Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
Results for Rorc_Nr1d1
Z-value: 0.98
Transcription factors associated with Rorc_Nr1d1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
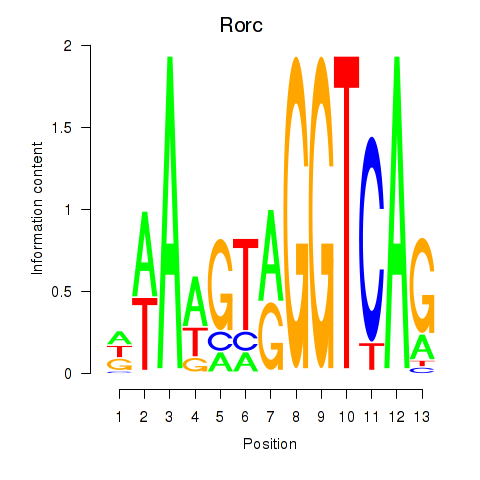
Rorc
|
ENSRNOG00000020836 | RAR-related orphan receptor C |
|
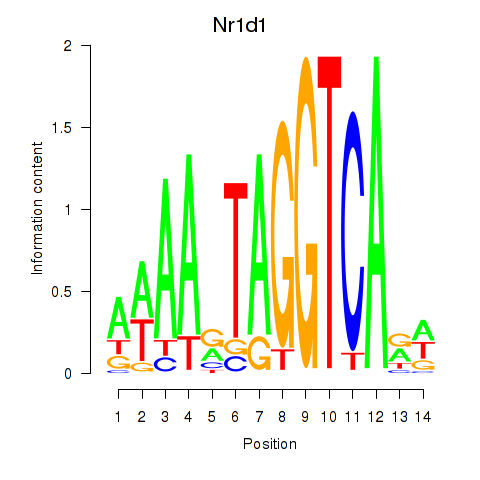
Nr1d1
|
ENSRNOG00000009329 | nuclear receptor subfamily 1, group D, member 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nr1d1 | rn6_v1_chr10_-_86688730_86688730 | 0.54 | 5.8e-26 | Click! |
| Rorc | rn6_v1_chr2_+_195617021_195617044 | -0.48 | 3.7e-20 | Click! |
Activity profile of Rorc_Nr1d1 motif
Sorted Z-values of Rorc_Nr1d1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_79713567 | 107.57 |
ENSRNOT00000012110
|
C1qtnf4
|
C1q and tumor necrosis factor related protein 4 |
| chr10_+_86303727 | 74.74 |
ENSRNOT00000037752
|
Ppp1r1b
|
protein phosphatase 1, regulatory (inhibitor) subunit 1B |
| chr3_+_98297554 | 57.06 |
ENSRNOT00000006524
|
Kcna4
|
potassium voltage-gated channel subfamily A member 4 |
| chr11_+_66316606 | 49.38 |
ENSRNOT00000041715
|
Stxbp5l
|
syntaxin binding protein 5-like |
| chr11_+_87366621 | 42.71 |
ENSRNOT00000040978
|
Aifm3
|
apoptosis inducing factor, mitochondria associated 3 |
| chr2_-_189096785 | 41.60 |
ENSRNOT00000028200
|
Chrnb2
|
cholinergic receptor nicotinic beta 2 subunit |
| chr8_+_33239139 | 38.13 |
ENSRNOT00000011589
|
Arhgap32
|
Rho GTPase activating protein 32 |
| chr7_+_64672722 | 32.74 |
ENSRNOT00000064448
ENSRNOT00000005539 |
Grip1
|
glutamate receptor interacting protein 1 |
| chr8_-_82641536 | 32.15 |
ENSRNOT00000014491
|
Scg3
|
secretogranin III |
| chr7_-_136853154 | 29.13 |
ENSRNOT00000087376
|
Nell2
|
neural EGFL like 2 |
| chrX_+_6791136 | 27.78 |
ENSRNOT00000003984
|
LOC100909913
|
norrin-like |
| chr4_-_155275161 | 26.84 |
ENSRNOT00000032690
|
Rimklb
|
ribosomal modification protein rimK-like family member B |
| chr10_-_86688730 | 25.99 |
ENSRNOT00000055333
|
Nr1d1
|
nuclear receptor subfamily 1, group D, member 1 |
| chr4_+_136512201 | 23.62 |
ENSRNOT00000051645
|
Cntn6
|
contactin 6 |
| chrX_+_33895769 | 22.80 |
ENSRNOT00000085993
ENSRNOT00000079365 |
Reps2
|
RALBP1 associated Eps domain containing protein 2 |
| chr1_+_101687855 | 21.57 |
ENSRNOT00000028546
ENSRNOT00000091597 |
Dbp
|
D-box binding PAR bZIP transcription factor |
| chr3_+_101899068 | 20.79 |
ENSRNOT00000079600
ENSRNOT00000006256 |
Muc15
|
mucin 15, cell surface associated |
| chr18_+_30527705 | 19.70 |
ENSRNOT00000027168
|
Pcdhb14
|
protocadherin beta 14 |
| chr1_-_170404056 | 19.58 |
ENSRNOT00000024402
|
Apbb1
|
amyloid beta precursor protein binding family B member 1 |
| chr4_-_124338176 | 18.51 |
ENSRNOT00000016628
|
Prickle2
|
prickle planar cell polarity protein 2 |
| chr12_-_2174131 | 15.62 |
ENSRNOT00000001313
|
Pcp2
|
Purkinje cell protein 2 |
| chr15_-_8183682 | 15.26 |
ENSRNOT00000011438
|
Nkiras1
|
NFKB inhibitor interacting Ras-like 1 |
| chr9_-_40008680 | 14.39 |
ENSRNOT00000016578
|
Khdrbs2
|
KH RNA binding domain containing, signal transduction associated 2 |
| chr5_+_172825072 | 13.96 |
ENSRNOT00000068525
|
Cfap74
|
cilia and flagella associated protein 74 |
| chr5_+_43603043 | 13.64 |
ENSRNOT00000009899
|
Epha7
|
Eph receptor A7 |
| chr1_-_67065797 | 13.23 |
ENSRNOT00000048152
|
Vom1r46
|
vomeronasal 1 receptor 46 |
| chrX_+_14019961 | 12.45 |
ENSRNOT00000004785
|
Sytl5
|
synaptotagmin-like 5 |
| chr1_-_90520012 | 12.30 |
ENSRNOT00000028698
|
Kctd15
|
potassium channel tetramerization domain containing 15 |
| chr18_+_70548607 | 12.27 |
ENSRNOT00000085318
|
Myo5b
|
myosin Vb |
| chr1_+_16478127 | 12.08 |
ENSRNOT00000019076
|
Ahi1
|
Abelson helper integration site 1 |
| chr3_-_104502471 | 11.96 |
ENSRNOT00000040306
|
Ryr3
|
ryanodine receptor 3 |
| chr17_-_2278613 | 11.91 |
ENSRNOT00000046525
|
Cntnap3b
|
contactin associated protein-like 3B |
| chr19_-_25062934 | 11.86 |
ENSRNOT00000060117
|
Gm10644
|
predicted gene 10644 |
| chr4_+_199916 | 11.68 |
ENSRNOT00000009317
|
Htr5a
|
5-hydroxytryptamine receptor 5A |
| chr3_+_56056925 | 11.32 |
ENSRNOT00000088351
ENSRNOT00000010508 |
Klhl23
|
kelch-like family member 23 |
| chr1_-_88066101 | 11.21 |
ENSRNOT00000079473
ENSRNOT00000027893 |
Ryr1
|
ryanodine receptor 1 |
| chr14_+_100311173 | 10.75 |
ENSRNOT00000031275
|
Ppp3r1
|
protein phosphatase 3, regulatory subunit B, alpha |
| chr1_+_219199158 | 10.28 |
ENSRNOT00000024034
|
LOC688778
|
similar to fatty aldehyde dehydrogenase-like |
| chr1_-_214414763 | 9.75 |
ENSRNOT00000025240
ENSRNOT00000077776 |
LOC100911440
|
mitochondrial glutamate carrier 1-like |
| chr13_-_49169918 | 9.69 |
ENSRNOT00000000036
|
Tmcc2
|
transmembrane and coiled-coil domain family 2 |
| chr8_-_71055969 | 9.69 |
ENSRNOT00000075734
|
Ankdd1a
|
ankyrin repeat and death domain containing 1A |
| chr20_-_14573519 | 9.60 |
ENSRNOT00000001772
|
Rab36
|
RAB36, member RAS oncogene family |
| chr14_-_16452890 | 9.00 |
ENSRNOT00000067222
ENSRNOT00000091577 |
Sept11
|
septin 11 |
| chr8_+_48443767 | 8.80 |
ENSRNOT00000010127
|
C1qtnf5
|
C1q and tumor necrosis factor related protein 5 |
| chr1_+_170205591 | 8.57 |
ENSRNOT00000071063
|
LOC686660
|
similar to olfactory receptor 692 |
| chr8_+_107882219 | 8.47 |
ENSRNOT00000019902
|
Dzip1l
|
DAZ interacting zinc finger protein 1-like |
| chr7_-_139223116 | 8.31 |
ENSRNOT00000086559
|
Endou
|
endonuclease, poly(U)-specific |
| chrX_+_106791333 | 8.18 |
ENSRNOT00000050302
|
Tceal9
|
transcription elongation factor A like 9 |
| chr1_-_90520344 | 8.06 |
ENSRNOT00000078598
|
Kctd15
|
potassium channel tetramerization domain containing 15 |
| chr1_+_177093387 | 7.93 |
ENSRNOT00000021858
|
Mical2
|
microtubule associated monooxygenase, calponin and LIM domain containing 2 |
| chr1_+_221756286 | 7.92 |
ENSRNOT00000028636
|
Pygm
|
glycogen phosphorylase, muscle associated |
| chr2_-_166255302 | 7.74 |
ENSRNOT00000016012
|
B3galnt1
|
beta-1,3-N-acetylgalactosaminyltransferase 1 (globoside blood group) |
| chr4_+_49369296 | 7.31 |
ENSRNOT00000007822
|
Wnt16
|
wingless-type MMTV integration site family, member 16 |
| chr3_+_139894331 | 6.99 |
ENSRNOT00000064695
|
Rin2
|
Ras and Rab interactor 2 |
| chrX_-_64908682 | 6.24 |
ENSRNOT00000084107
|
Zc4h2
|
zinc finger C4H2-type containing |
| chr5_-_88612626 | 5.94 |
ENSRNOT00000089560
|
Tle1
|
transducin like enhancer of split 1 |
| chr8_+_48569328 | 5.48 |
ENSRNOT00000084030
|
Ccdc153
|
coiled-coil domain containing 153 |
| chr3_-_112789282 | 5.26 |
ENSRNOT00000090680
ENSRNOT00000066181 |
Ttbk2
|
tau tubulin kinase 2 |
| chr6_-_146470456 | 5.22 |
ENSRNOT00000018479
|
RGD1560883
|
similar to KIAA0825 protein |
| chr19_+_42983869 | 4.88 |
ENSRNOT00000025570
|
Glg1
|
golgi glycoprotein 1 |
| chr7_+_37018459 | 4.80 |
ENSRNOT00000087297
|
Eea1
|
early endosome antigen 1 |
| chr7_-_114573900 | 4.76 |
ENSRNOT00000011219
|
Ptk2
|
protein tyrosine kinase 2 |
| chr1_-_22281788 | 4.56 |
ENSRNOT00000021318
|
Stx7
|
syntaxin 7 |
| chr10_+_14248399 | 4.55 |
ENSRNOT00000077689
|
Spsb3
|
splA/ryanodine receptor domain and SOCS box containing 3 |
| chr14_+_80403001 | 4.29 |
ENSRNOT00000012109
|
Cpz
|
carboxypeptidase Z |
| chr13_-_51955735 | 4.04 |
ENSRNOT00000007597
|
Ptprv
|
protein tyrosine phosphatase, receptor type, V |
| chr20_+_1647441 | 3.93 |
ENSRNOT00000000986
|
Olr1730
|
olfactory receptor 1730 |
| chr6_-_76552559 | 3.93 |
ENSRNOT00000065230
|
Ralgapa1
|
Ral GTPase activating protein catalytic alpha subunit 1 |
| chrX_+_136488691 | 3.91 |
ENSRNOT00000093432
|
Arhgap36
|
Rho GTPase activating protein 36 |
| chr1_-_169598835 | 3.81 |
ENSRNOT00000072996
|
Olr156
|
olfactory receptor 156 |
| chr9_-_71651512 | 3.75 |
ENSRNOT00000032782
|
Plekhm3
|
pleckstrin homology domain containing M3 |
| chr3_+_172155496 | 3.61 |
ENSRNOT00000066279
|
Stx16
|
syntaxin 16 |
| chr13_+_103171047 | 3.48 |
ENSRNOT00000003216
|
Rab3gap2
|
RAB3 GTPase activating non-catalytic protein subunit 2 |
| chr15_-_32925673 | 3.41 |
ENSRNOT00000081045
|
Olr1646
|
olfactory receptor 1646 |
| chr14_-_84334066 | 3.38 |
ENSRNOT00000006160
|
Mtfp1
|
mitochondrial fission process 1 |
| chr10_-_72417070 | 3.24 |
ENSRNOT00000036814
|
Usp32
|
ubiquitin specific peptidase 32 |
| chr7_-_12405022 | 3.20 |
ENSRNOT00000021648
|
Cirbp
|
cold inducible RNA binding protein |
| chr11_-_82664554 | 3.08 |
ENSRNOT00000002425
ENSRNOT00000087105 |
Senp2
|
Sumo1/sentrin/SMT3 specific peptidase 2 |
| chr8_+_82380757 | 2.94 |
ENSRNOT00000013512
|
Leo1
|
LEO1 homolog, Paf1/RNA polymerase II complex component |
| chr11_-_39767802 | 2.92 |
ENSRNOT00000072806
|
RGD1565472
|
similar to RIKEN cDNA 2310003P10 |
| chr17_+_68477446 | 2.85 |
ENSRNOT00000086236
ENSRNOT00000068174 |
Pitrm1
|
pitrilysin metallopeptidase 1 |
| chr14_-_72970329 | 2.76 |
ENSRNOT00000006325
|
Pp2d1
|
protein phosphatase 2C-like domain containing 1 |
| chr2_-_60461458 | 2.60 |
ENSRNOT00000024285
|
Brix1
|
BRX1, biogenesis of ribosomes |
| chrX_+_40286592 | 2.56 |
ENSRNOT00000087898
|
Mbtps2
|
membrane-bound transcription factor peptidase, site 2 |
| chr19_-_37725623 | 2.16 |
ENSRNOT00000024197
|
Gfod2
|
glucose-fructose oxidoreductase domain containing 2 |
| chr1_+_66663965 | 2.09 |
ENSRNOT00000040738
|
LOC108348135
|
vomeronasal type-1 receptor 2-like |
| chr14_+_69800156 | 2.07 |
ENSRNOT00000072746
|
Lcorl
|
ligand dependent nuclear receptor corepressor-like |
| chr8_-_43443531 | 2.06 |
ENSRNOT00000007901
|
LOC103693060
|
olfactory receptor 148-like |
| chr17_+_45801528 | 2.05 |
ENSRNOT00000089221
|
Olr1664
|
olfactory receptor 1664 |
| chr1_-_72339395 | 2.00 |
ENSRNOT00000021772
|
Zfp580
|
zinc finger protein 580 |
| chr18_-_74059533 | 1.97 |
ENSRNOT00000038767
|
RGD1308601
|
similar to hypothetical protein |
| chr3_+_44025300 | 1.97 |
ENSRNOT00000006319
|
Galnt5
|
polypeptide N-acetylgalactosaminyltransferase 5 |
| chr14_-_115352562 | 1.93 |
ENSRNOT00000050321
|
Erlec1
|
endoplasmic reticulum lectin 1 |
| chr1_-_172465446 | 1.87 |
ENSRNOT00000088386
|
LOC103690410
|
olfactory receptor 484-like |
| chr1_-_20962526 | 1.86 |
ENSRNOT00000061332
ENSRNOT00000017322 ENSRNOT00000017412 ENSRNOT00000079688 ENSRNOT00000017417 |
Epb41l2
|
erythrocyte membrane protein band 4.1-like 2 |
| chr19_-_26194198 | 1.85 |
ENSRNOT00000005917
|
Wdr83
|
WD repeat domain 83 |
| chr18_-_79258570 | 1.81 |
ENSRNOT00000022401
|
Galr1
|
galanin receptor 1 |
| chr8_+_69484174 | 1.69 |
ENSRNOT00000077290
ENSRNOT00000056518 |
RGD1562747
|
similar to RIKEN cDNA 1110012L19 |
| chr1_+_102017263 | 1.61 |
ENSRNOT00000028680
|
Nomo1
|
nodal modulator 1 |
| chr8_-_42793484 | 1.54 |
ENSRNOT00000072871
|
AABR07069942.1
|
|
| chr13_+_34498749 | 1.45 |
ENSRNOT00000093127
|
Clasp1
|
cytoplasmic linker associated protein 1 |
| chr10_-_12818137 | 1.43 |
ENSRNOT00000060972
|
Olr1382
|
olfactory receptor 1382 |
| chr12_+_25430464 | 1.43 |
ENSRNOT00000002020
|
Gtf2i
|
general transcription factor II I |
| chr13_-_52834134 | 1.33 |
ENSRNOT00000038874
|
Igfn1
|
immunoglobulin-like and fibronectin type III domain containing 1 |
| chr16_+_81153489 | 1.29 |
ENSRNOT00000024999
|
Grk1
|
G protein-coupled receptor kinase 1 |
| chr20_+_55594676 | 1.29 |
ENSRNOT00000057010
|
Sim1
|
single-minded family bHLH transcription factor 1 |
| chr3_-_76708430 | 1.25 |
ENSRNOT00000007890
|
Olr630
|
olfactory receptor 630 |
| chr4_+_87516218 | 1.22 |
ENSRNOT00000090182
|
Vom1r66
|
vomeronasal 1 receptor 66 |
| chr1_-_171162912 | 1.16 |
ENSRNOT00000074866
|
Olr229
|
olfactory receptor 229 |
| chr10_-_15610826 | 1.11 |
ENSRNOT00000027851
|
Hbz
|
hemoglobin subunit zeta |
| chr5_-_50684409 | 0.98 |
ENSRNOT00000013181
|
RGD1359108
|
similar to RIKEN cDNA 3110043O21 |
| chr10_-_18131562 | 0.92 |
ENSRNOT00000006676
|
Tlx3
|
T-cell leukemia, homeobox 3 |
| chr2_-_35192515 | 0.92 |
ENSRNOT00000086631
|
LOC100909937
|
olfactory receptor 149-like |
| chr1_-_63457134 | 0.86 |
ENSRNOT00000083436
|
AABR07071874.1
|
|
| chr7_-_101140308 | 0.80 |
ENSRNOT00000006279
|
Fam84b
|
family with sequence similarity 84, member B |
| chr9_-_88534710 | 0.80 |
ENSRNOT00000020880
|
Tm4sf20
|
transmembrane 4 L six family member 20 |
| chr15_-_26780965 | 0.75 |
ENSRNOT00000041733
|
Olr1624
|
olfactory receptor 1624 |
| chr4_-_125929002 | 0.74 |
ENSRNOT00000083271
|
Magi1
|
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
| chr4_-_123510217 | 0.65 |
ENSRNOT00000080734
|
Slc41a3
|
solute carrier family 41, member 3 |
| chr5_+_113725717 | 0.55 |
ENSRNOT00000032248
|
Tek
|
TEK receptor tyrosine kinase |
| chr9_+_66952720 | 0.54 |
ENSRNOT00000023678
|
Nbeal1
|
neurobeachin-like 1 |
| chr3_-_33075685 | 0.50 |
ENSRNOT00000006937
|
Orc4
|
origin recognition complex, subunit 4 |
| chr3_+_59408546 | 0.49 |
ENSRNOT00000031418
|
AABR07052523.1
|
|
| chr18_+_35136132 | 0.49 |
ENSRNOT00000078731
|
Spink5
|
serine peptidase inhibitor, Kazal type 5 |
| chr3_+_77018186 | 0.40 |
ENSRNOT00000074443
|
Olr646
|
olfactory receptor 646 |
| chr8_+_42833019 | 0.39 |
ENSRNOT00000071050
|
LOC100910227
|
olfactory receptor 149-like |
| chrX_+_13989401 | 0.38 |
ENSRNOT00000004790
|
Hypm
|
huntingtin interacting protein M |
| chr8_-_43470377 | 0.34 |
ENSRNOT00000050312
|
LOC103690273
|
olfactory receptor 149 |
| chr8_+_19548488 | 0.32 |
ENSRNOT00000008605
|
Olr1148
|
olfactory receptor 1148 |
| chrX_-_72133692 | 0.29 |
ENSRNOT00000004263
|
Cited1
|
Cbp/p300-interacting transactivator with Glu/Asp-rich carboxy-terminal domain 1 |
| chr11_-_45616429 | 0.28 |
ENSRNOT00000046587
|
Olr1533
|
olfactory receptor 1533 |
| chrX_-_14756047 | 0.26 |
ENSRNOT00000060737
|
LOC102552474
|
protein FAM47E-like |
| chr2_-_264293010 | 0.20 |
ENSRNOT00000082532
ENSRNOT00000067843 |
LOC103691744
|
cystathionine gamma-lyase |
| chr10_-_94365049 | 0.17 |
ENSRNOT00000011894
|
Strada
|
STE20-related kinase adaptor alpha |
| chr4_-_117751329 | 0.13 |
ENSRNOT00000080096
|
LOC100910829
|
probable N-acetyltransferase CML2-like |
Network of associatons between targets according to the STRING database.
First level regulatory network of Rorc_Nr1d1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 13.9 | 41.6 | GO:0060084 | synaptic transmission involved in micturition(GO:0060084) |
| 10.7 | 74.7 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 8.7 | 26.0 | GO:0060086 | circadian temperature homeostasis(GO:0060086) |
| 3.1 | 18.4 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 3.0 | 12.1 | GO:0039020 | pronephric nephron tubule development(GO:0039020) |
| 2.6 | 29.1 | GO:0046543 | development of secondary female sexual characteristics(GO:0046543) |
| 2.6 | 7.9 | GO:0019417 | sulfur oxidation(GO:0019417) |
| 2.1 | 16.9 | GO:0016056 | rhodopsin mediated signaling pathway(GO:0016056) |
| 2.0 | 12.3 | GO:0032439 | endosome localization(GO:0032439) |
| 1.9 | 11.7 | GO:0007198 | adenylate cyclase-inhibiting serotonin receptor signaling pathway(GO:0007198) |
| 1.6 | 11.2 | GO:0071313 | cellular response to caffeine(GO:0071313) |
| 1.2 | 3.5 | GO:1903373 | positive regulation of endoplasmic reticulum tubular network organization(GO:1903373) |
| 1.1 | 4.6 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.9 | 10.7 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.9 | 2.6 | GO:0036500 | ATF6-mediated unfolded protein response(GO:0036500) |
| 0.8 | 49.4 | GO:0046676 | negative regulation of insulin secretion(GO:0046676) |
| 0.7 | 18.5 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
| 0.6 | 7.3 | GO:0090399 | replicative senescence(GO:0090399) |
| 0.6 | 13.4 | GO:0051481 | negative regulation of cytosolic calcium ion concentration(GO:0051481) |
| 0.6 | 1.8 | GO:0043400 | cortisol secretion(GO:0043400) regulation of cortisol secretion(GO:0051462) positive regulation of cortisol secretion(GO:0051464) |
| 0.6 | 23.6 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.6 | 9.7 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.6 | 7.7 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.5 | 42.7 | GO:0097194 | execution phase of apoptosis(GO:0097194) |
| 0.5 | 5.9 | GO:2000811 | negative regulation of anoikis(GO:2000811) |
| 0.5 | 3.2 | GO:0070417 | cellular response to cold(GO:0070417) |
| 0.4 | 5.3 | GO:1990403 | embryonic brain development(GO:1990403) |
| 0.4 | 2.9 | GO:0033523 | histone H2B ubiquitination(GO:0033523) |
| 0.4 | 19.3 | GO:0045739 | positive regulation of DNA repair(GO:0045739) |
| 0.4 | 7.9 | GO:0005980 | glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.3 | 3.1 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.2 | 12.5 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.2 | 57.1 | GO:0071805 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.2 | 4.8 | GO:0039703 | viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) |
| 0.2 | 3.6 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.2 | 0.5 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.2 | 1.9 | GO:0070862 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.2 | 1.5 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.2 | 13.7 | GO:0048024 | regulation of mRNA splicing, via spliceosome(GO:0048024) |
| 0.1 | 19.7 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.1 | 8.8 | GO:0070206 | protein trimerization(GO:0070206) |
| 0.1 | 0.5 | GO:1900425 | negative regulation of defense response to bacterium(GO:1900425) |
| 0.1 | 4.9 | GO:1903318 | negative regulation of protein processing(GO:0010955) negative regulation of protein maturation(GO:1903318) |
| 0.1 | 1.1 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.1 | 2.6 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.1 | 20.2 | GO:0001889 | liver development(GO:0001889) |
| 0.1 | 0.9 | GO:0002087 | regulation of respiratory gaseous exchange by neurological system process(GO:0002087) |
| 0.1 | 5.3 | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.1 | 20.4 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.1 | 9.2 | GO:0051291 | protein heterooligomerization(GO:0051291) |
| 0.1 | 24.7 | GO:0016358 | dendrite development(GO:0016358) |
| 0.1 | 0.3 | GO:0003340 | negative regulation of mesenchymal to epithelial transition involved in metanephros morphogenesis(GO:0003340) |
| 0.1 | 28.7 | GO:0007264 | small GTPase mediated signal transduction(GO:0007264) |
| 0.1 | 1.9 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 2.0 | GO:0032757 | positive regulation of interleukin-8 production(GO:0032757) |
| 0.0 | 1.6 | GO:0031648 | protein destabilization(GO:0031648) |
| 0.0 | 4.0 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 8.5 | GO:0060271 | cilium morphogenesis(GO:0060271) |
| 0.0 | 1.0 | GO:0016239 | positive regulation of macroautophagy(GO:0016239) |
| 0.0 | 11.3 | GO:0016567 | protein ubiquitination(GO:0016567) |
| 0.0 | 27.3 | GO:0007606 | sensory perception of chemical stimulus(GO:0007606) |
| 0.0 | 0.5 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 2.8 | GO:0006470 | protein dephosphorylation(GO:0006470) |
| 0.0 | 2.2 | GO:0030198 | extracellular matrix organization(GO:0030198) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.9 | 19.6 | GO:1990761 | growth cone lamellipodium(GO:1990761) growth cone filopodium(GO:1990812) |
| 2.6 | 23.2 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 2.1 | 41.6 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 1.8 | 10.7 | GO:0005955 | calcineurin complex(GO:0005955) |
| 1.2 | 12.3 | GO:0045179 | apical cortex(GO:0045179) |
| 0.8 | 4.8 | GO:0044308 | axonal spine(GO:0044308) |
| 0.8 | 18.5 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.7 | 12.1 | GO:0036038 | MKS complex(GO:0036038) |
| 0.6 | 4.6 | GO:0070820 | tertiary granule(GO:0070820) |
| 0.5 | 57.1 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.5 | 23.6 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.4 | 4.9 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.4 | 2.9 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.4 | 1.5 | GO:0031592 | centrosomal corona(GO:0031592) |
| 0.2 | 196.7 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.2 | 32.1 | GO:0030658 | transport vesicle membrane(GO:0030658) |
| 0.2 | 5.3 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.2 | 28.9 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.2 | 11.3 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.2 | 8.8 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.2 | 3.6 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.2 | 7.9 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.1 | 11.5 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.1 | 12.5 | GO:0070382 | exocytic vesicle(GO:0070382) |
| 0.1 | 55.7 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.1 | 0.5 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.1 | 1.1 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.1 | 1.0 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.1 | 3.2 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.1 | 0.5 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.1 | 1.9 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 6.2 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 3.1 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 1.3 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 11.5 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 24.9 | 74.7 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 8.9 | 26.8 | GO:0072590 | N-acetyl-L-aspartate-L-glutamate ligase activity(GO:0072590) |
| 5.8 | 23.2 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 3.0 | 57.1 | GO:0030955 | potassium ion binding(GO:0030955) |
| 2.6 | 7.9 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 2.4 | 41.6 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 1.9 | 9.7 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 1.7 | 42.7 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 1.6 | 32.7 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 1.5 | 7.7 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 1.5 | 10.7 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 1.4 | 13.6 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 1.3 | 26.0 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 1.0 | 23.6 | GO:0005112 | Notch binding(GO:0005112) |
| 0.9 | 19.6 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.9 | 14.4 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.8 | 11.7 | GO:0051378 | serotonin binding(GO:0051378) |
| 0.7 | 2.9 | GO:1990269 | RNA polymerase II C-terminal domain phosphoserine binding(GO:1990269) |
| 0.6 | 3.1 | GO:0070137 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.5 | 68.5 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.5 | 29.1 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.5 | 4.8 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.5 | 1.9 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.5 | 3.2 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.3 | 8.3 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.3 | 7.9 | GO:0071949 | FAD binding(GO:0071949) |
| 0.3 | 12.3 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.2 | 8.2 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.2 | 1.8 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.2 | 2.0 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.2 | 4.8 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.2 | 5.9 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.2 | 7.3 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.2 | 38.1 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
| 0.2 | 16.5 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.2 | 4.9 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.2 | 1.3 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.2 | 1.5 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.2 | 4.3 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 4.5 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 17.5 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.1 | 15.6 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.1 | 1.1 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.1 | 24.0 | GO:0005525 | GTP binding(GO:0005525) |
| 0.0 | 2.8 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 5.4 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 1.4 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 3.2 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 4.0 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.5 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.3 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.0 | 11.3 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 1.9 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.7 | GO:0051393 | alpha-actinin binding(GO:0051393) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 137.2 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.8 | 8.3 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.5 | 4.8 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.4 | 36.7 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.4 | 11.2 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.3 | 13.6 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.3 | 23.6 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.3 | 10.7 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.2 | 19.6 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.2 | 5.9 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.1 | 10.4 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.1 | 29.1 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 1.3 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 0.5 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.5 | 41.6 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 2.8 | 85.5 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 1.4 | 20.8 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 1.1 | 57.1 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.8 | 11.7 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.5 | 20.6 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.5 | 7.9 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.4 | 8.2 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.3 | 4.8 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.3 | 28.9 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.3 | 12.3 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.2 | 23.6 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.2 | 2.6 | REACTOME ACTIVATION OF CHAPERONES BY ATF6 ALPHA | Genes involved in Activation of Chaperones by ATF6-alpha |
| 0.1 | 5.9 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.1 | 7.3 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.1 | 2.0 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.5 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.0 | 4.8 | REACTOME TOLL RECEPTOR CASCADES | Genes involved in Toll Receptor Cascades |
| 0.0 | 0.5 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 1.5 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.2 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |