | GO:0060672 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 1.6 | 6.5 | GO:0032627 | interleukin-23 production(GO:0032627) regulation of interleukin-23 production(GO:0032667) negative regulation of interleukin-23 production(GO:0032707) |
| 1.6 | 6.3 | GO:0071718 | sodium-independent icosanoid transport(GO:0071718) |
| 1.6 | 9.5 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 1.5 | 6.2 | GO:0061073 | ciliary body morphogenesis(GO:0061073) |
| 1.5 | 3.1 | GO:0090579 | transcriptional activation by promoter-enhancer looping(GO:0071733) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 1.5 | 12.0 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 1.5 | 7.5 | GO:0002920 | regulation of humoral immune response(GO:0002920) |
| 1.5 | 5.9 | GO:0023021 | termination of signal transduction(GO:0023021) |
| 1.5 | 24.9 | GO:0045653 | negative regulation of megakaryocyte differentiation(GO:0045653) |
| 1.4 | 7.2 | GO:0006787 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 1.4 | 5.8 | GO:1902608 | regulation of large conductance calcium-activated potassium channel activity(GO:1902606) positive regulation of large conductance calcium-activated potassium channel activity(GO:1902608) |
| 1.4 | 7.2 | GO:0042535 | positive regulation of tumor necrosis factor biosynthetic process(GO:0042535) |
| 1.4 | 8.6 | GO:0060263 | regulation of respiratory burst(GO:0060263) |
| 1.4 | 7.0 | GO:0035617 | stress granule disassembly(GO:0035617) |
| 1.4 | 15.2 | GO:0042738 | exogenous drug catabolic process(GO:0042738) |
| 1.4 | 4.1 | GO:0021678 | third ventricle development(GO:0021678) |
| 1.3 | 2.6 | GO:0060838 | lymphatic endothelial cell fate commitment(GO:0060838) regulation of transcription involved in lymphatic endothelial cell fate commitment(GO:0060849) |
| 1.3 | 10.5 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 1.3 | 5.1 | GO:0060744 | thelarche(GO:0042695) mammary gland branching involved in thelarche(GO:0060744) |
| 1.3 | 15.2 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 1.3 | 7.6 | GO:0033567 | DNA replication, Okazaki fragment processing(GO:0033567) |
| 1.3 | 360.4 | GO:0016485 | protein processing(GO:0016485) |
| 1.3 | 7.5 | GO:0061052 | negative regulation of cell growth involved in cardiac muscle cell development(GO:0061052) |
| 1.3 | 23.8 | GO:0071157 | negative regulation of cell cycle arrest(GO:0071157) |
| 1.2 | 8.7 | GO:0034625 | fatty acid elongation, monounsaturated fatty acid(GO:0034625) |
| 1.2 | 37.3 | GO:0033006 | regulation of mast cell activation involved in immune response(GO:0033006) regulation of mast cell degranulation(GO:0043304) |
| 1.2 | 10.8 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 1.2 | 3.5 | GO:1901844 | regulation of cell communication by electrical coupling involved in cardiac conduction(GO:1901844) |
| 1.2 | 8.2 | GO:0035878 | nail development(GO:0035878) |
| 1.1 | 3.4 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 1.1 | 6.7 | GO:0072675 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 1.1 | 22.0 | GO:0097320 | membrane tubulation(GO:0097320) |
| 1.1 | 6.5 | GO:0048478 | replication fork protection(GO:0048478) |
| 1.1 | 4.3 | GO:0009115 | xanthine catabolic process(GO:0009115) xanthine metabolic process(GO:0046110) |
| 1.1 | 7.4 | GO:0002361 | CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0002361) |
| 1.1 | 5.3 | GO:1901724 | miRNA catabolic process(GO:0010587) positive regulation of cell proliferation involved in kidney development(GO:1901724) |
| 1.1 | 83.2 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 1.1 | 3.2 | GO:0060490 | serotonergic neuron axon guidance(GO:0036515) dichotomous subdivision of terminal units involved in lung branching(GO:0060448) orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 1.0 | 9.4 | GO:0085020 | protein K6-linked ubiquitination(GO:0085020) |
| 1.0 | 11.5 | GO:1902255 | positive regulation of intrinsic apoptotic signaling pathway by p53 class mediator(GO:1902255) |
| 1.0 | 3.1 | GO:0060295 | regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) positive regulation of feeding behavior(GO:2000253) |
| 1.0 | 6.1 | GO:0015677 | copper ion import(GO:0015677) |
| 1.0 | 5.0 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 1.0 | 7.0 | GO:0051451 | myoblast migration(GO:0051451) |
| 1.0 | 15.9 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 1.0 | 4.0 | GO:0009597 | detection of virus(GO:0009597) |
| 1.0 | 21.2 | GO:0048485 | sympathetic nervous system development(GO:0048485) |
| 0.9 | 2.8 | GO:0033505 | floor plate formation(GO:0021508) floor plate morphogenesis(GO:0033505) |
| 0.9 | 19.7 | GO:0010592 | positive regulation of lamellipodium assembly(GO:0010592) |
| 0.9 | 3.7 | GO:0002101 | tRNA wobble cytosine modification(GO:0002101) |
| 0.9 | 2.7 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 0.9 | 8.1 | GO:0035814 | negative regulation of renal sodium excretion(GO:0035814) |
| 0.9 | 13.3 | GO:1902042 | negative regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902042) |
| 0.9 | 2.6 | GO:0016240 | autophagosome docking(GO:0016240) |
| 0.9 | 4.3 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.9 | 1.7 | GO:0030969 | mRNA splicing via endonucleolytic cleavage and ligation involved in unfolded protein response(GO:0030969) mRNA splicing, via endonucleolytic cleavage and ligation(GO:0070054) mRNA endonucleolytic cleavage involved in unfolded protein response(GO:0070055) |
| 0.9 | 24.9 | GO:0045576 | mast cell activation(GO:0045576) |
| 0.9 | 6.8 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.8 | 4.2 | GO:1900034 | regulation of cellular response to heat(GO:1900034) |
| 0.8 | 5.8 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.8 | 13.9 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.8 | 2.4 | GO:2000342 | negative regulation of chemokine (C-X-C motif) ligand 2 production(GO:2000342) |
| 0.8 | 17.4 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.8 | 34.7 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.8 | 12.6 | GO:0050901 | leukocyte tethering or rolling(GO:0050901) |
| 0.8 | 3.2 | GO:0090204 | protein localization to nuclear pore(GO:0090204) |
| 0.8 | 3.1 | GO:0070366 | regulation of hepatocyte differentiation(GO:0070366) |
| 0.8 | 27.1 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.8 | 10.8 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.8 | 6.2 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.8 | 6.1 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.8 | 4.6 | GO:1900115 | extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.7 | 8.2 | GO:0045779 | negative regulation of bone resorption(GO:0045779) |
| 0.7 | 12.7 | GO:0002716 | negative regulation of natural killer cell mediated immunity(GO:0002716) |
| 0.7 | 3.7 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.7 | 1.5 | GO:0097037 | heme export(GO:0097037) |
| 0.7 | 17.0 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.7 | 21.0 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.7 | 2.1 | GO:0021569 | rhombomere 3 development(GO:0021569) |
| 0.7 | 3.4 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.7 | 10.6 | GO:0042573 | retinoic acid metabolic process(GO:0042573) |
| 0.7 | 3.3 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.7 | 7.3 | GO:1901299 | negative regulation of hydrogen peroxide-mediated programmed cell death(GO:1901299) |
| 0.7 | 2.6 | GO:0032620 | interleukin-17 production(GO:0032620) |
| 0.7 | 9.8 | GO:0043129 | surfactant homeostasis(GO:0043129) |
| 0.6 | 5.8 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.6 | 8.9 | GO:0035066 | positive regulation of histone acetylation(GO:0035066) |
| 0.6 | 6.4 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.6 | 1.3 | GO:1900368 | regulation of RNA interference(GO:1900368) |
| 0.6 | 18.2 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.6 | 14.4 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.6 | 2.5 | GO:0035965 | cardiolipin acyl-chain remodeling(GO:0035965) |
| 0.6 | 12.1 | GO:0032611 | interleukin-1 beta production(GO:0032611) |
| 0.6 | 3.5 | GO:0090367 | negative regulation of mRNA modification(GO:0090367) |
| 0.6 | 10.8 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.6 | 2.8 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.6 | 5.5 | GO:0040018 | positive regulation of multicellular organism growth(GO:0040018) |
| 0.5 | 10.9 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.5 | 2.2 | GO:0036265 | RNA (guanine-N7)-methylation(GO:0036265) |
| 0.5 | 3.2 | GO:1990573 | membrane repolarization during atrial cardiac muscle cell action potential(GO:0098914) potassium ion import across plasma membrane(GO:1990573) |
| 0.5 | 4.2 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.5 | 2.1 | GO:2000348 | protein linear polyubiquitination(GO:0097039) regulation of CD40 signaling pathway(GO:2000348) |
| 0.5 | 2.6 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 0.5 | 1.5 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.5 | 10.7 | GO:0002548 | monocyte chemotaxis(GO:0002548) |
| 0.5 | 1.0 | GO:0048298 | immunoglobulin production in mucosal tissue(GO:0002426) positive regulation of isotype switching to IgA isotypes(GO:0048298) |
| 0.5 | 16.7 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.5 | 4.5 | GO:0046549 | photoreceptor cell outer segment organization(GO:0035845) retinal rod cell development(GO:0046548) retinal cone cell development(GO:0046549) |
| 0.5 | 2.5 | GO:0034627 | 'de novo' NAD biosynthetic process(GO:0034627) |
| 0.5 | 7.2 | GO:0032674 | regulation of interleukin-5 production(GO:0032674) |
| 0.5 | 11.9 | GO:0060512 | prostate gland morphogenesis(GO:0060512) |
| 0.5 | 4.7 | GO:0038180 | nerve growth factor signaling pathway(GO:0038180) |
| 0.5 | 3.6 | GO:0090179 | planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.5 | 7.7 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.4 | 11.6 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.4 | 29.3 | GO:0045766 | positive regulation of angiogenesis(GO:0045766) |
| 0.4 | 19.2 | GO:1901998 | toxin transport(GO:1901998) |
| 0.4 | 35.3 | GO:0019882 | antigen processing and presentation(GO:0019882) |
| 0.4 | 1.3 | GO:0097681 | base-excision repair, DNA ligation(GO:0006288) double-strand break repair via alternative nonhomologous end joining(GO:0097681) |
| 0.4 | 2.6 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.4 | 5.9 | GO:0042100 | B cell proliferation(GO:0042100) |
| 0.4 | 5.0 | GO:0035278 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.4 | 10.0 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.4 | 2.5 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.4 | 1.2 | GO:0060903 | positive regulation of meiosis I(GO:0060903) |
| 0.4 | 1.2 | GO:0045541 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.4 | 2.8 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.4 | 3.2 | GO:0010586 | miRNA metabolic process(GO:0010586) |
| 0.4 | 1.2 | GO:0097298 | regulation of nucleus size(GO:0097298) |
| 0.4 | 2.7 | GO:0002934 | desmosome organization(GO:0002934) |
| 0.4 | 3.1 | GO:1901550 | regulation of endothelial cell development(GO:1901550) regulation of establishment of endothelial barrier(GO:1903140) |
| 0.4 | 28.1 | GO:0051092 | positive regulation of NF-kappaB transcription factor activity(GO:0051092) |
| 0.4 | 4.5 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.4 | 2.6 | GO:0000972 | transcription-dependent tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000972) |
| 0.4 | 1.1 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.4 | 45.6 | GO:0090501 | RNA phosphodiester bond hydrolysis(GO:0090501) |
| 0.4 | 3.6 | GO:2000010 | positive regulation of protein localization to cell surface(GO:2000010) |
| 0.4 | 13.3 | GO:0002250 | adaptive immune response(GO:0002250) |
| 0.4 | 5.4 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.4 | 12.3 | GO:0035904 | aorta development(GO:0035904) |
| 0.3 | 1.7 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.3 | 2.4 | GO:0060837 | blood vessel endothelial cell differentiation(GO:0060837) |
| 0.3 | 3.8 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.3 | 9.3 | GO:0070534 | protein K63-linked ubiquitination(GO:0070534) |
| 0.3 | 3.1 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.3 | 1.6 | GO:0007207 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.3 | 1.0 | GO:0002023 | reduction of food intake in response to dietary excess(GO:0002023) |
| 0.3 | 5.7 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.3 | 1.3 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.3 | 1.9 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.3 | 6.1 | GO:0046037 | GMP metabolic process(GO:0046037) |
| 0.3 | 1.2 | GO:0071376 | response to corticotropin-releasing hormone(GO:0043435) general adaptation syndrome(GO:0051866) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.3 | 0.9 | GO:0035522 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.3 | 1.8 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.3 | 4.7 | GO:0048873 | homeostasis of number of cells within a tissue(GO:0048873) |
| 0.3 | 1.7 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.3 | 1.4 | GO:0046628 | positive regulation of insulin receptor signaling pathway(GO:0046628) |
| 0.3 | 23.6 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.3 | 9.0 | GO:2000036 | regulation of stem cell population maintenance(GO:2000036) |
| 0.3 | 4.4 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.3 | 4.4 | GO:0060712 | spongiotrophoblast layer development(GO:0060712) |
| 0.3 | 2.4 | GO:0045616 | regulation of keratinocyte differentiation(GO:0045616) |
| 0.3 | 1.3 | GO:0042976 | activation of Janus kinase activity(GO:0042976) |
| 0.3 | 2.0 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.2 | 5.4 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.2 | 9.0 | GO:0006879 | cellular iron ion homeostasis(GO:0006879) |
| 0.2 | 2.1 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
| 0.2 | 0.7 | GO:1902416 | positive regulation of mRNA binding(GO:1902416) positive regulation of RNA binding(GO:1905216) |
| 0.2 | 1.6 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.2 | 3.0 | GO:0009396 | folic acid-containing compound biosynthetic process(GO:0009396) |
| 0.2 | 1.4 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.2 | 14.2 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.2 | 1.1 | GO:0009732 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.2 | 4.5 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.2 | 11.7 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.2 | 7.7 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.2 | 2.4 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.2 | 1.1 | GO:1903392 | negative regulation of focal adhesion assembly(GO:0051895) negative regulation of adherens junction organization(GO:1903392) |
| 0.2 | 7.5 | GO:0050715 | positive regulation of cytokine secretion(GO:0050715) |
| 0.2 | 12.2 | GO:0000045 | autophagosome assembly(GO:0000045) |
| 0.2 | 10.7 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.2 | 8.8 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.2 | 3.6 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.2 | 38.3 | GO:0006898 | receptor-mediated endocytosis(GO:0006898) |
| 0.2 | 7.0 | GO:0097421 | liver regeneration(GO:0097421) |
| 0.2 | 7.5 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.2 | 39.0 | GO:0042110 | T cell activation(GO:0042110) T cell aggregation(GO:0070489) |
| 0.2 | 11.2 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) |
| 0.2 | 0.6 | GO:0060965 | negative regulation of posttranscriptional gene silencing(GO:0060149) negative regulation of gene silencing by miRNA(GO:0060965) negative regulation of gene silencing by RNA(GO:0060967) negative regulation of gene silencing(GO:0060969) |
| 0.2 | 16.8 | GO:0009615 | response to virus(GO:0009615) |
| 0.2 | 5.8 | GO:0048146 | positive regulation of fibroblast proliferation(GO:0048146) |
| 0.2 | 1.5 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) |
| 0.2 | 13.6 | GO:0008203 | cholesterol metabolic process(GO:0008203) |
| 0.2 | 1.4 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.2 | 6.9 | GO:0030819 | positive regulation of cAMP biosynthetic process(GO:0030819) |
| 0.2 | 6.5 | GO:0033138 | positive regulation of peptidyl-serine phosphorylation(GO:0033138) |
| 0.2 | 0.9 | GO:0007198 | adenylate cyclase-inhibiting serotonin receptor signaling pathway(GO:0007198) |
| 0.2 | 1.1 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.2 | 3.5 | GO:0046825 | regulation of protein export from nucleus(GO:0046825) |
| 0.2 | 48.9 | GO:0007169 | transmembrane receptor protein tyrosine kinase signaling pathway(GO:0007169) |
| 0.2 | 2.1 | GO:0007625 | grooming behavior(GO:0007625) |
| 0.2 | 0.6 | GO:0043101 | purine-containing compound salvage(GO:0043101) |
| 0.2 | 3.5 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.1 | 7.5 | GO:0006414 | translational elongation(GO:0006414) |
| 0.1 | 1.8 | GO:0019835 | cytolysis(GO:0019835) |
| 0.1 | 1.0 | GO:0046579 | positive regulation of Ras protein signal transduction(GO:0046579) |
| 0.1 | 3.1 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.1 | 0.2 | GO:0019747 | regulation of isoprenoid metabolic process(GO:0019747) |
| 0.1 | 6.6 | GO:0060349 | bone morphogenesis(GO:0060349) |
| 0.1 | 4.7 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.1 | 1.5 | GO:0009086 | methionine biosynthetic process(GO:0009086) |
| 0.1 | 0.3 | GO:1904781 | positive regulation of protein localization to centrosome(GO:1904781) |
| 0.1 | 1.5 | GO:0021527 | spinal cord association neuron differentiation(GO:0021527) |
| 0.1 | 5.7 | GO:0001895 | retina homeostasis(GO:0001895) |
| 0.1 | 1.6 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.1 | 0.4 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.1 | 0.5 | GO:0061526 | acetylcholine secretion(GO:0061526) |
| 0.1 | 0.3 | GO:0090241 | negative regulation of histone H4 acetylation(GO:0090241) |
| 0.1 | 2.5 | GO:0043507 | positive regulation of JUN kinase activity(GO:0043507) |
| 0.1 | 2.6 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.1 | 0.7 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.1 | 7.3 | GO:0007059 | chromosome segregation(GO:0007059) |
| 0.1 | 0.4 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.1 | 3.3 | GO:0051703 | social behavior(GO:0035176) intraspecies interaction between organisms(GO:0051703) |
| 0.1 | 1.3 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.1 | 2.3 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.1 | 2.5 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.1 | 3.5 | GO:0008277 | regulation of G-protein coupled receptor protein signaling pathway(GO:0008277) |
| 0.1 | 1.6 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.1 | 1.1 | GO:0000187 | activation of MAPK activity(GO:0000187) |
| 0.1 | 2.5 | GO:0051298 | centrosome duplication(GO:0051298) |
| 0.1 | 0.2 | GO:0090435 | protein localization to nuclear envelope(GO:0090435) |
| 0.1 | 0.3 | GO:0046532 | regulation of photoreceptor cell differentiation(GO:0046532) |
| 0.1 | 0.4 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.1 | 1.5 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) DNA-templated transcriptional preinitiation complex assembly(GO:0070897) |
| 0.1 | 2.8 | GO:0006400 | tRNA modification(GO:0006400) |
| 0.1 | 1.0 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.1 | 3.4 | GO:0007088 | regulation of mitotic nuclear division(GO:0007088) |
| 0.0 | 0.2 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.0 | 0.5 | GO:0045672 | positive regulation of osteoclast differentiation(GO:0045672) |
| 0.0 | 0.3 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.0 | 2.7 | GO:0000724 | double-strand break repair via homologous recombination(GO:0000724) |
| 0.0 | 2.1 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 0.3 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.0 | 0.3 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.0 | 0.6 | GO:0061053 | somite development(GO:0061053) |
| 0.0 | 0.7 | GO:0090305 | nucleic acid phosphodiester bond hydrolysis(GO:0090305) |
| 0.0 | 0.1 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.0 | 0.1 | GO:0001573 | ganglioside metabolic process(GO:0001573) |
| 0.0 | 0.9 | GO:0007127 | meiosis I(GO:0007127) |
| 0.0 | 0.3 | GO:0046676 | negative regulation of insulin secretion(GO:0046676) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 22.0 | 65.9 | GO:0089717 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 19.7 | 78.8 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 14.5 | 43.6 | GO:0097233 | lamellar body membrane(GO:0097232) alveolar lamellar body membrane(GO:0097233) |
| 13.5 | 107.7 | GO:0044194 | cytolytic granule(GO:0044194) |
| 11.4 | 34.2 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 9.4 | 103.4 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 7.2 | 21.5 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 7.1 | 71.5 | GO:0032009 | early phagosome(GO:0032009) |
| 7.0 | 21.0 | GO:1990423 | RZZ complex(GO:1990423) |
| 6.5 | 77.6 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 5.7 | 28.7 | GO:0072557 | IPAF inflammasome complex(GO:0072557) NLRP1 inflammasome complex(GO:0072558) AIM2 inflammasome complex(GO:0097169) |
| 5.3 | 32.0 | GO:0071547 | piP-body(GO:0071547) |
| 5.3 | 26.6 | GO:0034455 | t-UTP complex(GO:0034455) |
| 5.2 | 20.7 | GO:0009330 | DNA topoisomerase complex (ATP-hydrolyzing)(GO:0009330) |
| 5.1 | 66.2 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 5.0 | 15.0 | GO:0070821 | tertiary granule membrane(GO:0070821) |
| 4.7 | 14.0 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 4.6 | 18.5 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 4.5 | 27.1 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 4.5 | 179.6 | GO:0098636 | protein complex involved in cell adhesion(GO:0098636) |
| 4.4 | 17.6 | GO:0031436 | BRCA1-BARD1 complex(GO:0031436) |
| 4.0 | 40.2 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 3.7 | 14.8 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 3.5 | 10.5 | GO:0055087 | Ski complex(GO:0055087) |
| 2.9 | 35.0 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 2.7 | 13.5 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 2.6 | 10.5 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 2.6 | 20.9 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 2.5 | 7.5 | GO:0032798 | Swi5-Sfr1 complex(GO:0032798) |
| 2.3 | 9.4 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 2.3 | 11.5 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 2.2 | 6.6 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 2.2 | 39.9 | GO:0030056 | hemidesmosome(GO:0030056) |
| 2.2 | 15.4 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 2.0 | 6.0 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 2.0 | 11.8 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 1.9 | 7.8 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 1.9 | 17.1 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 1.8 | 542.9 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 1.7 | 5.0 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 1.7 | 13.3 | GO:0000796 | condensin complex(GO:0000796) |
| 1.6 | 9.9 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 1.6 | 46.6 | GO:0042588 | zymogen granule(GO:0042588) |
| 1.6 | 338.1 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
| 1.5 | 23.0 | GO:0072687 | meiotic spindle(GO:0072687) |
| 1.5 | 20.4 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 1.3 | 5.4 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 1.3 | 9.0 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 1.2 | 25.7 | GO:0001891 | phagocytic cup(GO:0001891) |
| 1.2 | 23.6 | GO:0044217 | other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) |
| 1.2 | 10.6 | GO:0031209 | SCAR complex(GO:0031209) |
| 1.2 | 4.7 | GO:0000938 | GARP complex(GO:0000938) |
| 1.1 | 15.3 | GO:0005766 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 1.1 | 6.3 | GO:0033010 | paranodal junction(GO:0033010) |
| 1.1 | 13.7 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 1.1 | 3.2 | GO:0060187 | cell pole(GO:0060187) |
| 1.0 | 6.3 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 1.0 | 8.3 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 1.0 | 22.0 | GO:0001772 | immunological synapse(GO:0001772) |
| 1.0 | 5.9 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 1.0 | 2.9 | GO:0005745 | m-AAA complex(GO:0005745) |
| 1.0 | 6.7 | GO:0042382 | paraspeckles(GO:0042382) |
| 1.0 | 4.8 | GO:0031501 | mannosyltransferase complex(GO:0031501) |
| 0.9 | 24.9 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.9 | 12.4 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.9 | 3.5 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.9 | 96.1 | GO:0005884 | actin filament(GO:0005884) |
| 0.8 | 10.9 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.8 | 0.8 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.8 | 6.0 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.7 | 5.1 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.7 | 4.3 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.7 | 10.0 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.7 | 14.0 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.7 | 5.4 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.7 | 11.5 | GO:0001741 | XY body(GO:0001741) |
| 0.7 | 2.7 | GO:0098553 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.7 | 7.3 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.7 | 12.5 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.6 | 9.0 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.6 | 10.8 | GO:0032039 | integrator complex(GO:0032039) |
| 0.6 | 10.8 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.6 | 31.8 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.6 | 3.6 | GO:0000243 | commitment complex(GO:0000243) |
| 0.6 | 4.7 | GO:0045261 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.6 | 10.9 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.5 | 8.2 | GO:0030057 | desmosome(GO:0030057) |
| 0.5 | 2.1 | GO:0031417 | NatC complex(GO:0031417) |
| 0.5 | 2.1 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.5 | 2.6 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.5 | 3.1 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.5 | 3.5 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.5 | 25.5 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.5 | 33.6 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.5 | 3.4 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.5 | 46.6 | GO:0005604 | basement membrane(GO:0005604) |
| 0.4 | 9.9 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.4 | 37.5 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.4 | 1.3 | GO:0070421 | DNA ligase III-XRCC1 complex(GO:0070421) |
| 0.4 | 1.3 | GO:0031074 | nucleocytoplasmic shuttling complex(GO:0031074) |
| 0.4 | 9.4 | GO:0002102 | podosome(GO:0002102) |
| 0.4 | 488.5 | GO:0005615 | extracellular space(GO:0005615) |
| 0.4 | 38.4 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.4 | 80.1 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.4 | 12.4 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.4 | 17.3 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.4 | 9.1 | GO:0019897 | extrinsic component of plasma membrane(GO:0019897) |
| 0.4 | 4.8 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.4 | 46.9 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.4 | 2.2 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.4 | 10.6 | GO:0005921 | gap junction(GO:0005921) |
| 0.4 | 16.3 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.4 | 7.5 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.4 | 2.5 | GO:0031415 | NatA complex(GO:0031415) |
| 0.3 | 46.3 | GO:0005938 | cell cortex(GO:0005938) |
| 0.3 | 5.9 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.3 | 20.0 | GO:0000786 | nucleosome(GO:0000786) DNA packaging complex(GO:0044815) |
| 0.3 | 5.2 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.3 | 3.2 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.3 | 22.2 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.3 | 15.7 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.3 | 26.8 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.3 | 3.1 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.3 | 13.8 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.3 | 8.1 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.3 | 6.3 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.3 | 91.1 | GO:0009986 | cell surface(GO:0009986) |
| 0.3 | 63.8 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.3 | 5.4 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.3 | 16.1 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.3 | 3.3 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.2 | 1.2 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.2 | 0.7 | GO:0044609 | DBIRD complex(GO:0044609) |
| 0.2 | 1.0 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.2 | 3.1 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.2 | 15.3 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.2 | 49.9 | GO:0000323 | lytic vacuole(GO:0000323) lysosome(GO:0005764) |
| 0.2 | 2.1 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.2 | 3.0 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.2 | 3.3 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.2 | 7.2 | GO:0015030 | Cajal body(GO:0015030) |
| 0.2 | 5.2 | GO:0005901 | caveola(GO:0005901) |
| 0.1 | 5.3 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.1 | 0.9 | GO:0016591 | DNA-directed RNA polymerase II, holoenzyme(GO:0016591) |
| 0.1 | 7.1 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.1 | 4.1 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.1 | 2.6 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.1 | 0.5 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.1 | 3.2 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.1 | 0.7 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.1 | 0.9 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.1 | 9.5 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.1 | 0.8 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.1 | 1.6 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.1 | 0.5 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.1 | 1.1 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.1 | 0.7 | GO:0043189 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.1 | 0.7 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.1 | 1.0 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 1.1 | GO:0046930 | pore complex(GO:0046930) |
| 0.0 | 1.9 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.5 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 1.6 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.0 | 0.3 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.4 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.0 | 0.1 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.0 | 0.5 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 35.8 | 107.3 | GO:0004698 | calcium-dependent protein kinase C activity(GO:0004698) |
| 15.9 | 47.8 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 13.5 | 94.5 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 13.2 | 65.9 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 10.9 | 32.8 | GO:0002114 | interleukin-33 receptor activity(GO:0002114) |
| 10.7 | 85.8 | GO:0019958 | C-X-C chemokine binding(GO:0019958) |
| 8.4 | 33.6 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 8.0 | 32.0 | GO:0034584 | piRNA binding(GO:0034584) |
| 7.8 | 194.4 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 7.4 | 37.2 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 6.8 | 20.4 | GO:0071791 | chemokine (C-C motif) ligand 5 binding(GO:0071791) |
| 6.3 | 82.3 | GO:0035325 | Toll-like receptor binding(GO:0035325) |
| 6.3 | 31.4 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 6.2 | 18.6 | GO:0008384 | NF-kappaB-inducing kinase activity(GO:0004704) IkappaB kinase activity(GO:0008384) |
| 6.2 | 18.5 | GO:0031721 | haptoglobin binding(GO:0031720) hemoglobin alpha binding(GO:0031721) |
| 6.1 | 36.8 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 5.8 | 215.4 | GO:0008009 | chemokine activity(GO:0008009) |
| 5.7 | 51.3 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 5.3 | 26.6 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 5.2 | 15.6 | GO:0044715 | 8-oxo-dGDP phosphatase activity(GO:0044715) |
| 5.2 | 20.7 | GO:0003918 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 4.9 | 68.0 | GO:0033691 | sialic acid binding(GO:0033691) |
| 4.9 | 14.6 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 4.7 | 18.9 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 4.6 | 36.6 | GO:0032554 | purine deoxyribonucleotide binding(GO:0032554) |
| 4.3 | 47.8 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 4.3 | 30.1 | GO:0031433 | telethonin binding(GO:0031433) |
| 4.2 | 37.9 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 4.0 | 11.9 | GO:0008142 | oxysterol binding(GO:0008142) |
| 3.7 | 833.5 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 3.6 | 18.2 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 3.5 | 10.6 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 3.5 | 14.0 | GO:0047276 | N-acetyllactosaminide 3-alpha-galactosyltransferase activity(GO:0047276) |
| 3.4 | 30.7 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 3.4 | 23.6 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 3.4 | 10.1 | GO:0008413 | 8-oxo-7,8-dihydroguanosine triphosphate pyrophosphatase activity(GO:0008413) 8-oxo-7,8-dihydrodeoxyguanosine triphosphate pyrophosphatase activity(GO:0035539) |
| 3.3 | 183.1 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 3.2 | 37.9 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 3.1 | 65.6 | GO:0043274 | phospholipase binding(GO:0043274) |
| 3.1 | 9.2 | GO:0004066 | asparagine synthase (glutamine-hydrolyzing) activity(GO:0004066) |
| 3.0 | 39.1 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 3.0 | 6.0 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 3.0 | 20.9 | GO:0003896 | DNA primase activity(GO:0003896) |
| 3.0 | 11.8 | GO:0008309 | double-stranded DNA exodeoxyribonuclease activity(GO:0008309) |
| 2.9 | 8.7 | GO:0001847 | opsonin receptor activity(GO:0001847) |
| 2.8 | 14.0 | GO:0052813 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 2.8 | 8.4 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 2.8 | 16.6 | GO:0004340 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 2.8 | 11.0 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) serine-type exopeptidase activity(GO:0070008) |
| 2.7 | 93.1 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 2.7 | 29.4 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 2.6 | 10.5 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 2.6 | 7.8 | GO:0070976 | TIR domain binding(GO:0070976) |
| 2.4 | 56.0 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 2.4 | 11.9 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 2.4 | 59.2 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 2.4 | 9.4 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 2.3 | 23.4 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 2.3 | 7.0 | GO:0030977 | taurine binding(GO:0030977) |
| 2.2 | 20.2 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 2.2 | 11.0 | GO:0070052 | collagen V binding(GO:0070052) |
| 2.2 | 17.4 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 2.2 | 15.2 | GO:0016803 | ether hydrolase activity(GO:0016803) |
| 2.1 | 21.5 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 2.1 | 8.4 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 2.1 | 6.3 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) bradykinin receptor binding(GO:0031711) |
| 2.1 | 39.3 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 2.1 | 12.4 | GO:0000150 | recombinase activity(GO:0000150) |
| 2.0 | 153.9 | GO:1990782 | protein tyrosine kinase binding(GO:1990782) |
| 2.0 | 9.9 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 2.0 | 3.9 | GO:0035403 | histone kinase activity (H3-T6 specific)(GO:0035403) |
| 1.9 | 7.8 | GO:0030172 | troponin C binding(GO:0030172) |
| 1.9 | 15.2 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 1.9 | 41.3 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 1.8 | 24.7 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 1.8 | 22.9 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 1.8 | 5.3 | GO:0004500 | dopamine beta-monooxygenase activity(GO:0004500) |
| 1.7 | 7.0 | GO:0034618 | arginine binding(GO:0034618) |
| 1.7 | 31.3 | GO:0031996 | thioesterase binding(GO:0031996) |
| 1.7 | 5.2 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 1.7 | 11.9 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 1.7 | 5.0 | GO:1990763 | arrestin family protein binding(GO:1990763) |
| 1.6 | 11.5 | GO:0008296 | 3'-5'-exodeoxyribonuclease activity(GO:0008296) |
| 1.6 | 28.8 | GO:0005522 | profilin binding(GO:0005522) |
| 1.6 | 12.6 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 1.6 | 31.4 | GO:0019865 | immunoglobulin binding(GO:0019865) |
| 1.6 | 12.4 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 1.6 | 6.2 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 1.5 | 5.9 | GO:0005534 | galactose binding(GO:0005534) |
| 1.4 | 10.1 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 1.3 | 18.3 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 1.3 | 1.3 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 1.3 | 7.6 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 1.3 | 3.8 | GO:0050656 | 3'-phosphoadenosine 5'-phosphosulfate binding(GO:0050656) |
| 1.3 | 151.5 | GO:0004519 | endonuclease activity(GO:0004519) |
| 1.2 | 7.5 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 1.2 | 7.4 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 1.2 | 6.1 | GO:0052851 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 1.2 | 6.1 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 1.2 | 4.8 | GO:0047619 | acylcarnitine hydrolase activity(GO:0047619) |
| 1.2 | 5.9 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 1.1 | 25.0 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 1.1 | 13.6 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 1.1 | 23.4 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 1.1 | 37.6 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 1.1 | 5.5 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 1.1 | 20.6 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 1.1 | 9.5 | GO:0055104 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 1.1 | 24.3 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 1.0 | 3.0 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 1.0 | 14.0 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 1.0 | 21.9 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 1.0 | 9.9 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 1.0 | 8.9 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 1.0 | 6.8 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) |
| 1.0 | 5.8 | GO:0035197 | siRNA binding(GO:0035197) |
| 1.0 | 4.8 | GO:0051033 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 1.0 | 10.5 | GO:0043995 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.9 | 256.2 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.9 | 4.7 | GO:0004031 | aldehyde oxidase activity(GO:0004031) oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.9 | 5.3 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.9 | 2.6 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.9 | 9.5 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.8 | 5.0 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.8 | 4.2 | GO:0048019 | receptor antagonist activity(GO:0048019) |
| 0.8 | 6.7 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.8 | 3.3 | GO:0070568 | guanylyltransferase activity(GO:0070568) |
| 0.8 | 7.2 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.8 | 3.2 | GO:0086089 | voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization(GO:0086089) |
| 0.8 | 19.6 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.8 | 26.6 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.8 | 3.8 | GO:0004920 | interleukin-10 receptor activity(GO:0004920) |
| 0.8 | 6.0 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.7 | 4.5 | GO:0004471 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.7 | 12.4 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.7 | 54.7 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.7 | 2.9 | GO:0016428 | tRNA (cytosine-5-)-methyltransferase activity(GO:0016428) |
| 0.7 | 3.6 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.7 | 9.4 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.7 | 4.2 | GO:0050786 | N-formyl peptide receptor activity(GO:0004982) RAGE receptor binding(GO:0050786) |
| 0.7 | 6.3 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) |
| 0.7 | 10.6 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.7 | 45.8 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.6 | 2.6 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.6 | 20.0 | GO:0016860 | intramolecular oxidoreductase activity(GO:0016860) |
| 0.6 | 5.1 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.6 | 19.6 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.6 | 35.3 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.6 | 6.2 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.6 | 1.8 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.6 | 13.5 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.6 | 4.1 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.6 | 4.1 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.5 | 14.2 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.5 | 4.7 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.5 | 9.3 | GO:0031005 | filamin binding(GO:0031005) |
| 0.5 | 2.6 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.5 | 1.5 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.5 | 59.1 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.5 | 32.5 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.5 | 4.3 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.5 | 17.1 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.5 | 4.7 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.5 | 3.3 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.5 | 2.8 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 0.5 | 2.3 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.4 | 3.1 | GO:0019104 | DNA N-glycosylase activity(GO:0019104) |
| 0.4 | 3.1 | GO:0048185 | activin binding(GO:0048185) |
| 0.4 | 1.3 | GO:0004956 | prostaglandin D receptor activity(GO:0004956) |
| 0.4 | 13.2 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) |
| 0.4 | 1.6 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.4 | 11.0 | GO:0008237 | metallopeptidase activity(GO:0008237) |
| 0.4 | 13.4 | GO:0022829 | wide pore channel activity(GO:0022829) |
| 0.4 | 13.7 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.4 | 13.6 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.4 | 3.4 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.4 | 5.5 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.4 | 1.5 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 0.4 | 33.8 | GO:0019887 | protein kinase regulator activity(GO:0019887) |
| 0.4 | 8.0 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.4 | 2.1 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.3 | 1.7 | GO:0004586 | ornithine decarboxylase activity(GO:0004586) |
| 0.3 | 8.1 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.3 | 4.6 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.3 | 1.0 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.3 | 1.9 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.3 | 1.2 | GO:0004963 | follicle-stimulating hormone receptor activity(GO:0004963) |
| 0.3 | 7.9 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.3 | 6.0 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.3 | 2.7 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.3 | 10.6 | GO:0061733 | histone acetyltransferase activity(GO:0004402) peptide-lysine-N-acetyltransferase activity(GO:0061733) |
| 0.3 | 1.2 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.3 | 26.8 | GO:0035326 | enhancer binding(GO:0035326) |
| 0.3 | 58.4 | GO:0004866 | endopeptidase inhibitor activity(GO:0004866) |
| 0.3 | 5.5 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.3 | 0.9 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.3 | 1.4 | GO:0071253 | connexin binding(GO:0071253) |
| 0.3 | 8.8 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.3 | 2.5 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.3 | 2.2 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.3 | 67.2 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.3 | 6.8 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.2 | 1.5 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) chondroitin sulfotransferase activity(GO:0034481) |
| 0.2 | 5.1 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.2 | 5.8 | GO:0042923 | neuropeptide binding(GO:0042923) |
| 0.2 | 26.1 | GO:0008201 | heparin binding(GO:0008201) |
| 0.2 | 3.5 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.2 | 1.8 | GO:0001848 | complement binding(GO:0001848) |
| 0.2 | 1.1 | GO:0001032 | RNA polymerase III type 3 promoter DNA binding(GO:0001032) |
| 0.2 | 5.6 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.2 | 11.4 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.2 | 48.8 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.2 | 0.6 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.2 | 5.9 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.2 | 5.0 | GO:0019212 | phosphatase inhibitor activity(GO:0019212) |
| 0.2 | 10.8 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.2 | 8.5 | GO:0005518 | collagen binding(GO:0005518) |
| 0.2 | 6.7 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.2 | 3.0 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.2 | 1.5 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.2 | 3.0 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.1 | 3.5 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 6.3 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.1 | 0.6 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.1 | 11.5 | GO:0001047 | core promoter binding(GO:0001047) |
| 0.1 | 3.1 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.1 | 3.7 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.1 | 5.8 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 2.5 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.1 | 2.5 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.1 | 0.7 | GO:0008430 | selenium binding(GO:0008430) |
| 0.1 | 4.9 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.1 | 0.8 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.1 | 3.1 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.1 | 1.7 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.1 | 9.3 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.1 | 2.5 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.1 | 0.3 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.1 | 2.4 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.1 | 1.0 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 30.5 | GO:0008047 | enzyme activator activity(GO:0008047) |
| 0.1 | 6.5 | GO:0070851 | growth factor receptor binding(GO:0070851) |
| 0.1 | 2.7 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.1 | 0.5 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.1 | 1.7 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.1 | 0.7 | GO:0019841 | retinol binding(GO:0019841) |
| 0.1 | 1.5 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.1 | 2.4 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.1 | 4.3 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.1 | 1.8 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.1 | 0.7 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.1 | 1.0 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 3.5 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 1.0 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.1 | GO:0052795 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.0 | 0.4 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 1.6 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.3 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.6 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.6 | 94.2 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 5.0 | 114.8 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 3.8 | 30.8 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 3.7 | 29.3 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 3.5 | 52.2 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 3.4 | 256.5 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 3.4 | 124.1 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 3.1 | 137.6 | PID BCR 5PATHWAY | BCR signaling pathway |
| 3.0 | 87.7 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 2.8 | 123.0 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 2.7 | 72.1 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 2.6 | 41.1 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 2.1 | 97.9 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 1.9 | 90.6 | NABA COLLAGENS | Genes encoding collagen proteins |
| 1.8 | 7.2 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 1.7 | 19.2 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 1.7 | 16.9 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 1.6 | 26.2 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 1.6 | 19.3 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 1.6 | 6.3 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 1.6 | 31.1 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 1.5 | 138.9 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 1.5 | 35.2 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 1.4 | 44.3 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 1.3 | 45.6 | PID ARF6 PATHWAY | Arf6 signaling events |
| 1.3 | 23.8 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 1.2 | 49.8 | PID AURORA B PATHWAY | Aurora B signaling |
| 1.2 | 42.6 | PID BARD1 PATHWAY | BARD1 signaling events |
| 1.1 | 49.5 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 1.0 | 12.5 | PID IFNG PATHWAY | IFN-gamma pathway |
| 1.0 | 9.8 | PID GMCSF PATHWAY | GMCSF-mediated signaling events |
| 0.9 | 19.4 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.9 | 12.3 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.8 | 9.7 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.8 | 19.5 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.7 | 10.5 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.6 | 14.6 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.6 | 181.3 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.6 | 28.2 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.6 | 6.1 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.6 | 10.0 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.6 | 11.0 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.5 | 11.1 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.5 | 20.8 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.5 | 5.3 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.5 | 7.2 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.5 | 5.1 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.5 | 5.5 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.4 | 4.8 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.4 | 8.7 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.4 | 4.1 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.4 | 4.1 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.4 | 12.2 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.4 | 1.4 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.3 | 2.4 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.3 | 14.0 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.3 | 2.4 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.3 | 5.1 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.3 | 8.2 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.3 | 6.4 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.3 | 12.8 | PID E2F PATHWAY | E2F transcription factor network |
| 0.3 | 2.7 | ST GA13 PATHWAY | G alpha 13 Pathway |
| 0.3 | 1.2 | PID ERBB1 RECEPTOR PROXIMAL PATHWAY | EGF receptor (ErbB1) signaling pathway |
| 0.3 | 7.9 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.2 | 8.7 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.2 | 4.7 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.2 | 3.4 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.2 | 2.8 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.1 | 1.8 | PID MYC PATHWAY | C-MYC pathway |
| 0.1 | 4.0 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.1 | 6.1 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.1 | 5.4 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 4.4 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.1 | 3.6 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.1 | 2.6 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 1.3 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 0.4 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.0 | 1.2 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.8 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.3 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 9.0 | 450.9 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 8.9 | 240.0 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 8.0 | 159.4 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 6.2 | 80.3 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 5.2 | 78.7 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 3.6 | 43.6 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 3.5 | 51.9 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 3.4 | 51.7 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 3.2 | 126.2 | REACTOME COSTIMULATION BY THE CD28 FAMILY | Genes involved in Costimulation by the CD28 family |
| 3.0 | 75.6 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 3.0 | 83.6 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 2.9 | 41.2 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 2.9 | 5.8 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 2.9 | 20.3 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 2.6 | 37.1 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 2.6 | 94.6 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 2.5 | 34.7 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 2.5 | 27.1 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 2.5 | 31.9 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 2.5 | 46.6 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 2.4 | 68.4 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 2.4 | 100.1 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 2.4 | 28.7 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 2.3 | 4.7 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 2.2 | 80.7 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 2.2 | 107.2 | REACTOME LIPID DIGESTION MOBILIZATION AND TRANSPORT | Genes involved in Lipid digestion, mobilization, and transport |
| 2.1 | 22.8 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 1.8 | 106.7 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 1.3 | 43.0 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 1.3 | 5.3 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 1.3 | 16.5 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 1.3 | 13.8 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 1.2 | 39.4 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 1.2 | 22.5 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 1.1 | 19.3 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 1.1 | 16.5 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 1.1 | 18.3 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 1.0 | 11.5 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 0.9 | 20.5 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.9 | 8.7 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.9 | 11.2 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.8 | 30.7 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.8 | 46.1 | REACTOME EXTRACELLULAR MATRIX ORGANIZATION | Genes involved in Extracellular matrix organization |
| 0.8 | 2.4 | REACTOME AUTODEGRADATION OF THE E3 UBIQUITIN LIGASE COP1 | Genes involved in Autodegradation of the E3 ubiquitin ligase COP1 |
| 0.8 | 15.6 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.7 | 12.2 | REACTOME NUCLEAR EVENTS KINASE AND TRANSCRIPTION FACTOR ACTIVATION | Genes involved in Nuclear Events (kinase and transcription factor activation) |
| 0.7 | 65.9 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.6 | 14.6 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.6 | 5.5 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.6 | 16.3 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.6 | 18.5 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.6 | 50.7 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.6 | 45.7 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.6 | 10.1 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.6 | 8.9 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.6 | 10.5 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.5 | 4.6 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.4 | 5.4 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.4 | 16.2 | REACTOME RIG I MDA5 MEDIATED INDUCTION OF IFN ALPHA BETA PATHWAYS | Genes involved in RIG-I/MDA5 mediated induction of IFN-alpha/beta pathways |
| 0.4 | 18.1 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.4 | 7.3 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.4 | 6.9 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.4 | 7.2 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.4 | 16.5 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.4 | 5.7 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.4 | 7.7 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.4 | 7.7 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.3 | 5.0 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.3 | 42.3 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.3 | 3.1 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.3 | 10.9 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.3 | 5.2 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.3 | 4.4 | REACTOME BASE EXCISION REPAIR | Genes involved in Base Excision Repair |
| 0.2 | 6.3 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.2 | 1.3 | REACTOME REGULATORY RNA PATHWAYS | Genes involved in Regulatory RNA pathways |
| 0.2 | 3.6 | REACTOME PROCESSING OF CAPPED INTRONLESS PRE MRNA | Genes involved in Processing of Capped Intronless Pre-mRNA |
| 0.2 | 44.5 | REACTOME CLASS A1 RHODOPSIN LIKE RECEPTORS | Genes involved in Class A/1 (Rhodopsin-like receptors) |
| 0.2 | 2.3 | REACTOME SIGNALING BY FGFR1 MUTANTS | Genes involved in Signaling by FGFR1 mutants |
| 0.2 | 17.9 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.2 | 0.9 | REACTOME FORMATION OF THE HIV1 EARLY ELONGATION COMPLEX | Genes involved in Formation of the HIV-1 Early Elongation Complex |
| 0.2 | 13.9 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.2 | 2.5 | REACTOME GPCR LIGAND BINDING | Genes involved in GPCR ligand binding |
| 0.1 | 27.2 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.1 | 1.5 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.1 | 2.4 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.1 | 7.9 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.1 | 3.8 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.1 | 1.5 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.1 | 0.7 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 1.5 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.7 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.6 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.3 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 1.0 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.0 | 1.0 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.0 | 1.3 | REACTOME UNFOLDED PROTEIN RESPONSE | Genes involved in Unfolded Protein Response |
| 0.0 | 1.5 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.3 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.3 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.4 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.5 | REACTOME SPHINGOLIPID METABOLISM | Genes involved in Sphingolipid metabolism |