Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
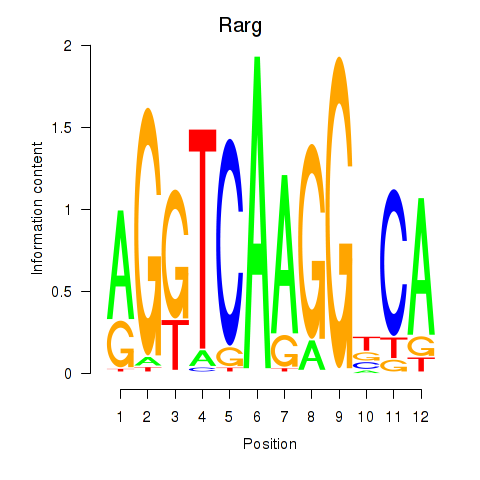
Results for Rarg
Z-value: 1.22
Transcription factors associated with Rarg
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Rarg
|
ENSRNOG00000012499 | retinoic acid receptor, gamma |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Rarg | rn6_v1_chr7_-_143863186_143863186 | -0.07 | 1.8e-01 | Click! |
Activity profile of Rarg motif
Sorted Z-values of Rarg motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr20_-_4489281 | 28.92 |
ENSRNOT00000031548
|
Cyp21a1
|
cytochrome P450, family 21, subfamily a, polypeptide 1 |
| chr10_+_10774639 | 27.97 |
ENSRNOT00000061208
|
Sept12
|
septin 12 |
| chr1_-_88066101 | 26.48 |
ENSRNOT00000079473
ENSRNOT00000027893 |
Ryr1
|
ryanodine receptor 1 |
| chr1_-_170431073 | 24.96 |
ENSRNOT00000024710
|
Hpx
|
hemopexin |
| chr1_+_81763614 | 23.68 |
ENSRNOT00000027254
|
Cd79a
|
CD79a molecule |
| chr2_-_183031214 | 23.01 |
ENSRNOT00000013260
|
LOC679811
|
similar to RIKEN cDNA D930015E06 |
| chr8_-_128711221 | 21.93 |
ENSRNOT00000055888
|
Xirp1
|
xin actin-binding repeat containing 1 |
| chr3_-_80012750 | 21.91 |
ENSRNOT00000018154
|
Nr1h3
|
nuclear receptor subfamily 1, group H, member 3 |
| chr11_-_81639872 | 21.70 |
ENSRNOT00000047595
ENSRNOT00000090031 ENSRNOT00000081864 |
Hrg
|
histidine-rich glycoprotein |
| chr1_+_165482155 | 21.14 |
ENSRNOT00000024005
|
Ucp3
|
uncoupling protein 3 |
| chr3_+_171213936 | 20.59 |
ENSRNOT00000031586
|
Pck1
|
phosphoenolpyruvate carboxykinase 1 |
| chr1_-_197821936 | 20.55 |
ENSRNOT00000055027
|
Cd19
|
CD19 molecule |
| chr9_-_16795887 | 19.99 |
ENSRNOT00000071609
|
Cd79al
|
Cd79a molecule-like |
| chr8_-_50539331 | 19.76 |
ENSRNOT00000088997
|
AABR07073400.1
|
|
| chr9_+_10956056 | 19.51 |
ENSRNOT00000073930
|
Plin5
|
perilipin 5 |
| chr12_+_16912249 | 18.68 |
ENSRNOT00000085936
|
Tmem184a
|
transmembrane protein 184A |
| chr4_+_122244711 | 18.24 |
ENSRNOT00000038251
|
Uroc1
|
urocanate hydratase 1 |
| chr20_-_4316715 | 17.88 |
ENSRNOT00000031704
|
C4b
|
complement C4B (Chido blood group) |
| chr7_-_29152442 | 17.81 |
ENSRNOT00000079774
|
Mybpc1
|
myosin binding protein C, slow type |
| chr13_+_78805347 | 17.69 |
ENSRNOT00000003748
|
Serpinc1
|
serpin family C member 1 |
| chrX_+_15273933 | 17.63 |
ENSRNOT00000075082
|
LOC108348091
|
erythroid transcription factor |
| chr6_-_127816055 | 17.59 |
ENSRNOT00000013175
|
Serpina3m
|
serine (or cysteine) proteinase inhibitor, clade A, member 3M |
| chr4_-_165629996 | 16.71 |
ENSRNOT00000068185
ENSRNOT00000007427 |
Ybx3
|
Y box binding protein 3 |
| chr3_-_80543031 | 16.69 |
ENSRNOT00000022233
|
F2
|
coagulation factor II |
| chr4_-_181348799 | 16.59 |
ENSRNOT00000033743
|
LOC690784
|
hypothetical protein LOC690783 |
| chr1_+_248723397 | 16.25 |
ENSRNOT00000072188
|
LOC100911854
|
mannose-binding protein C-like |
| chr3_-_62524996 | 16.16 |
ENSRNOT00000002114
|
Nfe2l2
|
nuclear factor, erythroid 2-like 2 |
| chr1_-_80599572 | 15.47 |
ENSRNOT00000024832
|
Apoc4
|
apolipoprotein C4 |
| chr5_-_19368431 | 15.31 |
ENSRNOT00000012819
|
Cyp7a1
|
cytochrome P450, family 7, subfamily a, polypeptide 1 |
| chr20_-_2678141 | 15.28 |
ENSRNOT00000072377
ENSRNOT00000083833 |
C4a
|
complement component 4A (Rodgers blood group) |
| chr8_-_56393233 | 15.25 |
ENSRNOT00000016263
|
Fdx1
|
ferredoxin 1 |
| chr1_+_65576535 | 15.24 |
ENSRNOT00000026575
|
Slc27a5
|
solute carrier family 27 member 5 |
| chr1_-_98490315 | 15.08 |
ENSRNOT00000056490
|
Cldnd2
|
claudin domain containing 2 |
| chr12_+_22153983 | 14.92 |
ENSRNOT00000080775
ENSRNOT00000001894 |
Pcolce
|
procollagen C-endopeptidase enhancer |
| chr13_-_47397890 | 14.69 |
ENSRNOT00000005505
|
C4bpb
|
complement component 4 binding protein, beta |
| chrX_+_54734385 | 14.57 |
ENSRNOT00000005023
|
Nr0b1
|
nuclear receptor subfamily 0, group B, member 1 |
| chr1_+_219329574 | 14.48 |
ENSRNOT00000071109
|
Cabp2
|
calcium binding protein 2 |
| chr9_+_17734345 | 14.42 |
ENSRNOT00000067104
|
Capn11
|
calpain 11 |
| chr10_-_109729019 | 14.39 |
ENSRNOT00000054959
|
Ppp1r27
|
protein phosphatase 1, regulatory subunit 27 |
| chr5_-_59165160 | 14.34 |
ENSRNOT00000029035
|
Fam221b
|
family with sequence similarity 221, member B |
| chr4_+_61850348 | 14.31 |
ENSRNOT00000013423
|
Akr1b7
|
aldo-keto reductase family 1, member B7 |
| chr5_+_90338795 | 14.14 |
ENSRNOT00000077864
ENSRNOT00000058882 |
LOC298139
|
similar to RIKEN cDNA 2310003M01 |
| chr7_-_14435967 | 14.14 |
ENSRNOT00000074801
|
Pglyrp2
|
peptidoglycan recognition protein 2 |
| chr14_-_86164341 | 13.98 |
ENSRNOT00000086343
ENSRNOT00000080147 |
Gck
|
glucokinase |
| chr8_-_111761871 | 13.95 |
ENSRNOT00000056483
|
RGD1310507
|
similar to RIKEN cDNA 1300017J02 |
| chr18_+_56071478 | 13.83 |
ENSRNOT00000025344
ENSRNOT00000025354 |
Cd74
|
CD74 molecule |
| chr4_-_30338679 | 13.74 |
ENSRNOT00000012050
|
Pon3
|
paraoxonase 3 |
| chr6_+_128046780 | 13.38 |
ENSRNOT00000078064
|
LOC500712
|
Ab1-233 |
| chr8_+_106317124 | 13.25 |
ENSRNOT00000018411
|
Nmnat3
|
nicotinamide nucleotide adenylyltransferase 3 |
| chr8_+_110982777 | 13.17 |
ENSRNOT00000010992
|
Ky
|
kyphoscoliosis peptidase |
| chr2_+_198655437 | 13.07 |
ENSRNOT00000028781
|
Hfe2
|
hemochromatosis type 2 (juvenile) |
| chr6_-_135089409 | 12.93 |
ENSRNOT00000089238
|
RGD1305298
|
hypothetical LOC299330 |
| chr3_-_2727616 | 12.92 |
ENSRNOT00000061904
|
C8g
|
complement C8 gamma chain |
| chr15_-_57651041 | 12.80 |
ENSRNOT00000072138
|
Spert
|
spermatid associated |
| chr16_-_49003246 | 12.75 |
ENSRNOT00000089501
|
Acsl1
|
acyl-CoA synthetase long-chain family member 1 |
| chr3_-_156905892 | 12.69 |
ENSRNOT00000022424
|
Emilin3
|
elastin microfibril interfacer 3 |
| chr2_+_114423533 | 12.65 |
ENSRNOT00000091221
|
Slc2a2
|
solute carrier family 2 member 2 |
| chr1_+_257076221 | 12.52 |
ENSRNOT00000044895
|
Slc35g1
|
solute carrier family 35, member G1 |
| chr4_-_30556814 | 12.51 |
ENSRNOT00000012760
|
Pdk4
|
pyruvate dehydrogenase kinase 4 |
| chr13_+_52625441 | 12.47 |
ENSRNOT00000012095
|
Tnni1
|
troponin I1, slow skeletal type |
| chr1_-_89539210 | 12.42 |
ENSRNOT00000077462
ENSRNOT00000028644 |
Hpn
|
hepsin |
| chr1_+_80777014 | 12.41 |
ENSRNOT00000079758
|
Gm19345
|
predicted gene, 19345 |
| chr2_-_23289266 | 12.36 |
ENSRNOT00000061708
|
Bhmt2
|
betaine-homocysteine S-methyltransferase 2 |
| chr10_+_4950484 | 12.22 |
ENSRNOT00000003450
|
Prm2
|
protamine 2 |
| chr1_+_193424812 | 12.20 |
ENSRNOT00000019939
|
Aqp8
|
aquaporin 8 |
| chr20_-_45259928 | 12.12 |
ENSRNOT00000087226
|
Slc16a10
|
solute carrier family 16 member 10 |
| chr11_+_30904733 | 12.10 |
ENSRNOT00000036027
|
Mrap
|
melanocortin 2 receptor accessory protein |
| chr8_+_116857684 | 12.10 |
ENSRNOT00000026711
|
Mst1
|
macrophage stimulating 1 |
| chr5_+_160306727 | 12.03 |
ENSRNOT00000016648
|
Agmat
|
agmatinase |
| chr3_-_153952108 | 12.03 |
ENSRNOT00000050465
|
Adig
|
adipogenin |
| chr5_-_29601748 | 11.95 |
ENSRNOT00000011330
|
Decr1
|
2,4-dienoyl-CoA reductase 1 |
| chrX_-_15693473 | 11.80 |
ENSRNOT00000013758
|
Prickle3
|
prickle planar cell polarity protein 3 |
| chr19_-_55284663 | 11.63 |
ENSRNOT00000018387
|
Snai3
|
snail family transcriptional repressor 3 |
| chr20_-_6195463 | 11.48 |
ENSRNOT00000092474
|
Pxt1
|
peroxisomal, testis specific 1 |
| chrX_-_71169865 | 11.45 |
ENSRNOT00000050415
|
Il2rg
|
interleukin 2 receptor subunit gamma |
| chr1_+_215610368 | 11.19 |
ENSRNOT00000078903
ENSRNOT00000087781 |
Tnni2
|
troponin I2, fast skeletal type |
| chr13_+_75105615 | 11.15 |
ENSRNOT00000076653
|
Tp53i3
|
tumor protein p53 inducible protein 3 |
| chr8_-_114449956 | 11.13 |
ENSRNOT00000056414
|
Col6a6
|
collagen type VI alpha 6 chain |
| chr14_-_86796378 | 11.12 |
ENSRNOT00000092021
|
Myo1g
|
myosin IG |
| chr16_+_18736154 | 11.02 |
ENSRNOT00000015723
|
Mbl1
|
mannose-binding lectin (protein A) 1 |
| chr5_-_166282831 | 10.92 |
ENSRNOT00000021348
|
Rbp7
|
retinol binding protein 7 |
| chr8_+_96564877 | 10.82 |
ENSRNOT00000017879
|
Mthfs
|
5,10-methenyltetrahydrofolate synthetase (5-formyltetrahydrofolate cyclo-ligase) |
| chr5_-_147784311 | 10.81 |
ENSRNOT00000074172
|
Fam167b
|
family with sequence similarity 167, member B |
| chr1_+_261281543 | 10.78 |
ENSRNOT00000018890
|
Ankrd2
|
ankyrin repeat domain 2 |
| chr11_+_74057361 | 10.78 |
ENSRNOT00000048746
|
Cpn2
|
carboxypeptidase N subunit 2 |
| chr2_-_197860699 | 10.67 |
ENSRNOT00000028740
|
Ecm1
|
extracellular matrix protein 1 |
| chr6_-_142903440 | 10.59 |
ENSRNOT00000075707
|
AABR07065823.2
|
|
| chr3_-_8981362 | 10.52 |
ENSRNOT00000086717
|
Crat
|
carnitine O-acetyltransferase |
| chr20_-_4698718 | 10.51 |
ENSRNOT00000047527
|
RT1-CE7
|
RT1 class I, locus CE7 |
| chr13_-_68360664 | 10.50 |
ENSRNOT00000030971
|
Hmcn1
|
hemicentin 1 |
| chr5_+_152290084 | 10.45 |
ENSRNOT00000089849
|
Aim1l
|
absent in melanoma 1-like |
| chr19_+_15094309 | 10.44 |
ENSRNOT00000083500
|
Ces1f
|
carboxylesterase 1F |
| chr16_+_50179458 | 10.29 |
ENSRNOT00000041946
|
F11
|
coagulation factor XI |
| chr16_-_60427474 | 10.28 |
ENSRNOT00000051720
|
Ppp1r3b
|
protein phosphatase 1, regulatory subunit 3B |
| chr14_-_80973456 | 10.25 |
ENSRNOT00000013257
|
Hgfac
|
HGF activator |
| chr3_-_127500709 | 10.19 |
ENSRNOT00000006330
|
Hao1
|
hydroxyacid oxidase 1 |
| chr9_-_61975640 | 10.16 |
ENSRNOT00000085744
|
Boll
|
boule homolog, RNA binding protein |
| chr10_-_105412627 | 10.15 |
ENSRNOT00000034817
|
Qrich2
|
glutamine rich 2 |
| chr6_+_128973956 | 10.12 |
ENSRNOT00000075399
|
Fam181a
|
family with sequence similarity 181, member A |
| chr1_+_167197549 | 10.11 |
ENSRNOT00000027427
|
Art1
|
ADP-ribosyltransferase 1 |
| chr5_+_79179417 | 10.08 |
ENSRNOT00000010454
|
Orm1
|
orosomucoid 1 |
| chr10_-_58973020 | 10.03 |
ENSRNOT00000020379
|
Smtnl2
|
smoothelin-like 2 |
| chr8_-_63155825 | 9.95 |
ENSRNOT00000011820
|
Tbc1d21
|
TBC1 domain family, member 21 |
| chr12_+_19196611 | 9.95 |
ENSRNOT00000001801
|
Azgp1
|
alpha-2-glycoprotein 1, zinc-binding |
| chr1_-_102255459 | 9.95 |
ENSRNOT00000067392
|
Ush1c
|
USH1 protein network component harmonin |
| chr1_-_219312240 | 9.78 |
ENSRNOT00000066691
|
RGD1307603
|
similar to hypothetical protein MGC37914 |
| chrX_-_64726210 | 9.68 |
ENSRNOT00000076012
ENSRNOT00000086265 |
Asb12
|
ankyrin repeat and SOCS box-containing 12 |
| chr1_-_80744831 | 9.59 |
ENSRNOT00000025913
|
Bcl3
|
B-cell CLL/lymphoma 3 |
| chr9_-_54457753 | 9.59 |
ENSRNOT00000020032
|
Stat1
|
signal transducer and activator of transcription 1 |
| chr1_-_221321459 | 9.58 |
ENSRNOT00000028406
ENSRNOT00000084800 |
Pola2
|
DNA polymerase alpha 2, accessory subunit |
| chr6_-_143195445 | 9.55 |
ENSRNOT00000078672
|
AABR07065837.1
|
|
| chr1_-_189359730 | 9.55 |
ENSRNOT00000066597
|
RGD1559600
|
RGD1559600 |
| chr16_-_71040847 | 9.53 |
ENSRNOT00000020606
|
Star
|
steroidogenic acute regulatory protein |
| chr4_-_114768029 | 9.51 |
ENSRNOT00000083404
|
Tlx2
|
T-cell leukemia homeobox 2 |
| chr15_+_24159647 | 9.47 |
ENSRNOT00000082675
|
Lgals3
|
galectin 3 |
| chr9_-_119332967 | 9.46 |
ENSRNOT00000021048
|
Myl12a
|
myosin light chain 12A |
| chr14_+_38030189 | 9.38 |
ENSRNOT00000035623
|
Txk
|
TXK tyrosine kinase |
| chr14_+_13192347 | 9.32 |
ENSRNOT00000000092
|
Antxr2
|
anthrax toxin receptor 2 |
| chr8_+_117486083 | 9.31 |
ENSRNOT00000027552
ENSRNOT00000088705 |
Prkar2a
|
protein kinase cAMP-dependent type 2 regulatory subunit alpha |
| chr2_+_252018597 | 9.28 |
ENSRNOT00000020452
|
Mcoln2
|
mucolipin 2 |
| chr4_+_56591715 | 9.28 |
ENSRNOT00000041207
|
Fam71f1
|
family with sequence similarity 71, member F1 |
| chr7_+_117351648 | 9.24 |
ENSRNOT00000056421
|
Smpd5
|
sphingomyelin phosphodiesterase 5 |
| chr16_-_7250924 | 9.20 |
ENSRNOT00000025394
|
Stab1
|
stabilin 1 |
| chr20_-_9876008 | 9.17 |
ENSRNOT00000001537
|
Tff2
|
trefoil factor 2 |
| chr1_+_64506735 | 9.14 |
ENSRNOT00000086331
|
Nlrp12
|
NLR family, pyrin domain containing 12 |
| chr15_+_33606124 | 9.10 |
ENSRNOT00000065210
|
AC115371.1
|
|
| chr6_-_143195143 | 9.09 |
ENSRNOT00000081337
|
AABR07065837.1
|
|
| chr16_+_21288876 | 9.08 |
ENSRNOT00000027990
|
Cilp2
|
cartilage intermediate layer protein 2 |
| chr8_-_63150753 | 9.06 |
ENSRNOT00000078666
|
Tbc1d21
|
TBC1 domain family, member 21 |
| chr10_+_104377748 | 9.01 |
ENSRNOT00000080714
|
Llgl2
|
LLGL2, scribble cell polarity complex component |
| chrX_+_158569056 | 9.00 |
ENSRNOT00000074170
|
LOC102550080
|
placenta-specific protein 1-like |
| chr4_-_144318580 | 9.00 |
ENSRNOT00000007591
|
Ssuh2
|
ssu-2 homolog |
| chr1_+_102924059 | 8.96 |
ENSRNOT00000078137
|
AC099150.1
|
|
| chr1_+_41325462 | 8.95 |
ENSRNOT00000081017
ENSRNOT00000078494 ENSRNOT00000088168 |
Esr1
|
estrogen receptor 1 |
| chr6_-_25211494 | 8.95 |
ENSRNOT00000009634
|
Xdh
|
xanthine dehydrogenase |
| chr6_+_104718512 | 8.91 |
ENSRNOT00000007947
|
Smoc1
|
SPARC related modular calcium binding 1 |
| chr17_+_4212890 | 8.81 |
ENSRNOT00000024467
|
Testin
|
testin gene |
| chr4_-_129619142 | 8.75 |
ENSRNOT00000047453
|
Lmod3
|
leiomodin 3 |
| chr16_-_54513349 | 8.71 |
ENSRNOT00000014809
ENSRNOT00000015127 |
Slc7a2
|
solute carrier family 7 member 2 |
| chr12_+_30603242 | 8.66 |
ENSRNOT00000068460
|
Sept14
|
septin 14 |
| chr9_-_9985630 | 8.66 |
ENSRNOT00000071780
|
Crb3
|
crumbs 3, cell polarity complex component |
| chr4_+_169161585 | 8.63 |
ENSRNOT00000079785
|
Emp1
|
epithelial membrane protein 1 |
| chr19_+_55094585 | 8.58 |
ENSRNOT00000068452
|
Zfpm1
|
zinc finger protein, multitype 1 |
| chr19_-_10976396 | 8.58 |
ENSRNOT00000073239
|
Nlrc5
|
NLR family, CARD domain containing 5 |
| chr1_-_198486157 | 8.57 |
ENSRNOT00000022758
|
Zg16
|
zymogen granule protein 16 |
| chr11_-_71419223 | 8.54 |
ENSRNOT00000002407
|
Tfrc
|
transferrin receptor |
| chr10_-_67479077 | 8.50 |
ENSRNOT00000090278
|
Tefm
|
transcription elongation factor, mitochondrial |
| chr8_+_50310405 | 8.44 |
ENSRNOT00000073507
|
Sik3
|
SIK family kinase 3 |
| chr17_+_22619891 | 8.38 |
ENSRNOT00000060403
|
Adtrp
|
androgen-dependent TFPI-regulating protein |
| chr8_-_23146689 | 8.36 |
ENSRNOT00000092200
|
Acp5
|
acid phosphatase 5, tartrate resistant |
| chr13_+_44424689 | 8.35 |
ENSRNOT00000005206
|
Acmsd
|
aminocarboxymuconate semialdehyde decarboxylase |
| chr10_-_5533695 | 8.29 |
ENSRNOT00000051564
|
Rpl39l
|
ribosomal protein L39-like |
| chr5_-_110706656 | 8.29 |
ENSRNOT00000033781
|
Izumo3
|
IZUMO family member 3 |
| chr5_+_154260062 | 8.27 |
ENSRNOT00000074998
|
Cnr2
|
cannabinoid receptor 2 |
| chr10_+_3655038 | 8.25 |
ENSRNOT00000045047
|
Cpped1
|
calcineurin-like phosphoesterase domain containing 1 |
| chr1_+_229066045 | 8.22 |
ENSRNOT00000016454
|
Glyat
|
glycine-N-acyltransferase |
| chr10_-_56426012 | 8.16 |
ENSRNOT00000092506
ENSRNOT00000084068 |
LOC497938
|
similar to RIKEN cDNA 4933402P03 |
| chr10_-_5260608 | 8.15 |
ENSRNOT00000003572
|
Ciita
|
class II, major histocompatibility complex, transactivator |
| chr10_-_29196563 | 8.14 |
ENSRNOT00000005205
|
Fabp6
|
fatty acid binding protein 6 |
| chr6_-_142534112 | 8.08 |
ENSRNOT00000073598
|
Ighv1-49
|
immunoglobulin heavy variable 1-49 |
| chr2_-_60657712 | 8.04 |
ENSRNOT00000040348
|
Rai14
|
retinoic acid induced 14 |
| chr14_-_17171580 | 8.03 |
ENSRNOT00000085525
|
Art3
|
ADP-ribosyltransferase 3 |
| chr8_-_60086403 | 8.00 |
ENSRNOT00000020544
|
Etfa
|
electron transfer flavoprotein alpha subunit |
| chr4_-_113902399 | 7.98 |
ENSRNOT00000087788
|
Tlx2
|
T-cell leukemia homeobox 2 |
| chr16_+_1979191 | 7.91 |
ENSRNOT00000014382
|
Ppif
|
peptidylprolyl isomerase F |
| chr9_-_71900044 | 7.86 |
ENSRNOT00000020322
|
Idh1
|
isocitrate dehydrogenase (NADP(+)) 1, cytosolic |
| chr8_+_115193146 | 7.85 |
ENSRNOT00000056391
|
Iqcf1
|
IQ motif containing F1 |
| chr3_+_143119687 | 7.84 |
ENSRNOT00000006608
|
Cst12
|
cystatin 12 |
| chr1_+_100199057 | 7.82 |
ENSRNOT00000025831
|
Klk1
|
kallikrein 1 |
| chr4_-_98995590 | 7.80 |
ENSRNOT00000008698
|
Thnsl2
|
threonine synthase-like 2 |
| chr10_+_103543895 | 7.79 |
ENSRNOT00000077882
|
Cd300ld
|
Cd300 molecule-like family member D |
| chr4_-_98735346 | 7.73 |
ENSRNOT00000008473
|
Tex37
|
testis expressed 37 |
| chr10_+_106785077 | 7.73 |
ENSRNOT00000075047
|
Tmc8
|
transmembrane channel-like 8 |
| chr20_-_13657943 | 7.69 |
ENSRNOT00000032290
|
Vpreb3
|
pre-B lymphocyte 3 |
| chr7_-_145048283 | 7.66 |
ENSRNOT00000055275
|
Gtsf1
|
gametocyte specific factor 1 |
| chr4_-_60358562 | 7.63 |
ENSRNOT00000018001
|
Chchd3
|
coiled-coil-helix-coiled-coil-helix domain containing 3 |
| chr5_-_136053210 | 7.61 |
ENSRNOT00000025903
|
Kif2c
|
kinesin family member 2C |
| chr9_-_44617210 | 7.57 |
ENSRNOT00000074343
|
Lyg1
|
lysozyme G1 |
| chr6_-_47848026 | 7.55 |
ENSRNOT00000011048
ENSRNOT00000090017 |
Allc
|
allantoicase |
| chr20_-_7307280 | 7.52 |
ENSRNOT00000000583
ENSRNOT00000079933 |
Spdef
|
SAM pointed domain containing ets transcription factor |
| chrX_-_157095274 | 7.51 |
ENSRNOT00000084088
|
Abcd1
|
ATP binding cassette subfamily D member 1 |
| chr7_+_11769400 | 7.48 |
ENSRNOT00000044417
|
Jsrp1
|
junctional sarcoplasmic reticulum protein 1 |
| chr4_-_169093135 | 7.47 |
ENSRNOT00000011461
|
Pbp2
|
phosphatidylethanolamine binding protein 2 |
| chr5_+_124442293 | 7.42 |
ENSRNOT00000041922
|
RGD1564074
|
similar to novel protein |
| chr2_+_189423559 | 7.40 |
ENSRNOT00000029076
|
Tpm3
|
tropomyosin 3 |
| chr4_+_148782479 | 7.35 |
ENSRNOT00000018133
|
LOC500300
|
similar to hypothetical protein MGC6835 |
| chr3_-_72447801 | 7.32 |
ENSRNOT00000088815
|
P2rx3
|
purinergic receptor P2X 3 |
| chr6_-_135026449 | 7.30 |
ENSRNOT00000009444
|
RGD1305298
|
hypothetical LOC299330 |
| chr15_-_34392066 | 7.26 |
ENSRNOT00000027315
|
Tgm1
|
transglutaminase 1 |
| chr4_-_6449949 | 7.25 |
ENSRNOT00000062026
|
Galntl5
|
polypeptide N-acetylgalactosaminyltransferase-like 5 |
| chr12_-_2007516 | 7.23 |
ENSRNOT00000037564
|
Pex11g
|
peroxisomal biogenesis factor 11 gamma |
| chr10_-_67478848 | 7.23 |
ENSRNOT00000005325
|
Tefm
|
transcription elongation factor, mitochondrial |
| chr15_+_61069581 | 7.21 |
ENSRNOT00000084333
|
Vwa8
|
von Willebrand factor A domain containing 8 |
| chr1_+_100708693 | 7.18 |
ENSRNOT00000071054
ENSRNOT00000071503 |
LOC100911976
|
izumo sperm-egg fusion protein 2-like |
| chr4_-_155051429 | 7.16 |
ENSRNOT00000020094
|
Klrg1
|
killer cell lectin like receptor G1 |
| chr20_+_4959294 | 7.13 |
ENSRNOT00000074223
|
Hspa1l
|
heat shock protein family A (Hsp70) member 1 like |
| chr1_+_100830064 | 7.06 |
ENSRNOT00000027320
|
Il4i1
|
interleukin 4 induced 1 |
| chr10_+_94471476 | 7.02 |
ENSRNOT00000088697
|
Tcam1
|
testicular cell adhesion molecule 1 |
| chr8_-_111850075 | 7.01 |
ENSRNOT00000082097
|
Cdv3
|
CDV3 homolog |
| chr1_-_107385257 | 7.00 |
ENSRNOT00000074296
|
Ccdc179
|
coiled-coil domain containing 179 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Rarg
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 10.7 | 42.9 | GO:0060335 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 7.2 | 21.7 | GO:0097037 | heme export(GO:0097037) |
| 6.5 | 19.5 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 5.6 | 16.7 | GO:1900738 | positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) response to thyrotropin-releasing hormone(GO:1905225) |
| 5.4 | 16.2 | GO:1902037 | negative regulation of hematopoietic stem cell differentiation(GO:1902037) |
| 4.9 | 24.6 | GO:0048550 | negative regulation of pinocytosis(GO:0048550) |
| 4.9 | 14.7 | GO:0045959 | negative regulation of complement activation, classical pathway(GO:0045959) |
| 4.8 | 19.4 | GO:0002325 | natural killer cell differentiation involved in immune response(GO:0002325) negative regulation of natural killer cell differentiation(GO:0032824) regulation of natural killer cell differentiation involved in immune response(GO:0032826) negative regulation of natural killer cell differentiation involved in immune response(GO:0032827) |
| 4.7 | 18.7 | GO:0018992 | germ-line sex determination(GO:0018992) |
| 4.4 | 17.7 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 4.4 | 13.2 | GO:0019355 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 4.4 | 17.6 | GO:0030221 | basophil differentiation(GO:0030221) |
| 4.4 | 17.5 | GO:0006114 | glycerol biosynthetic process(GO:0006114) |
| 4.3 | 8.6 | GO:0060375 | regulation of mast cell differentiation(GO:0060375) transcriptional activation by promoter-enhancer looping(GO:0071733) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 4.2 | 16.7 | GO:1902219 | negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902219) |
| 4.1 | 12.4 | GO:0034769 | basement membrane disassembly(GO:0034769) |
| 3.8 | 15.2 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 3.8 | 18.9 | GO:0009732 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 3.7 | 22.1 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 3.6 | 18.2 | GO:0006548 | histidine catabolic process(GO:0006548) |
| 3.6 | 10.8 | GO:0042560 | folic acid-containing compound catabolic process(GO:0009397) pteridine-containing compound catabolic process(GO:0042560) |
| 3.5 | 21.0 | GO:0045343 | MHC class I biosynthetic process(GO:0045341) regulation of MHC class I biosynthetic process(GO:0045343) positive regulation of MHC class I biosynthetic process(GO:0045345) |
| 3.5 | 10.5 | GO:0003162 | atrioventricular node development(GO:0003162) |
| 3.3 | 23.0 | GO:0033088 | negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 3.2 | 15.9 | GO:0015755 | fructose transport(GO:0015755) |
| 3.2 | 9.5 | GO:0017143 | insecticide metabolic process(GO:0017143) |
| 3.2 | 9.5 | GO:1903769 | negative regulation of cell proliferation in bone marrow(GO:1903769) |
| 3.1 | 12.5 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 2.9 | 8.7 | GO:0089718 | amino acid import across plasma membrane(GO:0089718) |
| 2.8 | 2.8 | GO:0006579 | amino-acid betaine catabolic process(GO:0006579) |
| 2.8 | 13.8 | GO:0002905 | mature B cell apoptotic process(GO:0002901) regulation of mature B cell apoptotic process(GO:0002905) negative regulation of mature B cell apoptotic process(GO:0002906) |
| 2.8 | 22.0 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 2.7 | 8.2 | GO:0045082 | positive regulation of interleukin-10 biosynthetic process(GO:0045082) |
| 2.6 | 7.9 | GO:1902445 | regulation of mitochondrial membrane permeability involved in programmed necrotic cell death(GO:1902445) |
| 2.6 | 10.5 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 2.6 | 15.5 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 2.5 | 10.2 | GO:0009441 | glycolate metabolic process(GO:0009441) |
| 2.5 | 7.5 | GO:0043605 | cellular amide catabolic process(GO:0043605) |
| 2.4 | 12.1 | GO:0015801 | aromatic amino acid transport(GO:0015801) |
| 2.4 | 7.2 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 2.4 | 12.0 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 2.4 | 9.6 | GO:0034240 | negative regulation of macrophage fusion(GO:0034240) |
| 2.3 | 4.6 | GO:2000259 | positive regulation of complement activation(GO:0045917) positive regulation of protein activation cascade(GO:2000259) |
| 2.3 | 9.0 | GO:0016332 | establishment or maintenance of polarity of embryonic epithelium(GO:0016332) |
| 2.2 | 9.0 | GO:1990375 | baculum development(GO:1990375) |
| 2.1 | 4.2 | GO:0071298 | cellular response to L-ascorbic acid(GO:0071298) |
| 2.1 | 6.3 | GO:0002554 | serotonin secretion by platelet(GO:0002554) |
| 2.1 | 2.1 | GO:0003130 | BMP signaling pathway involved in heart induction(GO:0003130) endodermal-mesodermal cell signaling(GO:0003133) endodermal-mesodermal cell signaling involved in heart induction(GO:0003134) |
| 2.1 | 28.9 | GO:0006705 | mineralocorticoid biosynthetic process(GO:0006705) |
| 2.0 | 6.1 | GO:0070904 | L-ascorbic acid transport(GO:0015882) transepithelial L-ascorbic acid transport(GO:0070904) |
| 2.0 | 6.0 | GO:0002462 | tolerance induction to nonself antigen(GO:0002462) |
| 2.0 | 9.9 | GO:1904970 | brush border assembly(GO:1904970) |
| 2.0 | 7.8 | GO:0060474 | positive regulation of sperm motility involved in capacitation(GO:0060474) |
| 1.9 | 27.3 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 1.9 | 5.8 | GO:1900739 | regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) protein localization to vacuolar membrane(GO:1903778) |
| 1.9 | 9.5 | GO:0002225 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antibacterial peptide production(GO:0002803) |
| 1.9 | 7.6 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 1.9 | 5.6 | GO:0007089 | traversing start control point of mitotic cell cycle(GO:0007089) |
| 1.9 | 5.6 | GO:1904139 | regulation of macrophage colony-stimulating factor signaling pathway(GO:1902226) regulation of response to macrophage colony-stimulating factor(GO:1903969) regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903972) microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 1.9 | 11.1 | GO:0038096 | immune response-regulating cell surface receptor signaling pathway involved in phagocytosis(GO:0002433) Fc-gamma receptor signaling pathway involved in phagocytosis(GO:0038096) |
| 1.8 | 10.9 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 1.8 | 5.4 | GO:0010037 | response to carbon dioxide(GO:0010037) |
| 1.8 | 9.0 | GO:0046110 | xanthine catabolic process(GO:0009115) xanthine metabolic process(GO:0046110) |
| 1.8 | 5.3 | GO:0042197 | chlorinated hydrocarbon metabolic process(GO:0042196) halogenated hydrocarbon metabolic process(GO:0042197) |
| 1.7 | 12.2 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 1.7 | 17.3 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 1.6 | 12.9 | GO:0071267 | amino acid salvage(GO:0043102) L-methionine salvage(GO:0071267) |
| 1.6 | 4.8 | GO:0030091 | protein repair(GO:0030091) |
| 1.6 | 4.8 | GO:0042509 | regulation of tyrosine phosphorylation of STAT protein(GO:0042509) positive regulation of tyrosine phosphorylation of STAT protein(GO:0042531) |
| 1.6 | 20.5 | GO:0006968 | cellular defense response(GO:0006968) |
| 1.6 | 3.2 | GO:0033087 | negative regulation of immature T cell proliferation(GO:0033087) |
| 1.6 | 12.5 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 1.6 | 7.8 | GO:0051790 | threonine metabolic process(GO:0006566) short-chain fatty acid biosynthetic process(GO:0051790) |
| 1.6 | 9.3 | GO:0097338 | response to clozapine(GO:0097338) |
| 1.5 | 7.6 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 1.5 | 15.2 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 1.5 | 4.6 | GO:0006742 | NADP catabolic process(GO:0006742) |
| 1.5 | 4.5 | GO:1901873 | regulation of post-translational protein modification(GO:1901873) |
| 1.5 | 4.5 | GO:1903923 | protein processing in phagocytic vesicle(GO:1900756) regulation of protein processing in phagocytic vesicle(GO:1903921) positive regulation of protein processing in phagocytic vesicle(GO:1903923) |
| 1.5 | 12.1 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) mammary duct terminal end bud growth(GO:0060763) |
| 1.5 | 7.5 | GO:0042758 | long-chain fatty acid catabolic process(GO:0042758) |
| 1.5 | 10.3 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 1.4 | 9.9 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 1.4 | 26.5 | GO:0043931 | ossification involved in bone maturation(GO:0043931) |
| 1.4 | 8.3 | GO:0046874 | quinolinate metabolic process(GO:0046874) |
| 1.4 | 4.1 | GO:0033512 | L-lysine catabolic process to acetyl-CoA via saccharopine(GO:0033512) |
| 1.4 | 6.8 | GO:0050917 | sensory perception of umami taste(GO:0050917) |
| 1.3 | 17.2 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 1.3 | 13.0 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 1.3 | 15.3 | GO:0070857 | regulation of bile acid biosynthetic process(GO:0070857) |
| 1.3 | 7.6 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 1.3 | 5.0 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 1.2 | 8.7 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 1.2 | 5.0 | GO:0050689 | negative regulation of defense response to virus by host(GO:0050689) |
| 1.2 | 3.7 | GO:0061206 | mesonephros morphogenesis(GO:0061206) mesonephric nephron development(GO:0061215) mesonephric nephron morphogenesis(GO:0061228) mesenchymal stem cell maintenance involved in mesonephric nephron morphogenesis(GO:0061235) regulation of mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:0061295) negative regulation of mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:0061296) mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:1901146) |
| 1.2 | 4.9 | GO:0060155 | apolipoprotein A-I-mediated signaling pathway(GO:0038027) platelet dense granule organization(GO:0060155) |
| 1.2 | 4.7 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 1.2 | 4.7 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 1.2 | 3.5 | GO:1990790 | response to glial cell derived neurotrophic factor(GO:1990790) cellular response to glial cell derived neurotrophic factor(GO:1990792) |
| 1.2 | 6.9 | GO:0035701 | hematopoietic stem cell migration(GO:0035701) |
| 1.1 | 9.2 | GO:0043031 | negative regulation of macrophage activation(GO:0043031) |
| 1.1 | 11.4 | GO:0032831 | regulation of CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0032829) positive regulation of CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0032831) |
| 1.1 | 4.5 | GO:0038158 | granulocyte colony-stimulating factor signaling pathway(GO:0038158) |
| 1.1 | 14.6 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 1.1 | 21.1 | GO:1990845 | adaptive thermogenesis(GO:1990845) |
| 1.1 | 3.3 | GO:0045065 | cytotoxic T cell differentiation(GO:0045065) |
| 1.1 | 3.3 | GO:0090158 | endoplasmic reticulum membrane organization(GO:0090158) positive regulation of ER to Golgi vesicle-mediated transport(GO:1902953) |
| 1.1 | 8.7 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 1.1 | 8.6 | GO:0002439 | chronic inflammatory response to antigenic stimulus(GO:0002439) |
| 1.1 | 7.5 | GO:0060480 | lung goblet cell differentiation(GO:0060480) |
| 1.0 | 3.1 | GO:0045085 | negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 1.0 | 7.3 | GO:0061368 | behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 1.0 | 14.6 | GO:0043619 | regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0043619) |
| 1.0 | 12.5 | GO:0014883 | transition between fast and slow fiber(GO:0014883) |
| 1.0 | 17.5 | GO:0048484 | enteric nervous system development(GO:0048484) |
| 1.0 | 2.0 | GO:2000669 | negative regulation of dendritic cell apoptotic process(GO:2000669) |
| 1.0 | 7.1 | GO:0007060 | male meiosis chromosome segregation(GO:0007060) |
| 1.0 | 4.0 | GO:0030449 | regulation of complement activation(GO:0030449) |
| 1.0 | 4.0 | GO:0021730 | trigeminal sensory nucleus development(GO:0021730) principal sensory nucleus of trigeminal nerve development(GO:0021740) |
| 1.0 | 3.0 | GO:0070268 | cuticle development(GO:0042335) cornification(GO:0070268) |
| 1.0 | 3.9 | GO:1990164 | histone H2A phosphorylation(GO:1990164) |
| 1.0 | 5.8 | GO:1990928 | response to amino acid starvation(GO:1990928) |
| 1.0 | 4.8 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
| 1.0 | 18.1 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 1.0 | 1.9 | GO:0090081 | regulation of heart induction by regulation of canonical Wnt signaling pathway(GO:0090081) |
| 0.9 | 8.5 | GO:0033572 | transferrin transport(GO:0033572) |
| 0.9 | 3.8 | GO:0070537 | histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.9 | 8.3 | GO:0038171 | cannabinoid signaling pathway(GO:0038171) |
| 0.9 | 3.7 | GO:0036118 | hyaluranon cable assembly(GO:0036118) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 0.9 | 4.6 | GO:0042590 | antigen processing and presentation of exogenous peptide antigen via MHC class I(GO:0042590) |
| 0.9 | 1.8 | GO:0002434 | immune complex clearance(GO:0002434) |
| 0.9 | 8.0 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.9 | 6.2 | GO:1901299 | negative regulation of hydrogen peroxide-mediated programmed cell death(GO:1901299) |
| 0.9 | 10.3 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.8 | 18.1 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.8 | 12.3 | GO:0042407 | cristae formation(GO:0042407) |
| 0.8 | 9.8 | GO:0030238 | male sex determination(GO:0030238) |
| 0.8 | 4.8 | GO:0060721 | regulation of spongiotrophoblast cell proliferation(GO:0060721) regulation of cell proliferation involved in embryonic placenta development(GO:0060723) |
| 0.8 | 2.3 | GO:1904100 | regulation of protein O-linked glycosylation(GO:1904098) positive regulation of protein O-linked glycosylation(GO:1904100) |
| 0.8 | 2.3 | GO:0002277 | myeloid dendritic cell activation involved in immune response(GO:0002277) |
| 0.7 | 2.2 | GO:0001698 | gastrin-induced gastric acid secretion(GO:0001698) |
| 0.7 | 2.2 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.7 | 2.2 | GO:0042732 | D-xylose metabolic process(GO:0042732) |
| 0.7 | 4.3 | GO:0046490 | isopentenyl diphosphate biosynthetic process(GO:0009240) isopentenyl diphosphate metabolic process(GO:0046490) |
| 0.7 | 4.3 | GO:0006868 | glutamine transport(GO:0006868) |
| 0.7 | 5.0 | GO:0060631 | meiotic DNA double-strand break formation(GO:0042138) regulation of meiosis I(GO:0060631) |
| 0.7 | 6.8 | GO:0006086 | acetyl-CoA biosynthetic process from pyruvate(GO:0006086) |
| 0.7 | 6.1 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.7 | 2.7 | GO:0002084 | protein depalmitoylation(GO:0002084) |
| 0.7 | 2.0 | GO:0006288 | base-excision repair, DNA ligation(GO:0006288) |
| 0.6 | 3.2 | GO:0036233 | glycine import(GO:0036233) |
| 0.6 | 5.8 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.6 | 5.7 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.6 | 6.3 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.6 | 1.9 | GO:1990743 | protein sialylation(GO:1990743) |
| 0.6 | 6.8 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.6 | 1.9 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.6 | 3.1 | GO:0038094 | Fc-gamma receptor signaling pathway(GO:0038094) |
| 0.6 | 1.8 | GO:0097052 | L-kynurenine metabolic process(GO:0097052) |
| 0.6 | 1.8 | GO:1902568 | regulation of eosinophil degranulation(GO:0043309) positive regulation of eosinophil degranulation(GO:0043311) positive regulation of eosinophil activation(GO:1902568) |
| 0.6 | 3.6 | GO:1901843 | membrane depolarization during bundle of His cell action potential(GO:0086048) positive regulation of high voltage-gated calcium channel activity(GO:1901843) |
| 0.6 | 8.9 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.6 | 3.5 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.6 | 13.4 | GO:0008206 | bile acid metabolic process(GO:0008206) |
| 0.6 | 7.6 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.6 | 2.9 | GO:0075525 | viral translational termination-reinitiation(GO:0075525) |
| 0.6 | 2.3 | GO:0060178 | regulation of exocyst localization(GO:0060178) |
| 0.6 | 4.5 | GO:0035646 | endosome to melanosome transport(GO:0035646) pigment accumulation(GO:0043476) cellular pigment accumulation(GO:0043482) endosome to pigment granule transport(GO:0043485) pigment granule maturation(GO:0048757) regulation of rubidium ion transport(GO:2000680) |
| 0.6 | 1.7 | GO:1903847 | regulation of aorta morphogenesis(GO:1903847) positive regulation of aorta morphogenesis(GO:1903849) |
| 0.6 | 2.2 | GO:0002540 | leukotriene production involved in inflammatory response(GO:0002540) |
| 0.6 | 3.4 | GO:0070836 | caveola assembly(GO:0070836) |
| 0.5 | 1.6 | GO:1901227 | negative regulation of transcription from RNA polymerase II promoter involved in heart development(GO:1901227) |
| 0.5 | 7.5 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.5 | 7.4 | GO:0090266 | regulation of spindle checkpoint(GO:0090231) regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.5 | 5.8 | GO:0006777 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) |
| 0.5 | 3.1 | GO:0010528 | regulation of transposition(GO:0010528) negative regulation of transposition(GO:0010529) DNA hypermethylation(GO:0044026) negative regulation of DNA-templated transcription, initiation(GO:2000143) |
| 0.5 | 14.0 | GO:0019835 | cytolysis(GO:0019835) |
| 0.5 | 8.2 | GO:0006544 | glycine metabolic process(GO:0006544) |
| 0.5 | 2.0 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.5 | 6.0 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.5 | 1.0 | GO:2000041 | regulation of planar cell polarity pathway involved in axis elongation(GO:2000040) negative regulation of planar cell polarity pathway involved in axis elongation(GO:2000041) |
| 0.5 | 15.6 | GO:0030261 | chromosome condensation(GO:0030261) |
| 0.5 | 7.3 | GO:0010838 | positive regulation of keratinocyte proliferation(GO:0010838) |
| 0.5 | 10.2 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.5 | 1.9 | GO:0010832 | negative regulation of myotube differentiation(GO:0010832) |
| 0.5 | 13.4 | GO:0006779 | porphyrin-containing compound biosynthetic process(GO:0006779) |
| 0.5 | 1.4 | GO:0018364 | peptidyl-glutamine methylation(GO:0018364) |
| 0.5 | 3.3 | GO:0016560 | protein import into peroxisome matrix, docking(GO:0016560) |
| 0.5 | 2.4 | GO:0071955 | recycling endosome to Golgi transport(GO:0071955) |
| 0.5 | 2.8 | GO:0015705 | iodide transport(GO:0015705) |
| 0.5 | 3.2 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.5 | 3.2 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.5 | 5.0 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.5 | 3.6 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.5 | 1.4 | GO:0030222 | eosinophil differentiation(GO:0030222) |
| 0.4 | 17.5 | GO:0045214 | sarcomere organization(GO:0045214) |
| 0.4 | 5.3 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.4 | 17.8 | GO:0050853 | B cell receptor signaling pathway(GO:0050853) |
| 0.4 | 4.7 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.4 | 2.1 | GO:0051454 | pH elevation(GO:0045852) intracellular pH elevation(GO:0051454) |
| 0.4 | 2.1 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.4 | 3.4 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 0.4 | 10.7 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.4 | 20.6 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.4 | 5.3 | GO:0050961 | detection of temperature stimulus involved in sensory perception(GO:0050961) detection of temperature stimulus involved in sensory perception of pain(GO:0050965) |
| 0.4 | 2.4 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.4 | 3.2 | GO:0060346 | bone trabecula formation(GO:0060346) |
| 0.4 | 8.3 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.4 | 2.0 | GO:0060620 | regulation of cholesterol import(GO:0060620) regulation of sterol import(GO:2000909) |
| 0.4 | 2.3 | GO:0033629 | negative regulation of cell adhesion mediated by integrin(GO:0033629) |
| 0.4 | 1.2 | GO:0045041 | protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.4 | 3.1 | GO:0043922 | negative regulation by host of viral transcription(GO:0043922) |
| 0.4 | 2.7 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.4 | 4.5 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.4 | 4.5 | GO:0035745 | CD4-positive, alpha-beta T cell cytokine production(GO:0035743) T-helper 2 cell cytokine production(GO:0035745) |
| 0.4 | 3.7 | GO:0035900 | response to isolation stress(GO:0035900) |
| 0.4 | 2.6 | GO:0010041 | response to iron(III) ion(GO:0010041) |
| 0.4 | 4.0 | GO:0048642 | negative regulation of skeletal muscle tissue development(GO:0048642) |
| 0.4 | 6.9 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.4 | 1.1 | GO:0015680 | intracellular copper ion transport(GO:0015680) |
| 0.4 | 7.5 | GO:0060314 | regulation of ryanodine-sensitive calcium-release channel activity(GO:0060314) |
| 0.3 | 7.7 | GO:0002675 | positive regulation of acute inflammatory response(GO:0002675) |
| 0.3 | 4.5 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.3 | 7.1 | GO:0042026 | protein refolding(GO:0042026) |
| 0.3 | 11.2 | GO:0006739 | NADP metabolic process(GO:0006739) |
| 0.3 | 10.4 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.3 | 13.1 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.3 | 2.0 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.3 | 9.2 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.3 | 6.2 | GO:0097435 | fibril organization(GO:0097435) |
| 0.3 | 1.0 | GO:2001268 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001268) |
| 0.3 | 4.2 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.3 | 3.8 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.3 | 6.3 | GO:0046337 | phosphatidylethanolamine metabolic process(GO:0046337) |
| 0.3 | 0.9 | GO:0072076 | nephrogenic mesenchyme development(GO:0072076) |
| 0.3 | 1.2 | GO:0019918 | peptidyl-arginine methylation, to symmetrical-dimethyl arginine(GO:0019918) |
| 0.3 | 2.5 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.3 | 1.2 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.3 | 1.8 | GO:0033601 | positive regulation of mammary gland epithelial cell proliferation(GO:0033601) |
| 0.3 | 1.8 | GO:0090367 | negative regulation of mRNA modification(GO:0090367) |
| 0.3 | 2.1 | GO:1901727 | positive regulation of histone deacetylase activity(GO:1901727) |
| 0.3 | 8.4 | GO:0036075 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.3 | 2.4 | GO:0044598 | polyketide metabolic process(GO:0030638) daunorubicin metabolic process(GO:0044597) doxorubicin metabolic process(GO:0044598) |
| 0.3 | 6.3 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.3 | 4.5 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.3 | 6.4 | GO:1903846 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.3 | 1.1 | GO:0071718 | sodium-independent icosanoid transport(GO:0071718) |
| 0.3 | 0.9 | GO:0097503 | sialylation(GO:0097503) |
| 0.3 | 0.8 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.3 | 6.7 | GO:0034508 | centromere complex assembly(GO:0034508) |
| 0.3 | 2.1 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.3 | 13.2 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.3 | 4.4 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.3 | 4.2 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.3 | 4.7 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.3 | 3.1 | GO:0007028 | cytoplasm organization(GO:0007028) |
| 0.3 | 3.8 | GO:0072673 | lamellipodium morphogenesis(GO:0072673) |
| 0.3 | 1.5 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.3 | 3.8 | GO:0010269 | response to selenium ion(GO:0010269) |
| 0.3 | 1.0 | GO:0015811 | L-cystine transport(GO:0015811) |
| 0.2 | 6.6 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.2 | 3.6 | GO:0045745 | positive regulation of G-protein coupled receptor protein signaling pathway(GO:0045745) |
| 0.2 | 2.8 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.2 | 0.7 | GO:0032789 | saturated monocarboxylic acid metabolic process(GO:0032788) unsaturated monocarboxylic acid metabolic process(GO:0032789) |
| 0.2 | 1.4 | GO:2000823 | regulation of androgen receptor activity(GO:2000823) |
| 0.2 | 4.1 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.2 | 3.8 | GO:0010447 | response to acidic pH(GO:0010447) |
| 0.2 | 3.5 | GO:0002230 | positive regulation of defense response to virus by host(GO:0002230) |
| 0.2 | 2.1 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.2 | 7.4 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.2 | 1.0 | GO:0022615 | protein to membrane docking(GO:0022615) |
| 0.2 | 1.8 | GO:0090435 | protein localization to nuclear envelope(GO:0090435) |
| 0.2 | 5.8 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.2 | 7.4 | GO:0033627 | cell adhesion mediated by integrin(GO:0033627) |
| 0.2 | 0.8 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.2 | 4.6 | GO:0060046 | regulation of acrosome reaction(GO:0060046) |
| 0.2 | 1.9 | GO:0030643 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.2 | 0.7 | GO:0061370 | testosterone biosynthetic process(GO:0061370) |
| 0.2 | 8.3 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.2 | 5.9 | GO:0050892 | intestinal absorption(GO:0050892) |
| 0.2 | 1.3 | GO:0042492 | gamma-delta T cell differentiation(GO:0042492) |
| 0.2 | 2.2 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.2 | 7.4 | GO:0035383 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.2 | 1.4 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.2 | 1.4 | GO:0032025 | response to cobalt ion(GO:0032025) |
| 0.2 | 2.4 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.2 | 5.1 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.2 | 3.1 | GO:0030225 | macrophage differentiation(GO:0030225) |
| 0.2 | 2.9 | GO:0009414 | response to water deprivation(GO:0009414) |
| 0.2 | 0.7 | GO:1900118 | negative regulation of execution phase of apoptosis(GO:1900118) |
| 0.2 | 0.5 | GO:0090666 | scaRNA localization to Cajal body(GO:0090666) |
| 0.2 | 2.3 | GO:0071173 | mitotic spindle assembly checkpoint(GO:0007094) spindle checkpoint(GO:0031577) spindle assembly checkpoint(GO:0071173) mitotic spindle checkpoint(GO:0071174) |
| 0.2 | 7.3 | GO:1902041 | regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902041) |
| 0.2 | 3.2 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.2 | 1.7 | GO:1904707 | positive regulation of vascular smooth muscle cell proliferation(GO:1904707) |
| 0.2 | 6.4 | GO:0007140 | male meiosis(GO:0007140) |
| 0.2 | 3.7 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.2 | 2.5 | GO:0042832 | defense response to protozoan(GO:0042832) |
| 0.2 | 1.8 | GO:0051281 | positive regulation of release of sequestered calcium ion into cytosol(GO:0051281) |
| 0.2 | 1.2 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.2 | 8.3 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.2 | 2.9 | GO:0006458 | 'de novo' protein folding(GO:0006458) |
| 0.1 | 3.3 | GO:0030500 | regulation of bone mineralization(GO:0030500) |
| 0.1 | 3.8 | GO:0042775 | mitochondrial ATP synthesis coupled electron transport(GO:0042775) |
| 0.1 | 0.7 | GO:1903909 | regulation of receptor clustering(GO:1903909) regulation of AMPA glutamate receptor clustering(GO:1904717) |
| 0.1 | 0.9 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.1 | 1.0 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) glycerophospholipid catabolic process(GO:0046475) |
| 0.1 | 2.0 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.1 | 2.9 | GO:0090200 | positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.1 | 59.1 | GO:0007283 | spermatogenesis(GO:0007283) |
| 0.1 | 0.7 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.1 | 0.5 | GO:1903350 | response to dopamine(GO:1903350) cellular response to dopamine(GO:1903351) |
| 0.1 | 4.0 | GO:0051341 | regulation of oxidoreductase activity(GO:0051341) |
| 0.1 | 1.1 | GO:0072189 | ureter development(GO:0072189) |
| 0.1 | 7.7 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.1 | 2.8 | GO:0050891 | multicellular organismal water homeostasis(GO:0050891) |
| 0.1 | 2.0 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.1 | 1.0 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.1 | 0.7 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 0.1 | 1.6 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.1 | 1.0 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 0.1 | 2.7 | GO:0043403 | skeletal muscle tissue regeneration(GO:0043403) |
| 0.1 | 7.3 | GO:0045667 | regulation of osteoblast differentiation(GO:0045667) |
| 0.1 | 0.9 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.1 | 1.3 | GO:0070286 | axonemal dynein complex assembly(GO:0070286) |
| 0.1 | 0.4 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.1 | 0.3 | GO:0036451 | cap mRNA methylation(GO:0036451) |
| 0.1 | 0.9 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 0.1 | 2.0 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.1 | 3.3 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.1 | 1.7 | GO:0071353 | cellular response to interleukin-4(GO:0071353) |
| 0.1 | 0.7 | GO:0045602 | negative regulation of endothelial cell differentiation(GO:0045602) |
| 0.1 | 1.6 | GO:0032438 | melanosome organization(GO:0032438) pigment granule organization(GO:0048753) |
| 0.1 | 3.3 | GO:0035904 | aorta development(GO:0035904) |
| 0.1 | 6.5 | GO:0010952 | positive regulation of peptidase activity(GO:0010952) |
| 0.1 | 2.9 | GO:0034113 | heterotypic cell-cell adhesion(GO:0034113) |
| 0.1 | 0.7 | GO:0071498 | cellular response to fluid shear stress(GO:0071498) |
| 0.1 | 2.4 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.1 | 3.8 | GO:0007338 | single fertilization(GO:0007338) |
| 0.1 | 4.2 | GO:0009311 | oligosaccharide metabolic process(GO:0009311) |
| 0.1 | 3.2 | GO:0032680 | regulation of tumor necrosis factor production(GO:0032680) regulation of tumor necrosis factor superfamily cytokine production(GO:1903555) |
| 0.1 | 1.4 | GO:0051898 | negative regulation of protein kinase B signaling(GO:0051898) |
| 0.1 | 2.1 | GO:1901998 | toxin transport(GO:1901998) |
| 0.1 | 7.5 | GO:0042742 | defense response to bacterium(GO:0042742) |
| 0.1 | 0.1 | GO:0016557 | peroxisome membrane biogenesis(GO:0016557) |
| 0.1 | 1.8 | GO:0097421 | liver regeneration(GO:0097421) |
| 0.1 | 10.1 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.1 | 2.3 | GO:0006672 | ceramide metabolic process(GO:0006672) |
| 0.0 | 0.9 | GO:0006379 | mRNA cleavage(GO:0006379) |
| 0.0 | 1.4 | GO:0070534 | protein K63-linked ubiquitination(GO:0070534) |
| 0.0 | 1.7 | GO:0043507 | positive regulation of JUN kinase activity(GO:0043507) |
| 0.0 | 1.8 | GO:0050715 | positive regulation of cytokine secretion(GO:0050715) |
| 0.0 | 0.3 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.0 | 1.2 | GO:0006809 | nitric oxide biosynthetic process(GO:0006809) |
| 0.0 | 0.3 | GO:2001241 | positive regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001241) |
| 0.0 | 1.9 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 0.3 | GO:2000059 | negative regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000059) |
| 0.0 | 1.9 | GO:0051092 | positive regulation of NF-kappaB transcription factor activity(GO:0051092) |
| 0.0 | 1.0 | GO:0002011 | morphogenesis of an epithelial sheet(GO:0002011) |
| 0.0 | 0.5 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.0 | 0.1 | GO:0008594 | photoreceptor cell morphogenesis(GO:0008594) |
| 0.0 | 2.3 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 2.4 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
| 0.0 | 0.1 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.0 | 2.6 | GO:0001890 | placenta development(GO:0001890) |
| 0.0 | 0.5 | GO:0030728 | ovulation(GO:0030728) |
| 0.0 | 1.2 | GO:0014065 | phosphatidylinositol 3-kinase signaling(GO:0014065) |
| 0.0 | 0.7 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 14.5 | GO:0055114 | oxidation-reduction process(GO:0055114) |
| 0.0 | 0.2 | GO:0071157 | negative regulation of cell cycle arrest(GO:0071157) |
| 0.0 | 0.6 | GO:0006446 | regulation of translational initiation(GO:0006446) |
| 0.0 | 0.6 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 0.2 | GO:1904948 | midbrain dopaminergic neuron differentiation(GO:1904948) |
| 0.0 | 20.3 | GO:0007608 | sensory perception of smell(GO:0007608) |
| 0.0 | 0.1 | GO:0035196 | dsRNA fragmentation(GO:0031050) production of miRNAs involved in gene silencing by miRNA(GO:0035196) production of small RNA involved in gene silencing by RNA(GO:0070918) |
| 0.0 | 0.3 | GO:0042130 | negative regulation of T cell proliferation(GO:0042130) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.9 | 23.7 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 5.4 | 21.7 | GO:0061474 | phagolysosome membrane(GO:0061474) |
| 4.6 | 13.8 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 4.1 | 12.2 | GO:0046691 | intracellular canaliculus(GO:0046691) |
| 3.6 | 21.6 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 2.9 | 26.5 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 2.3 | 28.1 | GO:0005861 | troponin complex(GO:0005861) |
| 2.1 | 21.1 | GO:0031105 | septin complex(GO:0031105) |
| 2.0 | 44.1 | GO:0044217 | other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) |
| 1.7 | 29.4 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 1.7 | 5.0 | GO:0005745 | m-AAA complex(GO:0005745) |
| 1.5 | 20.6 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 1.5 | 14.6 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 1.3 | 15.5 | GO:0034385 | very-low-density lipoprotein particle(GO:0034361) triglyceride-rich lipoprotein particle(GO:0034385) |
| 1.2 | 8.6 | GO:0060205 | cytoplasmic membrane-bounded vesicle lumen(GO:0060205) |
| 1.2 | 3.7 | GO:0036117 | hyaluranon cable(GO:0036117) |
| 1.2 | 9.6 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 1.1 | 4.5 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 1.1 | 4.5 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 1.1 | 139.9 | GO:0072562 | blood microparticle(GO:0072562) |
| 1.1 | 7.6 | GO:0061617 | MICOS complex(GO:0061617) |
| 1.1 | 13.0 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 1.1 | 9.6 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 1.1 | 7.4 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 1.1 | 6.3 | GO:0030313 | cell outer membrane(GO:0009279) cell envelope(GO:0030313) external encapsulating structure part(GO:0044462) |
| 1.0 | 9.3 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 1.0 | 10.3 | GO:0042587 | glycogen granule(GO:0042587) |
| 1.0 | 8.6 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.9 | 14.9 | GO:0045180 | basal cortex(GO:0045180) |
| 0.9 | 5.4 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.9 | 9.0 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.9 | 3.6 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.9 | 2.6 | GO:0005962 | mitochondrial isocitrate dehydrogenase complex (NAD+)(GO:0005962) isocitrate dehydrogenase complex (NAD+)(GO:0045242) |
| 0.9 | 16.2 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.8 | 4.2 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.8 | 7.6 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.8 | 4.1 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.8 | 3.2 | GO:1990005 | granular vesicle(GO:1990005) |
| 0.8 | 6.8 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.8 | 3.8 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.7 | 2.0 | GO:0070421 | DNA ligase III-XRCC1 complex(GO:0070421) |
| 0.6 | 6.8 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) |
| 0.6 | 2.4 | GO:0035363 | histone locus body(GO:0035363) |
| 0.6 | 5.9 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.6 | 4.6 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.6 | 8.0 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.6 | 7.3 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.5 | 16.7 | GO:0005921 | gap junction(GO:0005921) |
| 0.5 | 3.6 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.5 | 2.0 | GO:0002947 | tumor necrosis factor receptor superfamily complex(GO:0002947) |
| 0.5 | 4.5 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.5 | 26.3 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.5 | 8.2 | GO:0033202 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.5 | 64.7 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.5 | 1.8 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.5 | 1.4 | GO:1990590 | ATF1-ATF4 transcription factor complex(GO:1990590) |
| 0.5 | 3.6 | GO:0000796 | condensin complex(GO:0000796) |
| 0.5 | 13.2 | GO:0005753 | mitochondrial proton-transporting ATP synthase complex(GO:0005753) |
| 0.5 | 3.2 | GO:0016942 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
| 0.5 | 2.7 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.4 | 7.0 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.4 | 3.8 | GO:0002177 | manchette(GO:0002177) |
| 0.4 | 3.8 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.4 | 4.9 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.4 | 3.3 | GO:0000243 | commitment complex(GO:0000243) |
| 0.4 | 11.1 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.4 | 4.0 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.4 | 1.2 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.4 | 3.8 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.4 | 7.1 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.4 | 1.9 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.4 | 4.4 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.4 | 26.5 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.3 | 6.3 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.3 | 12.5 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.3 | 1.7 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.3 | 24.8 | GO:0005811 | lipid particle(GO:0005811) |
| 0.3 | 1.0 | GO:0070820 | tertiary granule(GO:0070820) |
| 0.3 | 1.7 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.3 | 48.1 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.3 | 6.8 | GO:0008305 | integrin complex(GO:0008305) |
| 0.3 | 3.9 | GO:0000801 | central element(GO:0000801) |
| 0.3 | 3.6 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.3 | 1.8 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.3 | 4.7 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.3 | 2.0 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.3 | 7.6 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.3 | 2.0 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.3 | 25.8 | GO:0005604 | basement membrane(GO:0005604) |
| 0.3 | 1.8 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.3 | 1.8 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.3 | 0.8 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.3 | 32.2 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.2 | 11.9 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.2 | 6.7 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.2 | 5.8 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.2 | 1.4 | GO:0001651 | dense fibrillar component(GO:0001651) |
| 0.2 | 2.9 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.2 | 8.5 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.2 | 4.2 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.2 | 12.1 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.2 | 5.6 | GO:0031306 | intrinsic component of mitochondrial outer membrane(GO:0031306) |
| 0.2 | 7.8 | GO:0016235 | aggresome(GO:0016235) |
| 0.2 | 1.1 | GO:0070449 | elongin complex(GO:0070449) |
| 0.2 | 1.9 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.2 | 3.5 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.2 | 16.7 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.2 | 5.0 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.2 | 16.6 | GO:0016605 | PML body(GO:0016605) |
| 0.2 | 2.6 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.2 | 39.6 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.2 | 10.4 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.2 | 2.0 | GO:0030057 | desmosome(GO:0030057) |
| 0.2 | 8.2 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.2 | 38.0 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.1 | 8.2 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.1 | 3.4 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.1 | 1.3 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.1 | 21.7 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.1 | 5.4 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.1 | 6.9 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.1 | 4.9 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.1 | 2.8 | GO:0034706 | sodium channel complex(GO:0034706) |
| 0.1 | 0.5 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.1 | 7.2 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.1 | 1.4 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.1 | 6.6 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.1 | 3.3 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.1 | 3.8 | GO:0015030 | Cajal body(GO:0015030) |
| 0.1 | 0.8 | GO:0042555 | MCM complex(GO:0042555) |
| 0.1 | 3.8 | GO:0030141 | secretory granule(GO:0030141) |
| 0.1 | 3.6 | GO:0000776 | kinetochore(GO:0000776) |
| 0.1 | 13.6 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.1 | 82.2 | GO:0005615 | extracellular space(GO:0005615) |
| 0.1 | 13.6 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.1 | 5.2 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.1 | 0.3 | GO:0098576 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) lumenal side of membrane(GO:0098576) |
| 0.1 | 7.0 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.1 | 3.5 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.1 | 3.3 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 1.8 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.1 | 15.1 | GO:0045177 | apical part of cell(GO:0045177) |
| 0.0 | 2.8 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 4.7 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 2.5 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.4 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 0.2 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.0 | 36.6 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.0 | 0.6 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 1.0 | GO:0005840 | ribosome(GO:0005840) |
| 0.0 | 0.9 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.3 | GO:0001533 | cornified envelope(GO:0001533) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.3 | 21.9 | GO:0032810 | sterol response element binding(GO:0032810) |
| 5.3 | 21.1 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 5.2 | 15.7 | GO:0030337 | DNA polymerase processivity factor activity(GO:0030337) |
| 4.8 | 19.4 | GO:0016019 | peptidoglycan receptor activity(GO:0016019) |
| 4.6 | 13.7 | GO:0046573 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 4.4 | 13.2 | GO:0004515 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 4.1 | 12.2 | GO:0001716 | L-amino-acid oxidase activity(GO:0001716) |
| 4.0 | 12.1 | GO:0031783 | corticotropin hormone receptor binding(GO:0031780) type 5 melanocortin receptor binding(GO:0031783) |
| 3.6 | 14.6 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 3.6 | 10.8 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 3.4 | 20.2 | GO:0008865 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 3.3 | 19.5 | GO:0035473 | lipase binding(GO:0035473) |
| 3.2 | 12.8 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 3.1 | 12.5 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 3.0 | 12.0 | GO:0008670 | 2,4-dienoyl-CoA reductase (NADPH) activity(GO:0008670) |
| 3.0 | 26.9 | GO:0015232 | heme transporter activity(GO:0015232) |
| 2.8 | 17.0 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 2.7 | 10.8 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 2.6 | 7.9 | GO:0051990 | (R)-2-hydroxyglutarate dehydrogenase activity(GO:0051990) |
| 2.6 | 7.8 | GO:0070905 | serine binding(GO:0070905) |
| 2.6 | 17.9 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 2.5 | 10.2 | GO:0003973 | (S)-2-hydroxy-acid oxidase activity(GO:0003973) |
| 2.3 | 18.1 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 2.2 | 15.6 | GO:0031014 | troponin T binding(GO:0031014) |
| 2.2 | 13.1 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 2.1 | 10.7 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 2.1 | 59.5 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 2.1 | 16.7 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 2.1 | 8.3 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 2.1 | 6.2 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 2.0 | 6.1 | GO:0070890 | L-ascorbate:sodium symporter activity(GO:0008520) L-ascorbic acid transporter activity(GO:0015229) sodium-dependent L-ascorbate transmembrane transporter activity(GO:0070890) |
| 1.9 | 11.4 | GO:0019976 | interleukin-2 binding(GO:0019976) |
| 1.9 | 9.5 | GO:0050785 | advanced glycation end-product receptor activity(GO:0050785) |
| 1.9 | 22.7 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 1.9 | 5.6 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 1.9 | 11.2 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 1.8 | 9.0 | GO:0031798 | type 1 metabotropic glutamate receptor binding(GO:0031798) |
| 1.8 | 9.0 | GO:0016726 | xanthine dehydrogenase activity(GO:0004854) oxidoreductase activity, acting on CH or CH2 groups, NAD or NADP as acceptor(GO:0016726) |
| 1.8 | 15.9 | GO:0008494 | translation activator activity(GO:0008494) |
| 1.7 | 8.7 | GO:0001155 | TFIIIA-class transcription factor binding(GO:0001155) |
| 1.7 | 6.9 | GO:0004452 | isopentenyl-diphosphate delta-isomerase activity(GO:0004452) |
| 1.6 | 13.0 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 1.6 | 14.3 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 1.6 | 12.6 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 1.6 | 4.7 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 1.5 | 13.9 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 1.5 | 4.6 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 1.5 | 15.2 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 1.5 | 12.1 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 1.5 | 4.5 | GO:0019797 | procollagen-proline 3-dioxygenase activity(GO:0019797) |
| 1.5 | 4.5 | GO:0051916 | granulocyte colony-stimulating factor binding(GO:0051916) |
| 1.5 | 19.6 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 1.5 | 4.5 | GO:0031762 | follicle-stimulating hormone receptor binding(GO:0031762) |
| 1.5 | 8.7 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 1.4 | 5.8 | GO:0008265 | Mo-molybdopterin cofactor sulfurase activity(GO:0008265) |
| 1.4 | 4.2 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 1.4 | 4.1 | GO:0031714 | C5a anaphylatoxin chemotactic receptor binding(GO:0031714) |
| 1.4 | 4.1 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 1.4 | 8.2 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 1.3 | 6.7 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 1.3 | 9.3 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 1.3 | 9.2 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 1.3 | 10.5 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 1.3 | 5.2 | GO:0004365 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 1.3 | 9.1 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 1.3 | 13.0 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 1.3 | 3.8 | GO:0004874 | aryl hydrocarbon receptor activity(GO:0004874) |
| 1.3 | 5.1 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 1.3 | 7.5 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 1.2 | 12.2 | GO:0046870 | cadmium ion binding(GO:0046870) |
| 1.2 | 4.8 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 1.2 | 14.4 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 1.2 | 12.0 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 1.2 | 3.6 | GO:0086057 | voltage-gated calcium channel activity involved in bundle of His cell action potential(GO:0086057) |
| 1.2 | 12.9 | GO:0001848 | complement binding(GO:0001848) |
| 1.2 | 8.1 | GO:0032052 | bile acid binding(GO:0032052) |
| 1.2 | 9.2 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 1.1 | 20.6 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 1.1 | 7.9 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 1.1 | 6.7 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 1.1 | 8.8 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 1.1 | 6.4 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 1.1 | 8.5 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 1.0 | 4.1 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 1.0 | 22.5 | GO:0019865 | immunoglobulin binding(GO:0019865) |
| 1.0 | 4.9 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 1.0 | 5.8 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.9 | 3.7 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.9 | 11.7 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.9 | 12.2 | GO:0015250 | water channel activity(GO:0015250) |
| 0.9 | 17.0 | GO:0005537 | mannose binding(GO:0005537) |
| 0.8 | 6.5 | GO:0003996 | acyl-CoA ligase activity(GO:0003996) fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.8 | 14.4 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.8 | 2.4 | GO:0047025 | 3-oxoacyl-[acyl-carrier-protein] reductase (NADH) activity(GO:0047025) |
| 0.8 | 3.2 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.8 | 4.6 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.8 | 3.8 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.7 | 2.2 | GO:0004051 | arachidonate 5-lipoxygenase activity(GO:0004051) |
| 0.7 | 11.1 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.7 | 7.3 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.7 | 2.2 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.7 | 2.1 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.7 | 2.1 | GO:0038100 | nodal binding(GO:0038100) |
| 0.7 | 4.9 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.7 | 2.7 | GO:0034603 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.7 | 3.3 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.7 | 2.0 | GO:0008481 | sphinganine kinase activity(GO:0008481) |
| 0.7 | 9.2 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.7 | 2.6 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.6 | 3.2 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.6 | 3.8 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.6 | 6.3 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.6 | 1.8 | GO:0070548 | L-glutamine aminotransferase activity(GO:0070548) |
| 0.6 | 1.8 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.6 | 14.4 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.6 | 14.8 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.6 | 2.3 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.6 | 20.4 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.6 | 3.4 | GO:0036374 | gamma-glutamyltransferase activity(GO:0003840) glutathione hydrolase activity(GO:0036374) |
| 0.6 | 3.3 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.5 | 8.7 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.5 | 2.7 | GO:0001010 | transcription factor activity, sequence-specific DNA binding transcription factor recruiting(GO:0001010) |
| 0.5 | 3.7 | GO:0016717 | oxidoreductase activity, acting on paired donors, with oxidation of a pair of donors resulting in the reduction of molecular oxygen to two molecules of water(GO:0016717) |
| 0.5 | 3.6 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.5 | 2.1 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.5 | 21.1 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.5 | 5.0 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.5 | 7.0 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.5 | 4.0 | GO:0008430 | selenium binding(GO:0008430) |
| 0.5 | 3.9 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.5 | 14.9 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.5 | 1.9 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.5 | 24.0 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.5 | 1.4 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.4 | 6.2 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.4 | 9.1 | GO:0016638 | oxidoreductase activity, acting on the CH-NH2 group of donors(GO:0016638) |
| 0.4 | 9.0 | GO:0015106 | bicarbonate transmembrane transporter activity(GO:0015106) |
| 0.4 | 11.2 | GO:0008198 | ferrous iron binding(GO:0008198) |
| 0.4 | 5.8 | GO:0042171 | lysophosphatidic acid acyltransferase activity(GO:0042171) |
| 0.4 | 9.8 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.4 | 3.3 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.4 | 13.4 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.4 | 3.2 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.4 | 5.1 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.4 | 8.5 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.4 | 6.1 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.4 | 0.7 | GO:0070401 | NADP+ binding(GO:0070401) |
| 0.4 | 2.6 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.4 | 3.3 | GO:1990446 | U1 snRNA binding(GO:0030619) U1 snRNP binding(GO:1990446) |
| 0.4 | 18.4 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.4 | 4.3 | GO:0015194 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.4 | 2.8 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.3 | 10.0 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.3 | 2.0 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.3 | 1.0 | GO:0031766 | galanin receptor binding(GO:0031763) type 2 galanin receptor binding(GO:0031765) type 3 galanin receptor binding(GO:0031766) |
| 0.3 | 2.8 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.3 | 6.3 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.3 | 1.2 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.3 | 60.5 | GO:0004866 | endopeptidase inhibitor activity(GO:0004866) endopeptidase regulator activity(GO:0061135) |
| 0.3 | 3.2 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.3 | 2.3 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.3 | 0.9 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.3 | 4.4 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.3 | 1.7 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.3 | 3.8 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.3 | 19.7 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.3 | 4.2 | GO:0015926 | glucosidase activity(GO:0015926) |
| 0.3 | 2.1 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.3 | 0.8 | GO:0000171 | ribonuclease MRP activity(GO:0000171) |
| 0.3 | 3.6 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.3 | 5.3 | GO:0019825 | oxygen binding(GO:0019825) |
| 0.3 | 52.6 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.2 | 11.2 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.2 | 4.6 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 0.2 | 28.4 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.2 | 4.7 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.2 | 2.4 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.2 | 6.3 | GO:0005501 | retinoid binding(GO:0005501) |
| 0.2 | 4.6 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.2 | 6.2 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.2 | 17.2 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.2 | 1.1 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 0.2 | 15.0 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.2 | 9.4 | GO:0003823 | antigen binding(GO:0003823) |
| 0.2 | 2.5 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.2 | 1.4 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.2 | 10.4 | GO:0001158 | enhancer sequence-specific DNA binding(GO:0001158) |
| 0.2 | 3.4 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.2 | 4.0 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.2 | 1.2 | GO:0035014 | phosphatidylinositol 3-kinase regulator activity(GO:0035014) 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.2 | 1.9 | GO:0050750 | co-receptor binding(GO:0039706) low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.2 | 1.9 | GO:0019808 | polyamine binding(GO:0019808) |
| 0.2 | 2.6 | GO:0004579 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.2 | 0.9 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.2 | 6.2 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.2 | 2.4 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.2 | 15.5 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.2 | 9.4 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.2 | 0.7 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.2 | 3.1 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.2 | 5.7 | GO:0016667 | oxidoreductase activity, acting on a sulfur group of donors(GO:0016667) |
| 0.2 | 1.7 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.2 | 4.0 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.1 | 10.8 | GO:0050660 | flavin adenine dinucleotide binding(GO:0050660) |
| 0.1 | 1.0 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.1 | 1.8 | GO:0019104 | DNA N-glycosylase activity(GO:0019104) |
| 0.1 | 2.2 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.1 | 0.7 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.1 | 0.8 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.1 | 15.2 | GO:0008201 | heparin binding(GO:0008201) |
| 0.1 | 9.8 | GO:0002020 | protease binding(GO:0002020) |
| 0.1 | 6.1 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.1 | 3.0 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 3.2 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.1 | 1.1 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.1 | 3.5 | GO:0016655 | oxidoreductase activity, acting on NAD(P)H, quinone or similar compound as acceptor(GO:0016655) |
| 0.1 | 2.8 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.1 | 2.8 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.1 | 8.2 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.1 | 15.4 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.1 | 1.1 | GO:0004955 | prostaglandin receptor activity(GO:0004955) |
| 0.1 | 0.7 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.1 | 0.3 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.1 | 3.3 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.1 | 10.8 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.1 | 3.3 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.1 | 5.1 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.1 | 2.4 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 1.2 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.1 | 1.8 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.1 | 19.7 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.1 | 0.9 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.1 | 1.7 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 1.3 | GO:0016651 | oxidoreductase activity, acting on NAD(P)H(GO:0016651) |
| 0.1 | 1.0 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.1 | 1.7 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 0.1 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.1 | 0.8 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 0.5 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.1 | 0.5 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.1 | 0.7 | GO:0098918 | structural constituent of postsynaptic specialization(GO:0098879) structural constituent of synapse(GO:0098918) structural constituent of postsynaptic density(GO:0098919) |
| 0.1 | 0.3 | GO:0004161 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.0 | 3.3 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.0 | 0.9 | GO:0008373 | sialyltransferase activity(GO:0008373) |
| 0.0 | 0.7 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.0 | 0.5 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.0 | 1.4 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 0.6 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 1.8 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 0.5 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.7 | GO:0098811 | transcriptional activator activity, RNA polymerase II transcription factor binding(GO:0001190) transcriptional repressor activity, RNA polymerase II activating transcription factor binding(GO:0098811) |
| 0.0 | 9.5 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.6 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 0.1 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.0 | 1.4 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.0 | 0.0 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.0 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.7 | GO:0001047 | core promoter binding(GO:0001047) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 76.1 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 1.2 | 17.5 | PID ALK2 PATHWAY | ALK2 signaling events |
| 1.0 | 17.5 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.8 | 3.2 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.7 | 29.5 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.7 | 9.5 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.7 | 14.1 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.7 | 41.6 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.6 | 31.0 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.6 | 11.7 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.6 | 20.6 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.5 | 14.1 | PID GMCSF PATHWAY | GMCSF-mediated signaling events |
| 0.5 | 17.7 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.5 | 10.6 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.5 | 13.0 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.5 | 24.2 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.5 | 1.4 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.5 | 80.2 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.4 | 8.7 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.4 | 9.5 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.3 | 0.7 | PID S1P S1P3 PATHWAY | S1P3 pathway |
| 0.3 | 9.2 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.3 | 12.8 | PID BMP PATHWAY | BMP receptor signaling |
| 0.3 | 6.9 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.2 | 4.1 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.2 | 53.0 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.2 | 10.4 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.2 | 5.9 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.2 | 14.1 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.2 | 10.3 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.2 | 7.5 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.2 | 5.6 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.2 | 5.8 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.2 | 7.3 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.2 | 9.8 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.2 | 3.7 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.2 | 2.8 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.2 | 6.7 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.2 | 7.1 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.2 | 8.0 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.2 | 4.5 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.1 | 1.8 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.1 | 3.8 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.1 | 3.2 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.1 | 3.1 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.1 | 3.0 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.1 | 1.4 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.1 | 1.2 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.1 | 1.4 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.1 | 2.0 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 1.7 | PID ATM PATHWAY | ATM pathway |
| 0.1 | 0.7 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.1 | 0.1 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.1 | 2.0 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 0.3 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.7 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 34.1 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 2.4 | 24.0 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 2.3 | 30.5 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 2.3 | 34.4 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 2.3 | 20.6 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 1.7 | 15.0 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 1.4 | 53.3 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 1.3 | 12.0 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 1.3 | 18.4 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 1.3 | 19.3 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 1.1 | 4.4 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 1.1 | 6.3 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 1.0 | 12.2 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 1.0 | 26.6 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.9 | 8.3 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.9 | 35.3 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.9 | 11.4 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.9 | 9.6 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.9 | 12.0 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.8 | 6.6 | REACTOME DEFENSINS | Genes involved in Defensins |
| 0.8 | 4.0 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.8 | 54.9 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.7 | 22.2 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.6 | 8.1 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.6 | 12.9 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.6 | 9.6 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.5 | 10.3 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.5 | 9.4 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.5 | 5.7 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.5 | 17.6 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.5 | 19.2 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.5 | 29.7 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.5 | 40.1 | REACTOME RESPIRATORY ELECTRON TRANSPORT ATP SYNTHESIS BY CHEMIOSMOTIC COUPLING AND HEAT PRODUCTION BY UNCOUPLING PROTEINS | Genes involved in Respiratory electron transport, ATP synthesis by chemiosmotic coupling, and heat production by uncoupling proteins. |
| 0.4 | 9.3 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.4 | 32.2 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.4 | 2.3 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.3 | 10.3 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.3 | 3.7 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.3 | 4.3 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.3 | 3.6 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.3 | 6.0 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.3 | 4.6 | REACTOME APC C CDH1 MEDIATED DEGRADATION OF CDC20 AND OTHER APC C CDH1 TARGETED PROTEINS IN LATE MITOSIS EARLY G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |
| 0.3 | 7.9 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.3 | 5.9 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.3 | 2.8 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.3 | 11.0 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.3 | 3.5 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.3 | 4.3 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.2 | 15.3 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.2 | 3.8 | REACTOME RESOLUTION OF AP SITES VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the single-nucleotide replacement pathway |
| 0.2 | 2.8 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.2 | 6.0 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.2 | 5.3 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.2 | 1.7 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.2 | 2.7 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.2 | 3.2 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.2 | 5.2 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.2 | 15.4 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.2 | 2.8 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.2 | 27.0 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.1 | 2.4 | REACTOME TAK1 ACTIVATES NFKB BY PHOSPHORYLATION AND ACTIVATION OF IKKS COMPLEX | Genes involved in TAK1 activates NFkB by phosphorylation and activation of IKKs complex |
| 0.1 | 2.1 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 2.6 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.1 | 4.3 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.1 | 4.5 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.1 | 4.1 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.1 | 2.1 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 6.6 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.1 | 3.3 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.1 | 2.7 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 2.2 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.1 | 4.4 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.1 | 0.8 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.1 | 1.2 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.1 | 2.0 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.1 | 0.7 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.1 | 2.3 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.1 | 6.9 | REACTOME 3 UTR MEDIATED TRANSLATIONAL REGULATION | Genes involved in 3' -UTR-mediated translational regulation |
| 0.1 | 2.0 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.5 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 1.1 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.0 | 1.2 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.0 | 0.5 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 1.2 | REACTOME TRAF6 MEDIATED INDUCTION OF NFKB AND MAP KINASES UPON TLR7 8 OR 9 ACTIVATION | Genes involved in TRAF6 mediated induction of NFkB and MAP kinases upon TLR7/8 or 9 activation |
| 0.0 | 0.6 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 0.8 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.3 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |