Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
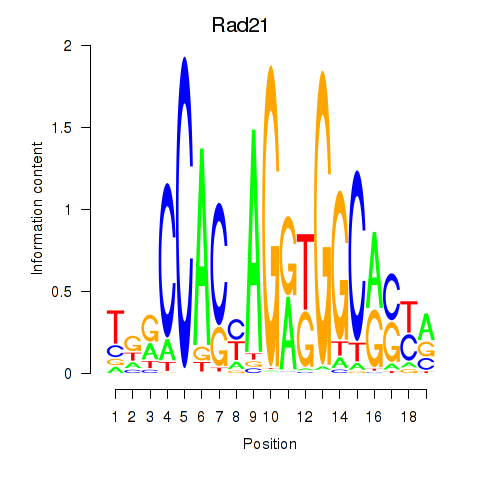
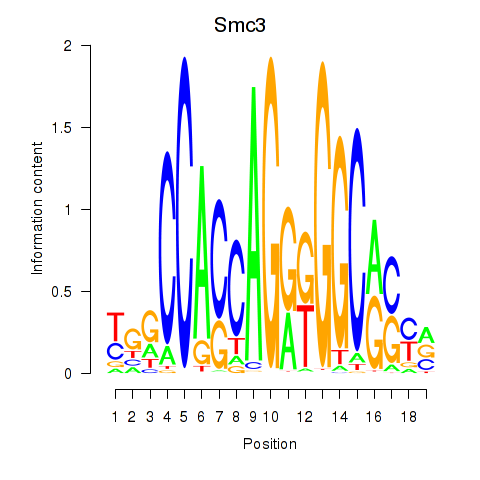
Results for Rad21_Smc3
Z-value: 1.83
Transcription factors associated with Rad21_Smc3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Rad21
|
ENSRNOG00000004420 | RAD21 cohesin complex component |
|
Smc3
|
ENSRNOG00000014173 | structural maintenance of chromosomes 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Rad21 | rn6_v1_chr7_-_91538673_91538673 | 0.29 | 7.7e-08 | Click! |
| Smc3 | rn6_v1_chr1_+_274309758_274309758 | 0.29 | 1.3e-07 | Click! |
Activity profile of Rad21_Smc3 motif
Sorted Z-values of Rad21_Smc3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_+_32244416 | 74.36 |
ENSRNOT00000012019
ENSRNOT00000060254 |
AABR07017902.1
|
|
| chr6_-_138662365 | 70.04 |
ENSRNOT00000066209
ENSRNOT00000084892 |
Ighm
|
immunoglobulin heavy constant mu |
| chr1_+_82480195 | 68.99 |
ENSRNOT00000028051
|
Tgfb1
|
transforming growth factor, beta 1 |
| chr15_+_57241968 | 65.57 |
ENSRNOT00000082191
|
Lcp1
|
lymphocyte cytosolic protein 1 |
| chr15_+_31948035 | 62.57 |
ENSRNOT00000071627
|
AABR07017868.1
|
|
| chr13_+_51958834 | 58.92 |
ENSRNOT00000007833
|
Ptpn7
|
protein tyrosine phosphatase, non-receptor type 7 |
| chr6_-_139041654 | 57.00 |
ENSRNOT00000075664
|
AABR07065656.1
|
|
| chr6_-_138900915 | 56.93 |
ENSRNOT00000075363
|
AABR07065656.3
|
|
| chr15_+_30270740 | 55.37 |
ENSRNOT00000070991
|
AABR07017707.1
|
|
| chr15_+_31950986 | 54.10 |
ENSRNOT00000080233
|
AABR07017868.4
|
|
| chr20_-_5212624 | 54.07 |
ENSRNOT00000074261
|
LOC103689996
|
antigen peptide transporter 2 |
| chr6_-_139747579 | 52.71 |
ENSRNOT00000075502
|
AABR07065705.3
|
|
| chr20_+_3995544 | 51.54 |
ENSRNOT00000000527
|
Tap2
|
transporter 2, ATP binding cassette subfamily B member |
| chr6_-_140215907 | 50.06 |
ENSRNOT00000086370
|
AABR07065750.1
|
|
| chr6_-_138536162 | 48.16 |
ENSRNOT00000083031
|
AABR07065643.1
|
|
| chr6_+_137959171 | 47.22 |
ENSRNOT00000006835
|
Crip1
|
cysteine rich protein 1 |
| chr15_+_31719866 | 46.57 |
ENSRNOT00000072127
|
AABR07017838.2
|
|
| chr6_-_138772736 | 46.13 |
ENSRNOT00000071492
|
AABR07065651.1
|
|
| chr15_-_29903755 | 46.01 |
ENSRNOT00000066802
|
AABR07017669.1
|
|
| chr6_-_141008427 | 45.58 |
ENSRNOT00000074472
|
AABR07065778.2
|
|
| chr12_-_22127021 | 45.52 |
ENSRNOT00000076261
|
Sap25
|
Sin3A-associated protein 25 |
| chr6_-_138948424 | 45.04 |
ENSRNOT00000072566
|
AABR07065656.2
|
|
| chr15_+_31579478 | 44.85 |
ENSRNOT00000071642
|
AABR07017830.1
|
|
| chr6_-_142372031 | 44.58 |
ENSRNOT00000063929
|
AABR07065814.3
|
|
| chr20_-_3374344 | 43.52 |
ENSRNOT00000082999
|
Ppp1r18
|
protein phosphatase 1, regulatory subunit 18 |
| chr9_+_40975836 | 42.59 |
ENSRNOT00000084470
|
Ptpn18
|
protein tyrosine phosphatase, non-receptor type 18 |
| chr8_-_62424303 | 42.31 |
ENSRNOT00000091223
|
Csk
|
c-src tyrosine kinase |
| chr6_-_138631997 | 42.28 |
ENSRNOT00000073304
|
AABR07065651.3
|
|
| chr6_+_139523495 | 42.17 |
ENSRNOT00000075467
|
AABR07065699.4
|
|
| chr3_+_114772603 | 42.14 |
ENSRNOT00000073569
|
MGC105649
|
hypothetical LOC302884 |
| chr12_-_22126858 | 42.12 |
ENSRNOT00000084480
|
Sap25
|
Sin3A-associated protein 25 |
| chr15_-_29341843 | 41.90 |
ENSRNOT00000047082
|
LOC100360997
|
rCG61261-like |
| chr6_-_138679506 | 41.32 |
ENSRNOT00000075376
|
AABR07065651.4
|
|
| chr9_-_92775816 | 40.51 |
ENSRNOT00000029635
|
RGD1563917
|
similar to Nuclear autoantigen Sp-100 (Speckled 100 kDa) |
| chr6_-_138507135 | 40.36 |
ENSRNOT00000071078
|
AABR07065640.1
|
|
| chr6_-_138931262 | 40.02 |
ENSRNOT00000078126
ENSRNOT00000042118 |
LOC100359993
|
Ighg protein-like |
| chr15_-_29987717 | 39.71 |
ENSRNOT00000071501
|
AABR07017676.2
|
|
| chr6_+_139486942 | 39.63 |
ENSRNOT00000080320
|
AABR07065699.3
|
|
| chr6_-_139710905 | 39.59 |
ENSRNOT00000077430
|
AABR07065705.2
|
|
| chr6_-_139004980 | 39.45 |
ENSRNOT00000085427
ENSRNOT00000087351 |
AABR07065656.4
|
|
| chr15_+_29157294 | 39.34 |
ENSRNOT00000085162
|
AABR07017604.1
|
|
| chr8_-_48564722 | 38.42 |
ENSRNOT00000067902
|
Cbl
|
Cbl proto-oncogene |
| chr15_+_31436302 | 38.14 |
ENSRNOT00000071012
|
AABR07017825.2
|
|
| chr6_-_138954577 | 37.98 |
ENSRNOT00000042728
|
AABR07065656.7
|
|
| chr6_-_138764901 | 37.40 |
ENSRNOT00000075175
|
AABR07065651.2
|
|
| chr15_-_30157685 | 37.40 |
ENSRNOT00000085082
|
AABR07017693.2
|
|
| chr10_+_55013703 | 37.08 |
ENSRNOT00000032785
|
Pik3r5
|
phosphoinositide-3-kinase, regulatory subunit 5 |
| chr6_+_139293294 | 36.52 |
ENSRNOT00000050297
ENSRNOT00000081817 |
LOC100359993
|
Ighg protein-like |
| chr6_-_140418831 | 36.25 |
ENSRNOT00000086301
|
AABR07065768.2
|
|
| chr19_+_37652969 | 36.09 |
ENSRNOT00000041970
|
Carmil2
|
capping protein regulator and myosin 1 linker 2 |
| chr15_+_31282047 | 35.92 |
ENSRNOT00000081826
|
AABR07017812.1
|
|
| chr15_-_30147793 | 35.46 |
ENSRNOT00000060399
|
AABR07017693.1
|
|
| chr15_+_31462806 | 34.52 |
ENSRNOT00000072946
|
AABR07017825.3
|
|
| chr15_+_31167860 | 34.44 |
ENSRNOT00000065401
|
LOC100360997
|
rCG61261-like |
| chr15_+_31895223 | 34.20 |
ENSRNOT00000092202
|
AABR07017855.1
|
|
| chr13_-_110257367 | 34.17 |
ENSRNOT00000005576
|
Dtl
|
denticleless E3 ubiquitin protein ligase homolog |
| chr15_+_31178339 | 33.85 |
ENSRNOT00000089064
|
AABR07017798.2
|
|
| chr15_-_29532988 | 33.76 |
ENSRNOT00000074782
|
AABR07017639.1
|
|
| chr1_-_1116926 | 33.58 |
ENSRNOT00000062027
|
Raet1l
|
retinoic acid early transcript 1L |
| chr15_+_31057655 | 33.49 |
ENSRNOT00000073618
|
AABR07017783.2
|
|
| chr6_-_142060032 | 32.76 |
ENSRNOT00000064717
|
AABR07065811.1
|
|
| chr14_-_45797887 | 32.72 |
ENSRNOT00000029209
|
Tbc1d1
|
TBC1 domain family member 1 |
| chr20_+_11365697 | 32.20 |
ENSRNOT00000001611
|
Aire
|
autoimmune regulator |
| chr2_+_248249468 | 31.60 |
ENSRNOT00000022648
|
Gbp4
|
guanylate binding protein 4 |
| chr9_+_98505259 | 31.45 |
ENSRNOT00000051810
|
Erfe
|
erythroferrone |
| chr5_-_150520884 | 31.20 |
ENSRNOT00000085482
|
Rcc1
|
regulator of chromosome condensation 1 |
| chr6_-_138685656 | 30.89 |
ENSRNOT00000041706
|
AABR07065651.7
|
|
| chr15_+_30462669 | 30.88 |
ENSRNOT00000060357
|
RGD1564212
|
similar to T cell receptor alpha chain |
| chr14_-_2867397 | 30.76 |
ENSRNOT00000030428
|
Rpl5
|
ribosomal protein L5 |
| chr20_-_10680283 | 30.64 |
ENSRNOT00000001579
|
Sik1
|
salt-inducible kinase 1 |
| chr15_-_29446332 | 29.97 |
ENSRNOT00000082901
|
AABR07017634.1
|
|
| chr4_+_170958196 | 29.76 |
ENSRNOT00000007905
|
Pde6h
|
phosphodiesterase 6H |
| chr15_+_32386816 | 29.57 |
ENSRNOT00000060322
|
AABR07017901.1
|
|
| chr15_+_31395772 | 29.37 |
ENSRNOT00000075087
|
AABR07017824.1
|
|
| chr6_-_138550417 | 29.28 |
ENSRNOT00000071945
|
AABR07065645.1
|
|
| chr20_+_3935529 | 29.21 |
ENSRNOT00000072458
|
LOC108348072
|
class II histocompatibility antigen, M alpha chain |
| chr15_-_30174197 | 28.09 |
ENSRNOT00000074419
|
LOC688340
|
T-cell receptor alpha chain V region PHDS58-like |
| chr15_-_29369504 | 28.04 |
ENSRNOT00000060297
|
AABR07017624.1
|
|
| chrX_-_105390580 | 27.65 |
ENSRNOT00000077547
|
Btk
|
Bruton tyrosine kinase |
| chr17_+_77224112 | 26.77 |
ENSRNOT00000024178
|
Mcm10
|
minichromosome maintenance 10 replication initiation factor |
| chr6_-_139637187 | 26.66 |
ENSRNOT00000089454
|
LOC100359993
|
Ighg protein-like |
| chr20_-_5244386 | 26.50 |
ENSRNOT00000070886
|
RT1-DMa
|
RT1 class II, locus DMa |
| chr15_+_30735921 | 26.26 |
ENSRNOT00000078887
|
AABR07017763.2
|
|
| chr10_-_84920886 | 26.24 |
ENSRNOT00000068083
|
Sp2
|
Sp2 transcription factor |
| chr15_+_31642169 | 26.15 |
ENSRNOT00000072362
|
AABR07017833.1
|
|
| chr15_-_29360198 | 26.15 |
ENSRNOT00000072745
|
LOC688340
|
T-cell receptor alpha chain V region PHDS58-like |
| chr11_-_55044581 | 25.82 |
ENSRNOT00000046330
|
Morc1
|
MORC family CW-type zinc finger 1 |
| chr10_+_4536180 | 25.18 |
ENSRNOT00000003309
|
Zc3h7a
|
zinc finger CCCH type containing 7 A |
| chr6_+_139531330 | 25.07 |
ENSRNOT00000083025
|
AABR07065699.1
|
|
| chr15_+_29108981 | 25.05 |
ENSRNOT00000073133
|
AABR07017597.1
|
|
| chr15_+_30802299 | 24.78 |
ENSRNOT00000072418
|
AABR07017768.3
|
|
| chr1_-_4653210 | 24.71 |
ENSRNOT00000019121
|
Rab32
|
RAB32, member RAS oncogene family |
| chr18_+_27576129 | 24.32 |
ENSRNOT00000070930
|
Kdm3b
|
lysine demethylase 3B |
| chr15_-_29922522 | 24.24 |
ENSRNOT00000081778
|
AABR07017669.2
|
|
| chr1_-_82004538 | 24.15 |
ENSRNOT00000087572
|
Pou2f2
|
POU class 2 homeobox 2 |
| chr15_-_29728771 | 23.68 |
ENSRNOT00000073056
|
AABR07017657.1
|
|
| chr15_-_29993639 | 23.58 |
ENSRNOT00000071822
|
AABR07017677.1
|
|
| chr1_+_221773254 | 23.43 |
ENSRNOT00000028646
|
Rasgrp2
|
RAS guanyl releasing protein 2 |
| chr15_+_30785248 | 23.40 |
ENSRNOT00000089085
|
AABR07017768.5
|
|
| chr10_-_70241254 | 23.00 |
ENSRNOT00000038376
|
Rad51d
|
RAD51 paralog D |
| chr17_-_71105286 | 22.94 |
ENSRNOT00000025901
|
Prkcq
|
protein kinase C, theta |
| chr15_-_29680444 | 22.92 |
ENSRNOT00000075008
|
AABR07017658.1
|
|
| chr15_-_30296451 | 22.63 |
ENSRNOT00000067263
|
AABR07017714.2
|
|
| chr1_-_198450688 | 22.61 |
ENSRNOT00000027392
|
Pagr1
|
Paxip1-associated glutamate-rich protein 1 |
| chr5_-_159602251 | 22.20 |
ENSRNOT00000011394
|
Necap2
|
NECAP endocytosis associated 2 |
| chrX_-_72078551 | 22.20 |
ENSRNOT00000076978
|
Rps4x
|
ribosomal protein S4, X-linked |
| chr15_+_29096865 | 21.78 |
ENSRNOT00000077709
|
AABR07017597.2
|
|
| chr15_+_31243097 | 21.75 |
ENSRNOT00000042721
|
LOC100360891
|
RGD1359684 protein-like |
| chr8_+_71822129 | 21.72 |
ENSRNOT00000089147
|
Dapk2
|
death-associated protein kinase 2 |
| chr10_-_19715832 | 21.52 |
ENSRNOT00000040278
|
RGD1564698
|
similar to ribosomal protein S10 |
| chr9_-_10441834 | 21.46 |
ENSRNOT00000043704
|
Rpl36
|
ribosomal protein L36 |
| chr5_+_122508388 | 21.44 |
ENSRNOT00000038410
|
Tctex1d1
|
Tctex1 domain containing 1 |
| chr15_+_29073755 | 21.19 |
ENSRNOT00000075409
|
AABR07017595.1
|
|
| chr1_-_198460126 | 21.19 |
ENSRNOT00000082940
ENSRNOT00000086019 |
Maz
|
MYC associated zinc finger protein |
| chr16_-_22411225 | 21.12 |
ENSRNOT00000016204
|
RGD1559708
|
similar to ribosomal protein S10 |
| chr13_-_110784209 | 20.85 |
ENSRNOT00000071698
|
Traf5
|
TNF receptor-associated factor 5 |
| chr15_+_31456265 | 20.85 |
ENSRNOT00000075092
|
AABR07017825.1
|
|
| chr6_-_139997537 | 20.59 |
ENSRNOT00000073207
|
AABR07065740.1
|
|
| chr20_-_32416044 | 20.49 |
ENSRNOT00000067004
|
Ccar1
|
cell division cycle and apoptosis regulator 1 |
| chr1_-_101131413 | 20.40 |
ENSRNOT00000093729
|
Flt3lg
|
fms-related tyrosine kinase 3 ligand |
| chr16_-_20890949 | 20.36 |
ENSRNOT00000081977
|
Homer3
|
homer scaffolding protein 3 |
| chr5_+_169506138 | 20.29 |
ENSRNOT00000014904
|
Rpl22
|
ribosomal protein L22 |
| chr9_-_82327534 | 20.28 |
ENSRNOT00000024444
|
Nhej1
|
nonhomologous end-joining factor 1 |
| chr4_-_170841187 | 20.13 |
ENSRNOT00000007485
|
Art4
|
ADP-ribosyltransferase 4 |
| chr12_-_46845079 | 20.07 |
ENSRNOT00000047466
ENSRNOT00000064115 |
Pxn
|
paxillin |
| chr15_+_30612409 | 20.05 |
ENSRNOT00000072977
|
AABR07017748.1
|
|
| chr1_-_198706852 | 20.02 |
ENSRNOT00000024074
|
Dctpp1
|
dCTP pyrophosphatase 1 |
| chr15_+_32188736 | 19.79 |
ENSRNOT00000081436
|
AABR07017878.1
|
|
| chr1_+_105113595 | 19.69 |
ENSRNOT00000020853
ENSRNOT00000079655 |
Prmt3
|
protein arginine methyltransferase 3 |
| chr4_+_145489869 | 19.60 |
ENSRNOT00000082618
|
Fancd2
|
Fanconi anemia, complementation group D2 |
| chr3_+_170994038 | 19.21 |
ENSRNOT00000081823
|
Spo11
|
SPO11, initiator of meiotic double stranded breaks |
| chr3_+_148150698 | 18.70 |
ENSRNOT00000087075
ENSRNOT00000010251 |
Hm13
|
histocompatibility minor 13 |
| chr1_-_157700064 | 18.48 |
ENSRNOT00000031974
|
Ddias
|
DNA damage-induced apoptosis suppressor |
| chr20_+_4020317 | 18.43 |
ENSRNOT00000000526
|
RT1-DOb
|
RT1 class II, locus DOb |
| chr17_+_9631925 | 17.77 |
ENSRNOT00000018114
|
Ddx41
|
DEAD-box helicase 41 |
| chr15_+_30750093 | 17.65 |
ENSRNOT00000071830
|
AABR07017763.1
|
|
| chr15_+_32296119 | 17.60 |
ENSRNOT00000072496
|
AABR07017892.1
|
|
| chr6_-_139033819 | 17.29 |
ENSRNOT00000091961
|
AABR07065656.6
|
|
| chr16_+_73584521 | 17.25 |
ENSRNOT00000024301
|
Gins4
|
GINS complex subunit 4 |
| chr16_+_1979191 | 17.15 |
ENSRNOT00000014382
|
Ppif
|
peptidylprolyl isomerase F |
| chr4_+_115024927 | 16.59 |
ENSRNOT00000090987
|
Mob1a
|
MOB kinase activator 1A |
| chr12_-_46794797 | 16.54 |
ENSRNOT00000001518
|
Rplp0
|
ribosomal protein lateral stalk subunit P0 |
| chr1_-_89258935 | 16.29 |
ENSRNOT00000003180
|
LOC100361079
|
ribosomal protein L36-like |
| chr19_-_15733412 | 16.25 |
ENSRNOT00000014831
|
Irx6
|
iroquois homeobox 6 |
| chr15_+_30910039 | 16.20 |
ENSRNOT00000072686
|
AABR07017772.1
|
|
| chr15_+_34282936 | 16.14 |
ENSRNOT00000026364
|
Irf9
|
interferon regulatory factor 9 |
| chr13_-_60496511 | 16.10 |
ENSRNOT00000004495
|
Cdc73
|
cell division cycle 73 |
| chr15_+_30862478 | 15.96 |
ENSRNOT00000071566
|
AABR07017769.1
|
|
| chr8_-_21661259 | 15.86 |
ENSRNOT00000068308
|
Fbxl12
|
F-box and leucine-rich repeat protein 12 |
| chr15_-_29623597 | 15.82 |
ENSRNOT00000074483
|
AABR07017647.1
|
|
| chr16_+_9563218 | 15.76 |
ENSRNOT00000035915
|
Arhgap22
|
Rho GTPase activating protein 22 |
| chr15_+_31852416 | 15.74 |
ENSRNOT00000060380
|
AABR07017849.1
|
|
| chr13_+_67611708 | 15.70 |
ENSRNOT00000063833
|
Tpr
|
translocated promoter region, nuclear basket protein |
| chr19_-_10620671 | 15.66 |
ENSRNOT00000021842
|
Ccl17
|
C-C motif chemokine ligand 17 |
| chr10_-_70259553 | 15.65 |
ENSRNOT00000011320
|
Nle1
|
notchless homolog 1 |
| chr9_+_16702460 | 15.48 |
ENSRNOT00000061432
|
Ptk7
|
protein tyrosine kinase 7 |
| chr1_+_219144205 | 15.27 |
ENSRNOT00000083942
|
Unc93b1
|
unc-93 homolog B1 (C. elegans) |
| chr12_-_42376426 | 15.25 |
ENSRNOT00000057258
|
LOC103689988
|
2'-5'-oligoadenylate synthase 1A-like |
| chr1_-_101426852 | 15.20 |
ENSRNOT00000028217
|
Ruvbl2
|
RuvB-like AAA ATPase 2 |
| chr10_+_55492404 | 15.06 |
ENSRNOT00000005588
ENSRNOT00000078038 |
Rpl26
|
ribosomal protein L26 |
| chr15_-_29841388 | 15.04 |
ENSRNOT00000084501
|
AABR07017662.4
|
|
| chr2_-_199771896 | 14.89 |
ENSRNOT00000043937
|
Chd1l
|
chromodomain helicase DNA binding protein 1-like |
| chr10_+_103703404 | 14.88 |
ENSRNOT00000086469
|
Rab37
|
RAB37, member RAS oncogene family |
| chr8_-_83693472 | 14.80 |
ENSRNOT00000015947
|
Fam83b
|
family with sequence similarity 83, member B |
| chr12_+_13299859 | 14.65 |
ENSRNOT00000039191
|
LOC683674
|
similar to Protein C7orf26 homolog |
| chr6_+_52751106 | 14.44 |
ENSRNOT00000014373
|
Twistnb
|
TWIST neighbor |
| chr18_+_36829062 | 14.43 |
ENSRNOT00000025467
ENSRNOT00000086979 |
Tcerg1
|
transcription elongation regulator 1 |
| chr15_+_32076297 | 14.30 |
ENSRNOT00000060310
|
AABR07017870.1
|
|
| chr7_+_34744483 | 14.28 |
ENSRNOT00000007465
|
Usp44
|
ubiquitin specific peptidase 44 |
| chr11_-_67756799 | 14.03 |
ENSRNOT00000030975
|
Parp9
|
poly (ADP-ribose) polymerase family, member 9 |
| chr3_-_80873887 | 13.98 |
ENSRNOT00000024280
|
Dgkz
|
diacylglycerol kinase zeta |
| chr8_+_132553218 | 13.76 |
ENSRNOT00000006554
|
Limd1
|
LIM domains containing 1 |
| chr15_+_30926938 | 13.73 |
ENSRNOT00000075625
|
AABR07017773.1
|
|
| chr3_-_72071895 | 13.67 |
ENSRNOT00000082857
|
Selenoh
|
selenoprotein H |
| chr20_+_7818289 | 13.61 |
ENSRNOT00000042539
|
Ppard
|
peroxisome proliferator-activated receptor delta |
| chr1_+_85386470 | 13.54 |
ENSRNOT00000093332
ENSRNOT00000044326 |
Plekhg2
|
pleckstrin homology and RhoGEF domain containing G2 |
| chr15_+_30827300 | 13.54 |
ENSRNOT00000073533
|
AABR07017768.2
|
|
| chr20_-_4316715 | 13.53 |
ENSRNOT00000031704
|
C4b
|
complement C4B (Chido blood group) |
| chr6_-_99576196 | 13.36 |
ENSRNOT00000040565
|
RGD1562381
|
similar to ribosomal protein S17 |
| chr3_+_103866144 | 13.32 |
ENSRNOT00000007688
|
NEWGENE_1582771
|
katanin regulatory subunit B1 like 1 |
| chr9_-_53732858 | 13.11 |
ENSRNOT00000029204
|
Nemp2
|
nuclear envelope integral membrane protein 2 |
| chr1_-_101131012 | 13.10 |
ENSRNOT00000082283
ENSRNOT00000093498 ENSRNOT00000093559 |
Flt3lg
Rpl13a
|
fms-related tyrosine kinase 3 ligand ribosomal protein L13A |
| chr1_-_21586910 | 12.79 |
ENSRNOT00000032424
|
Med23
|
mediator complex subunit 23 |
| chr1_+_44019059 | 12.76 |
ENSRNOT00000065733
ENSRNOT00000083425 |
Scaf8
|
SR-related CTD-associated factor 8 |
| chr12_+_23151180 | 12.76 |
ENSRNOT00000059486
|
Cux1
|
cut-like homeobox 1 |
| chr3_-_52998650 | 12.53 |
ENSRNOT00000009988
|
RGD1560831
|
similar to 40S ribosomal protein S3 |
| chr11_+_77815181 | 12.51 |
ENSRNOT00000002640
|
Cldn1
|
claudin 1 |
| chr12_+_660011 | 12.51 |
ENSRNOT00000040830
|
Pds5b
|
PDS5 cohesin associated factor B |
| chr10_-_85049331 | 12.46 |
ENSRNOT00000012538
|
Tbx21
|
T-box 21 |
| chr12_+_13716596 | 12.28 |
ENSRNOT00000080216
|
Actb
|
actin, beta |
| chr15_+_31141142 | 12.20 |
ENSRNOT00000060304
|
LOC103693854
|
uncharacterized LOC103693854 |
| chr16_+_19767264 | 12.18 |
ENSRNOT00000051802
ENSRNOT00000092073 |
Ocel1
|
occludin/ELL domain containing 1 |
| chr3_+_147772165 | 12.07 |
ENSRNOT00000008713
|
Tbc1d20
|
TBC1 domain family, member 20 |
| chr6_-_128989812 | 11.99 |
ENSRNOT00000085943
|
LOC100909439
|
ankyrin repeat and SOCS box protein 2-like |
| chr1_-_220867815 | 11.77 |
ENSRNOT00000028015
|
Mus81
|
MUS81 structure-specific endonuclease subunit |
| chr9_+_81402124 | 11.73 |
ENSRNOT00000046019
|
Rufy4
|
RUN and FYVE domain containing 4 |
| chr1_+_40529045 | 11.70 |
ENSRNOT00000026564
|
Mthfd1l
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1-like |
| chr15_+_19429508 | 11.64 |
ENSRNOT00000047113
|
LOC100361240
|
ribosomal protein S10-like |
| chr10_+_13084304 | 11.59 |
ENSRNOT00000066310
|
RGD1561157
|
RGD1561157 |
| chr3_-_83335960 | 11.52 |
ENSRNOT00000013914
|
Api5
|
apoptosis inhibitor 5 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Rad21_Smc3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 23.0 | 69.0 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 17.2 | 51.5 | GO:0046967 | antigen processing and presentation of exogenous peptide antigen via MHC class Ib(GO:0002477) antigen processing and presentation of exogenous protein antigen via MHC class Ib, TAP-dependent(GO:0002481) antigen processing and presentation of endogenous peptide antigen via MHC class Ib via ER pathway(GO:0002488) antigen processing and presentation of endogenous peptide antigen via MHC class Ib via ER pathway, TAP-dependent(GO:0002489) positive regulation of antigen processing and presentation of peptide antigen(GO:0002585) regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) cytosol to ER transport(GO:0046967) peptide antigen transport(GO:0046968) |
| 10.8 | 54.1 | GO:0002483 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 10.7 | 32.2 | GO:0002458 | peripheral T cell tolerance induction(GO:0002458) peripheral tolerance induction(GO:0002465) regulation of thymocyte migration(GO:2000410) |
| 10.3 | 30.8 | GO:1903630 | regulation of aminoacyl-tRNA ligase activity(GO:1903630) |
| 9.4 | 47.2 | GO:0060741 | prostate gland stromal morphogenesis(GO:0060741) |
| 9.2 | 18.4 | GO:0002583 | regulation of antigen processing and presentation of peptide antigen(GO:0002583) negative regulation of antigen processing and presentation of peptide antigen(GO:0002584) regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002586) |
| 9.0 | 36.1 | GO:2000813 | negative regulation of barbed-end actin filament capping(GO:2000813) |
| 8.4 | 33.5 | GO:0090290 | positive regulation of osteoclast proliferation(GO:0090290) |
| 8.0 | 24.1 | GO:0002380 | immunoglobulin secretion involved in immune response(GO:0002380) |
| 7.2 | 21.7 | GO:2000424 | regulation of eosinophil chemotaxis(GO:2000422) positive regulation of eosinophil chemotaxis(GO:2000424) |
| 7.1 | 42.3 | GO:0042997 | negative regulation of low-density lipoprotein particle clearance(GO:0010989) negative regulation of Golgi to plasma membrane protein transport(GO:0042997) |
| 7.0 | 7.0 | GO:0002578 | negative regulation of antigen processing and presentation(GO:0002578) regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002580) negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) |
| 6.9 | 27.6 | GO:0071226 | peripheral B cell selection(GO:0002343) B cell affinity maturation(GO:0002344) cellular response to molecule of fungal origin(GO:0071226) |
| 6.2 | 18.7 | GO:1904211 | membrane protein proteolysis involved in retrograde protein transport, ER to cytosol(GO:1904211) |
| 6.1 | 30.6 | GO:0032792 | negative regulation of CREB transcription factor activity(GO:0032792) |
| 5.8 | 29.0 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 5.8 | 23.0 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 5.7 | 22.9 | GO:2000318 | positive regulation of T-helper 17 type immune response(GO:2000318) |
| 5.7 | 17.2 | GO:1902445 | regulation of mitochondrial membrane permeability involved in programmed necrotic cell death(GO:1902445) |
| 5.5 | 65.6 | GO:0051639 | actin filament network formation(GO:0051639) |
| 5.4 | 16.3 | GO:0060489 | orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 5.3 | 21.2 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 5.2 | 15.7 | GO:0010847 | MAPK import into nucleus(GO:0000189) regulation of chromatin assembly(GO:0010847) positive regulation of chromatin assembly or disassembly(GO:0045799) |
| 5.1 | 15.2 | GO:0071733 | transcriptional activation by promoter-enhancer looping(GO:0071733) regulation of estrogen receptor binding(GO:0071898) negative regulation of estrogen receptor binding(GO:0071899) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 5.0 | 15.1 | GO:1902167 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902167) |
| 5.0 | 20.0 | GO:0009223 | pyrimidine deoxyribonucleotide catabolic process(GO:0009223) |
| 4.5 | 22.6 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 4.4 | 26.5 | GO:0002399 | MHC class II protein complex assembly(GO:0002399) peptide antigen assembly with MHC class II protein complex(GO:0002503) |
| 4.3 | 12.8 | GO:2000409 | positive regulation of T cell extravasation(GO:2000409) |
| 4.2 | 12.5 | GO:1903348 | positive regulation of bicellular tight junction assembly(GO:1903348) response to butyrate(GO:1903544) |
| 4.0 | 12.1 | GO:1902953 | positive regulation of ER to Golgi vesicle-mediated transport(GO:1902953) |
| 4.0 | 7.9 | GO:0006407 | rRNA export from nucleus(GO:0006407) |
| 3.9 | 11.7 | GO:0015942 | formate metabolic process(GO:0015942) |
| 3.8 | 19.2 | GO:1990918 | meiotic DNA double-strand break processing(GO:0000706) double-strand break repair involved in meiotic recombination(GO:1990918) |
| 3.7 | 25.8 | GO:2000143 | regulation of transposition(GO:0010528) negative regulation of transposition(GO:0010529) negative regulation of DNA-templated transcription, initiation(GO:2000143) |
| 3.5 | 10.6 | GO:2000359 | regulation of binding of sperm to zona pellucida(GO:2000359) negative regulation of binding of sperm to zona pellucida(GO:2000360) |
| 3.5 | 38.4 | GO:0090649 | response to oxygen-glucose deprivation(GO:0090649) cellular response to oxygen-glucose deprivation(GO:0090650) |
| 3.4 | 10.2 | GO:1990167 | protein K27-linked deubiquitination(GO:1990167) |
| 3.4 | 10.1 | GO:0032298 | positive regulation of DNA-dependent DNA replication initiation(GO:0032298) |
| 3.3 | 30.0 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 3.3 | 16.5 | GO:1904401 | cellular response to Thyroid stimulating hormone(GO:1904401) |
| 3.3 | 19.7 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 3.3 | 19.6 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 3.2 | 9.7 | GO:1904569 | regulation of selenocysteine incorporation(GO:1904569) |
| 3.1 | 24.7 | GO:1903232 | melanosome assembly(GO:1903232) |
| 3.0 | 3.0 | GO:0035711 | T-helper 1 cell activation(GO:0035711) |
| 2.7 | 2.7 | GO:0046078 | dUMP metabolic process(GO:0046078) |
| 2.6 | 44.5 | GO:0002504 | antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002504) |
| 2.6 | 7.8 | GO:0021898 | regulation of transcription from RNA polymerase II promoter involved in forebrain neuron fate commitment(GO:0021882) commitment of multipotent stem cells to neuronal lineage in forebrain(GO:0021898) |
| 2.6 | 7.8 | GO:0090164 | asymmetric Golgi ribbon formation(GO:0090164) |
| 2.6 | 35.8 | GO:0019985 | translesion synthesis(GO:0019985) |
| 2.5 | 10.1 | GO:1904690 | regulation of cap-independent translational initiation(GO:1903677) positive regulation of cap-independent translational initiation(GO:1903679) regulation of cytoplasmic translational initiation(GO:1904688) positive regulation of cytoplasmic translational initiation(GO:1904690) |
| 2.5 | 9.9 | GO:0034760 | regulation of iron ion transmembrane transport(GO:0034759) negative regulation of iron ion transmembrane transport(GO:0034760) regulation of cytochrome-c oxidase activity(GO:1904959) |
| 2.4 | 7.3 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 2.3 | 18.7 | GO:2000288 | positive regulation of myoblast proliferation(GO:2000288) |
| 2.3 | 13.8 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 2.3 | 9.1 | GO:0090625 | mRNA cleavage involved in gene silencing by siRNA(GO:0090625) |
| 2.2 | 15.7 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) negative regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001268) |
| 2.2 | 4.5 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 2.1 | 6.2 | GO:0098749 | cerebellar neuron development(GO:0098749) |
| 1.9 | 9.7 | GO:0016095 | polyprenol catabolic process(GO:0016095) |
| 1.9 | 19.4 | GO:0010891 | negative regulation of sequestering of triglyceride(GO:0010891) positive regulation of triglyceride catabolic process(GO:0010898) |
| 1.8 | 7.2 | GO:0051958 | methotrexate transport(GO:0051958) |
| 1.8 | 23.1 | GO:0090178 | regulation of establishment of planar polarity involved in neural tube closure(GO:0090178) |
| 1.7 | 13.5 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 1.6 | 8.1 | GO:0006030 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 1.5 | 4.5 | GO:0010909 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) |
| 1.5 | 16.3 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 1.5 | 8.8 | GO:0046602 | regulation of mitotic centrosome separation(GO:0046602) |
| 1.5 | 16.1 | GO:0045351 | type I interferon biosynthetic process(GO:0045351) |
| 1.5 | 16.1 | GO:0033523 | histone H2B ubiquitination(GO:0033523) |
| 1.5 | 4.4 | GO:0006990 | positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 1.4 | 5.8 | GO:0060431 | primary lung bud formation(GO:0060431) |
| 1.4 | 8.6 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 1.4 | 14.0 | GO:0060330 | regulation of response to interferon-gamma(GO:0060330) |
| 1.3 | 49.4 | GO:0035458 | cellular response to interferon-beta(GO:0035458) |
| 1.3 | 31.5 | GO:2000193 | positive regulation of fatty acid transport(GO:2000193) |
| 1.2 | 11.2 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 1.2 | 9.9 | GO:0009912 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 1.2 | 24.3 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 1.2 | 27.6 | GO:0035329 | hippo signaling(GO:0035329) |
| 1.2 | 14.3 | GO:0090231 | regulation of spindle checkpoint(GO:0090231) regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 1.2 | 10.7 | GO:1902414 | protein localization to cell junction(GO:1902414) |
| 1.1 | 12.5 | GO:0048304 | positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 1.1 | 9.1 | GO:0033299 | secretion of lysosomal enzymes(GO:0033299) |
| 1.1 | 26.8 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 1.1 | 22.9 | GO:0060396 | growth hormone receptor signaling pathway(GO:0060396) |
| 1.1 | 5.4 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 1.1 | 5.4 | GO:2000327 | regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000325) positive regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000327) |
| 1.1 | 29.8 | GO:0045745 | positive regulation of G-protein coupled receptor protein signaling pathway(GO:0045745) |
| 1.1 | 9.5 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 1.0 | 9.4 | GO:0000050 | urea cycle(GO:0000050) |
| 1.0 | 6.2 | GO:0010825 | positive regulation of centrosome duplication(GO:0010825) positive regulation of centrosome cycle(GO:0046607) |
| 1.0 | 6.0 | GO:0032202 | telomere assembly(GO:0032202) |
| 1.0 | 6.7 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 1.0 | 3.8 | GO:1904430 | negative regulation of t-circle formation(GO:1904430) |
| 0.9 | 3.8 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.9 | 9.0 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.9 | 9.0 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.9 | 0.9 | GO:0006649 | phospholipid transfer to membrane(GO:0006649) |
| 0.9 | 5.2 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.9 | 2.6 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 0.8 | 27.7 | GO:0006303 | double-strand break repair via nonhomologous end joining(GO:0006303) |
| 0.8 | 2.5 | GO:0018283 | metal incorporation into metallo-sulfur cluster(GO:0018282) iron incorporation into metallo-sulfur cluster(GO:0018283) |
| 0.8 | 10.4 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.8 | 18.7 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.8 | 45.0 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.8 | 1.5 | GO:0000957 | mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 0.7 | 3.0 | GO:0032919 | spermine acetylation(GO:0032919) |
| 0.7 | 19.1 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.7 | 9.4 | GO:0032957 | inositol trisphosphate metabolic process(GO:0032957) |
| 0.7 | 7.1 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.7 | 37.1 | GO:0043551 | regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.7 | 82.1 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.7 | 4.7 | GO:1901838 | positive regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901838) |
| 0.6 | 3.1 | GO:0042270 | protection from natural killer cell mediated cytotoxicity(GO:0042270) |
| 0.6 | 9.7 | GO:0051599 | response to hydrostatic pressure(GO:0051599) |
| 0.6 | 3.0 | GO:0071630 | nucleus-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071630) |
| 0.6 | 5.9 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.6 | 5.1 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.6 | 12.5 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.6 | 5.6 | GO:0071481 | cellular response to X-ray(GO:0071481) |
| 0.6 | 31.2 | GO:0051290 | protein heterotetramerization(GO:0051290) |
| 0.6 | 6.1 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.6 | 12.2 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.5 | 2.0 | GO:0071955 | recycling endosome to Golgi transport(GO:0071955) |
| 0.5 | 13.4 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.5 | 16.0 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.5 | 14.0 | GO:0031571 | mitotic G1 DNA damage checkpoint(GO:0031571) G1 DNA damage checkpoint(GO:0044783) |
| 0.4 | 2.2 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.4 | 2.2 | GO:0036233 | glycine import(GO:0036233) |
| 0.4 | 2.2 | GO:0060591 | chondroblast differentiation(GO:0060591) |
| 0.4 | 2.1 | GO:1903899 | positive regulation of PERK-mediated unfolded protein response(GO:1903899) |
| 0.4 | 13.2 | GO:0032508 | DNA duplex unwinding(GO:0032508) |
| 0.4 | 5.4 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.4 | 4.9 | GO:0007289 | spermatid nucleus differentiation(GO:0007289) |
| 0.4 | 1.2 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.4 | 2.0 | GO:0033625 | positive regulation of integrin activation(GO:0033625) |
| 0.4 | 5.1 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.4 | 24.7 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.4 | 4.5 | GO:0022615 | protein to membrane docking(GO:0022615) |
| 0.4 | 1.8 | GO:0071104 | response to interleukin-9(GO:0071104) |
| 0.4 | 1.8 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.3 | 2.3 | GO:0036089 | cleavage furrow formation(GO:0036089) |
| 0.3 | 12.3 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.3 | 2.6 | GO:0015886 | heme transport(GO:0015886) |
| 0.3 | 14.4 | GO:0006360 | transcription from RNA polymerase I promoter(GO:0006360) |
| 0.3 | 10.0 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.3 | 1.9 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.3 | 1.9 | GO:0015860 | purine nucleoside transmembrane transport(GO:0015860) |
| 0.3 | 0.9 | GO:2001137 | positive regulation of endocytic recycling(GO:2001137) |
| 0.3 | 4.3 | GO:0019184 | nonribosomal peptide biosynthetic process(GO:0019184) |
| 0.3 | 1.8 | GO:0045078 | interferon-gamma biosynthetic process(GO:0042095) regulation of interferon-gamma biosynthetic process(GO:0045072) positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.3 | 14.9 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.3 | 1.7 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.3 | 8.4 | GO:0000578 | embryonic axis specification(GO:0000578) |
| 0.3 | 8.6 | GO:0060065 | uterus development(GO:0060065) |
| 0.3 | 6.2 | GO:0051973 | positive regulation of telomerase activity(GO:0051973) |
| 0.3 | 1.9 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.3 | 5.5 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.3 | 4.1 | GO:2001241 | positive regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001241) |
| 0.3 | 1.5 | GO:0097490 | trunk segmentation(GO:0035290) trunk neural crest cell migration(GO:0036484) ventral trunk neural crest cell migration(GO:0036486) sympathetic neuron projection extension(GO:0097490) sympathetic neuron projection guidance(GO:0097491) |
| 0.3 | 3.3 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.2 | 2.0 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) |
| 0.2 | 6.6 | GO:0002717 | positive regulation of natural killer cell mediated immunity(GO:0002717) positive regulation of natural killer cell mediated cytotoxicity(GO:0045954) |
| 0.2 | 20.6 | GO:0048144 | fibroblast proliferation(GO:0048144) |
| 0.2 | 14.1 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.2 | 1.3 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.2 | 6.1 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.2 | 3.8 | GO:0030825 | positive regulation of cGMP metabolic process(GO:0030825) positive regulation of cGMP biosynthetic process(GO:0030828) |
| 0.2 | 3.5 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.2 | 2.4 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.2 | 1.8 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.2 | 2.5 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.2 | 5.5 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.2 | 5.8 | GO:0033119 | negative regulation of RNA splicing(GO:0033119) |
| 0.2 | 3.3 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.2 | 1.8 | GO:0097240 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.2 | 2.0 | GO:0007210 | serotonin receptor signaling pathway(GO:0007210) |
| 0.2 | 14.4 | GO:0045727 | positive regulation of translation(GO:0045727) |
| 0.2 | 3.1 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.2 | 10.5 | GO:0006338 | chromatin remodeling(GO:0006338) |
| 0.2 | 6.0 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.1 | 7.5 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.1 | 1.5 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.1 | 2.2 | GO:0042659 | regulation of cell fate specification(GO:0042659) |
| 0.1 | 13.0 | GO:0051092 | positive regulation of NF-kappaB transcription factor activity(GO:0051092) |
| 0.1 | 1.2 | GO:0048820 | hair follicle maturation(GO:0048820) |
| 0.1 | 5.1 | GO:0060976 | coronary vasculature development(GO:0060976) |
| 0.1 | 1.5 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.1 | 0.8 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.1 | 0.5 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.1 | 3.3 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 4.1 | GO:0035019 | somatic stem cell population maintenance(GO:0035019) |
| 0.1 | 4.8 | GO:0000380 | alternative mRNA splicing, via spliceosome(GO:0000380) |
| 0.1 | 4.6 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.1 | 1.6 | GO:0055070 | copper ion homeostasis(GO:0055070) |
| 0.1 | 5.3 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.1 | 1.4 | GO:1902230 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902230) |
| 0.1 | 7.0 | GO:0030317 | sperm motility(GO:0030317) |
| 0.1 | 2.2 | GO:2001238 | positive regulation of extrinsic apoptotic signaling pathway(GO:2001238) |
| 0.1 | 2.7 | GO:0007622 | rhythmic behavior(GO:0007622) circadian behavior(GO:0048512) |
| 0.1 | 0.5 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.1 | 13.1 | GO:0006417 | regulation of translation(GO:0006417) |
| 0.1 | 4.3 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.1 | 2.7 | GO:0090280 | positive regulation of calcium ion import(GO:0090280) |
| 0.1 | 5.8 | GO:0051091 | positive regulation of sequence-specific DNA binding transcription factor activity(GO:0051091) |
| 0.1 | 8.8 | GO:0051607 | defense response to virus(GO:0051607) |
| 0.1 | 2.8 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.1 | 2.1 | GO:0021762 | substantia nigra development(GO:0021762) |
| 0.1 | 8.4 | GO:0045471 | response to ethanol(GO:0045471) |
| 0.0 | 10.0 | GO:0008380 | RNA splicing(GO:0008380) |
| 0.0 | 1.4 | GO:0007131 | reciprocal meiotic recombination(GO:0007131) reciprocal DNA recombination(GO:0035825) |
| 0.0 | 1.4 | GO:0034629 | cellular protein complex localization(GO:0034629) |
| 0.0 | 0.1 | GO:0061304 | retinal blood vessel morphogenesis(GO:0061304) |
| 0.0 | 2.7 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.0 | 1.4 | GO:0001510 | RNA methylation(GO:0001510) |
| 0.0 | 0.1 | GO:1900620 | acetylcholine biosynthetic process(GO:0008292) acetate ester biosynthetic process(GO:1900620) |
| 0.0 | 0.7 | GO:0031639 | plasminogen activation(GO:0031639) |
| 0.0 | 0.3 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.0 | 1.1 | GO:0007173 | epidermal growth factor receptor signaling pathway(GO:0007173) |
| 0.0 | 0.4 | GO:0030517 | negative regulation of axon extension(GO:0030517) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 15.1 | 105.6 | GO:0042825 | TAP complex(GO:0042825) |
| 12.4 | 37.1 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 7.9 | 31.6 | GO:0020005 | symbiont-containing vacuole membrane(GO:0020005) |
| 6.8 | 34.2 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 6.8 | 20.3 | GO:0032807 | DNA ligase IV complex(GO:0032807) |
| 5.8 | 17.3 | GO:0000811 | GINS complex(GO:0000811) |
| 5.4 | 16.3 | GO:0060187 | cell pole(GO:0060187) |
| 4.6 | 74.1 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 4.6 | 23.0 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 4.1 | 12.4 | GO:0036488 | CHOP-C/EBP complex(GO:0036488) |
| 4.1 | 24.7 | GO:0031904 | endosome lumen(GO:0031904) |
| 3.7 | 11.0 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 3.6 | 36.1 | GO:0044354 | F-actin capping protein complex(GO:0008290) pinosome(GO:0044352) macropinosome(GO:0044354) |
| 3.4 | 10.1 | GO:0097124 | cyclin A2-CDK2 complex(GO:0097124) |
| 3.2 | 38.4 | GO:0016600 | flotillin complex(GO:0016600) |
| 3.1 | 25.1 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 2.9 | 11.8 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 2.7 | 18.7 | GO:0071556 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 2.6 | 42.1 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 2.5 | 15.2 | GO:0097255 | R2TP complex(GO:0097255) |
| 2.3 | 16.1 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 2.1 | 8.6 | GO:0002947 | tumor necrosis factor receptor superfamily complex(GO:0002947) |
| 1.9 | 54.1 | GO:0001891 | phagocytic cup(GO:0001891) |
| 1.9 | 25.1 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 1.9 | 3.8 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 1.9 | 13.1 | GO:0097452 | GAIT complex(GO:0097452) |
| 1.7 | 22.6 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 1.7 | 20.9 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 1.6 | 22.8 | GO:0031332 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 1.6 | 11.0 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 1.6 | 15.7 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 1.5 | 12.3 | GO:0097433 | dense body(GO:0097433) |
| 1.5 | 15.3 | GO:0032009 | early phagosome(GO:0032009) |
| 1.4 | 5.6 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 1.3 | 6.7 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 1.2 | 14.4 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 1.0 | 27.6 | GO:0042629 | mast cell granule(GO:0042629) |
| 1.0 | 20.4 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.9 | 4.7 | GO:0034455 | t-UTP complex(GO:0034455) |
| 0.9 | 8.3 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.9 | 7.2 | GO:0016589 | NURF complex(GO:0016589) |
| 0.9 | 19.6 | GO:0030057 | desmosome(GO:0030057) |
| 0.9 | 42.8 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.9 | 84.0 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.9 | 7.0 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.8 | 13.5 | GO:0044217 | other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) |
| 0.8 | 22.2 | GO:0030125 | clathrin vesicle coat(GO:0030125) |
| 0.8 | 4.5 | GO:0030681 | nucleolar ribonuclease P complex(GO:0005655) ribonuclease P complex(GO:0030677) multimeric ribonuclease P complex(GO:0030681) |
| 0.8 | 4.5 | GO:0070820 | tertiary granule(GO:0070820) |
| 0.7 | 2.2 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.7 | 25.5 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.7 | 74.4 | GO:0005902 | microvillus(GO:0005902) |
| 0.7 | 49.2 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.7 | 7.8 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.6 | 1.9 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 0.6 | 8.8 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.6 | 4.3 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.6 | 17.2 | GO:0005753 | mitochondrial proton-transporting ATP synthase complex(GO:0005753) |
| 0.6 | 6.1 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.5 | 9.7 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.5 | 5.0 | GO:0032797 | SMN complex(GO:0032797) |
| 0.4 | 8.8 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.4 | 3.2 | GO:0071541 | eukaryotic 43S preinitiation complex(GO:0016282) eukaryotic 48S preinitiation complex(GO:0033290) eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.4 | 2.3 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.4 | 4.9 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.4 | 5.9 | GO:0032039 | integrator complex(GO:0032039) |
| 0.4 | 3.6 | GO:0070187 | telosome(GO:0070187) |
| 0.3 | 7.6 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.3 | 11.6 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.3 | 1.2 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 0.3 | 47.1 | GO:0005840 | ribosome(GO:0005840) |
| 0.3 | 20.2 | GO:0005811 | lipid particle(GO:0005811) |
| 0.3 | 12.6 | GO:0016592 | mediator complex(GO:0016592) |
| 0.3 | 1.8 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.2 | 47.3 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.2 | 1.9 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.2 | 2.7 | GO:0090544 | BAF-type complex(GO:0090544) |
| 0.2 | 15.2 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.2 | 11.4 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.2 | 6.9 | GO:0005844 | polysome(GO:0005844) |
| 0.2 | 2.5 | GO:0030665 | clathrin-coated vesicle membrane(GO:0030665) |
| 0.2 | 17.5 | GO:0000781 | chromosome, telomeric region(GO:0000781) |
| 0.2 | 1.5 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.2 | 88.1 | GO:0000785 | chromatin(GO:0000785) |
| 0.2 | 7.1 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.2 | 1.4 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.1 | 15.2 | GO:0005814 | centriole(GO:0005814) |
| 0.1 | 9.0 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.1 | 0.5 | GO:0034774 | secretory granule lumen(GO:0034774) |
| 0.1 | 7.8 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.1 | 11.1 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.1 | 5.4 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 2.0 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.1 | 2.5 | GO:0015030 | Cajal body(GO:0015030) |
| 0.1 | 4.5 | GO:0001726 | ruffle(GO:0001726) |
| 0.1 | 0.3 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.1 | 4.7 | GO:0042641 | actomyosin(GO:0042641) |
| 0.1 | 43.0 | GO:0005730 | nucleolus(GO:0005730) |
| 0.1 | 1.7 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 1.4 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.8 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.0 | 1.8 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.9 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 1.9 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.2 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.1 | GO:0005606 | laminin-1 complex(GO:0005606) |
| 0.0 | 1.2 | GO:0045178 | basal part of cell(GO:0045178) |
| 0.0 | 2.9 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.6 | GO:0035097 | histone methyltransferase complex(GO:0035097) |
| 0.0 | 0.2 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 3.6 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 21.1 | 105.6 | GO:0015440 | peptide-transporting ATPase activity(GO:0015440) |
| 13.8 | 69.0 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 8.2 | 24.7 | GO:0035651 | AP-3 adaptor complex binding(GO:0035651) |
| 7.8 | 31.2 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 7.1 | 42.6 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 6.2 | 37.1 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 4.8 | 19.2 | GO:0061505 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 4.7 | 47.2 | GO:0003680 | AT DNA binding(GO:0003680) |
| 3.9 | 15.5 | GO:0071936 | coreceptor activity involved in Wnt signaling pathway(GO:0071936) |
| 3.8 | 23.0 | GO:0000150 | recombinase activity(GO:0000150) |
| 3.4 | 30.8 | GO:0055104 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 3.3 | 19.7 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 3.1 | 53.2 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 3.1 | 18.7 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 3.1 | 9.2 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 3.1 | 30.6 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 3.0 | 9.1 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 3.0 | 20.9 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 2.5 | 15.2 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 2.5 | 20.1 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 2.5 | 20.0 | GO:0032552 | deoxyribonucleotide binding(GO:0032552) |
| 2.5 | 17.2 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 2.4 | 9.7 | GO:0047751 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 2.3 | 11.7 | GO:0004488 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) methylenetetrahydrofolate dehydrogenase (NADP+) activity(GO:0004488) |
| 2.3 | 18.5 | GO:0034046 | poly(G) binding(GO:0034046) |
| 2.1 | 44.9 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 2.0 | 9.9 | GO:0032767 | copper-exporting ATPase activity(GO:0004008) copper-dependent protein binding(GO:0032767) copper-transporting ATPase activity(GO:0043682) |
| 1.9 | 26.8 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 1.9 | 7.5 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 1.8 | 56.7 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 1.8 | 7.2 | GO:0015350 | methotrexate transporter activity(GO:0015350) |
| 1.7 | 15.7 | GO:0043495 | protein anchor(GO:0043495) |
| 1.6 | 9.7 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 1.6 | 14.4 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 1.6 | 9.4 | GO:0001013 | RNA polymerase I regulatory region DNA binding(GO:0001013) |
| 1.6 | 17.3 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 1.5 | 6.1 | GO:0000405 | bubble DNA binding(GO:0000405) |
| 1.4 | 10.1 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 1.4 | 7.0 | GO:0070052 | collagen V binding(GO:0070052) |
| 1.4 | 15.3 | GO:0035325 | Toll-like receptor binding(GO:0035325) |
| 1.4 | 12.3 | GO:0030957 | Tat protein binding(GO:0030957) |
| 1.3 | 8.1 | GO:0004568 | chitinase activity(GO:0004568) |
| 1.3 | 39.9 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 1.3 | 11.8 | GO:0048256 | crossover junction endodeoxyribonuclease activity(GO:0008821) flap endonuclease activity(GO:0048256) |
| 1.3 | 19.6 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 1.3 | 37.9 | GO:0043175 | RNA polymerase core enzyme binding(GO:0043175) |
| 1.3 | 3.8 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 1.3 | 20.1 | GO:0051400 | BH domain binding(GO:0051400) |
| 1.2 | 6.1 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 1.2 | 24.3 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 1.2 | 17.0 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 1.2 | 14.0 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 1.2 | 5.8 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 1.1 | 22.9 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 1.1 | 22.6 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 1.0 | 5.1 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 1.0 | 20.9 | GO:0031996 | thioesterase binding(GO:0031996) |
| 1.0 | 3.0 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.9 | 15.1 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.9 | 9.1 | GO:0051766 | inositol trisphosphate kinase activity(GO:0051766) |
| 0.9 | 3.4 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.8 | 3.4 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.8 | 13.8 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.8 | 3.8 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.7 | 3.5 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.7 | 9.0 | GO:0005522 | profilin binding(GO:0005522) |
| 0.7 | 33.3 | GO:0045182 | translation regulator activity(GO:0045182) |
| 0.7 | 9.5 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.6 | 4.4 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.6 | 11.0 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 0.6 | 22.6 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.6 | 12.5 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.6 | 10.7 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.6 | 149.6 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.6 | 13.6 | GO:0036041 | long-chain fatty acid binding(GO:0036041) |
| 0.6 | 27.8 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.6 | 7.8 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.5 | 4.3 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.5 | 34.5 | GO:0004004 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) RNA-dependent ATPase activity(GO:0008186) |
| 0.5 | 27.6 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.5 | 3.4 | GO:0035375 | zymogen binding(GO:0035375) |
| 0.5 | 2.8 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.5 | 10.7 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.5 | 11.5 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.5 | 10.1 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.5 | 4.5 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.4 | 15.7 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.4 | 32.3 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.4 | 3.1 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.4 | 9.9 | GO:0005112 | Notch binding(GO:0005112) |
| 0.4 | 2.5 | GO:0016530 | metallochaperone activity(GO:0016530) |
| 0.4 | 3.3 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.4 | 3.6 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.4 | 8.3 | GO:0070001 | aspartic-type peptidase activity(GO:0070001) |
| 0.4 | 69.9 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.4 | 7.8 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.4 | 3.1 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.4 | 1.2 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.4 | 34.6 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.4 | 20.4 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.4 | 5.6 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.3 | 27.7 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.3 | 39.5 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.3 | 22.1 | GO:0043566 | structure-specific DNA binding(GO:0043566) |
| 0.3 | 2.6 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.3 | 13.9 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.3 | 8.8 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.3 | 8.3 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.3 | 17.4 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.2 | 2.0 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.2 | 3.3 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.2 | 5.8 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.2 | 7.2 | GO:0008094 | DNA-dependent ATPase activity(GO:0008094) |
| 0.2 | 1.5 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.2 | 9.6 | GO:0001046 | core promoter sequence-specific DNA binding(GO:0001046) |
| 0.2 | 7.7 | GO:0001047 | core promoter binding(GO:0001047) |
| 0.2 | 32.9 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.2 | 3.6 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.2 | 1.4 | GO:0019958 | C-X-C chemokine binding(GO:0019958) |
| 0.2 | 31.1 | GO:0005179 | hormone activity(GO:0005179) |
| 0.2 | 11.3 | GO:0002039 | p53 binding(GO:0002039) |
| 0.2 | 2.5 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.2 | 0.9 | GO:0090555 | phosphatidylethanolamine-translocating ATPase activity(GO:0090555) |
| 0.2 | 2.6 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.2 | 1.9 | GO:0005346 | adenine nucleotide transmembrane transporter activity(GO:0000295) purine ribonucleotide transmembrane transporter activity(GO:0005346) |
| 0.2 | 1.8 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.2 | 3.3 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.2 | 0.3 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.1 | 4.1 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.1 | 1.5 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.1 | 4.5 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 13.8 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.1 | 0.5 | GO:0016888 | endodeoxyribonuclease activity, producing 5'-phosphomonoesters(GO:0016888) |
| 0.1 | 1.2 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.1 | 5.5 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 4.9 | GO:0004402 | histone acetyltransferase activity(GO:0004402) peptide-lysine-N-acetyltransferase activity(GO:0061733) |
| 0.1 | 2.2 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.1 | 1.4 | GO:0008649 | rRNA methyltransferase activity(GO:0008649) |
| 0.1 | 2.1 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.1 | 2.2 | GO:0001158 | enhancer sequence-specific DNA binding(GO:0001158) |
| 0.1 | 3.3 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.1 | 1.2 | GO:0034483 | heparan sulfate sulfotransferase activity(GO:0034483) |
| 0.1 | 1.6 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.1 | 13.8 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.1 | 25.4 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.1 | 7.1 | GO:0042393 | histone binding(GO:0042393) |
| 0.1 | 6.9 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.1 | 0.9 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.0 | 8.6 | GO:0004866 | endopeptidase inhibitor activity(GO:0004866) |
| 0.0 | 2.9 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 1.5 | GO:0099589 | G-protein coupled serotonin receptor activity(GO:0004993) serotonin receptor activity(GO:0099589) |
| 0.0 | 1.3 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 0.5 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 2.0 | GO:0001948 | glycoprotein binding(GO:0001948) |
| 0.0 | 1.8 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 1.2 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 0.1 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.8 | 69.0 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 3.5 | 56.3 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 2.2 | 127.1 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 1.6 | 64.7 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 1.4 | 29.8 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 1.3 | 24.1 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.9 | 48.7 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.8 | 2.5 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.8 | 14.2 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.8 | 26.6 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.7 | 23.5 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.7 | 19.6 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.7 | 24.1 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.7 | 38.4 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.7 | 12.5 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.6 | 22.2 | SIG CHEMOTAXIS | Genes related to chemotaxis |
| 0.5 | 7.9 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.4 | 8.6 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.4 | 31.2 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.3 | 13.6 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.3 | 11.6 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.3 | 4.9 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.3 | 15.7 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.3 | 11.2 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.2 | 8.5 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.2 | 5.5 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.2 | 5.1 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.2 | 14.0 | PID P73PATHWAY | p73 transcription factor network |
| 0.2 | 18.6 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.2 | 4.1 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.2 | 7.8 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.2 | 9.5 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.2 | 2.2 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.2 | 7.2 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.1 | 2.2 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.1 | 3.3 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 3.6 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.1 | 3.1 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.1 | 1.7 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.1 | 26.1 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.1 | 2.8 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 2.2 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.1 | 2.7 | PID E2F PATHWAY | E2F transcription factor network |
| 0.1 | 1.2 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.3 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 1.4 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.8 | 42.3 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 3.4 | 60.4 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 3.0 | 38.4 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 3.0 | 56.1 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 2.8 | 19.6 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 1.6 | 158.0 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 1.6 | 17.3 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 1.6 | 21.7 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 1.4 | 23.4 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 1.4 | 36.9 | REACTOME ACTIVATION OF THE PRE REPLICATIVE COMPLEX | Genes involved in Activation of the pre-replicative complex |
| 1.2 | 24.4 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 1.2 | 37.1 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 1.2 | 15.3 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 1.1 | 9.9 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 1.1 | 20.5 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 1.1 | 27.6 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 1.0 | 22.9 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.9 | 47.7 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.8 | 9.7 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.8 | 16.6 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.8 | 3.1 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.7 | 9.6 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.6 | 34.3 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.6 | 8.6 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.5 | 14.5 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.5 | 5.1 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.4 | 12.5 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.4 | 3.6 | REACTOME TELOMERE MAINTENANCE | Genes involved in Telomere Maintenance |
| 0.4 | 11.0 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.4 | 9.5 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.4 | 5.8 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.4 | 4.4 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.4 | 28.0 | REACTOME LATE PHASE OF HIV LIFE CYCLE | Genes involved in Late Phase of HIV Life Cycle |
| 0.4 | 3.8 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.4 | 17.5 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.3 | 15.7 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.3 | 13.5 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.2 | 13.7 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.2 | 3.1 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.2 | 13.6 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.2 | 3.3 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.2 | 2.7 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.2 | 3.8 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.2 | 2.0 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.2 | 2.2 | REACTOME SIGNALING BY FGFR1 MUTANTS | Genes involved in Signaling by FGFR1 mutants |
| 0.2 | 3.0 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.2 | 9.0 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.1 | 2.2 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.1 | 11.5 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 12.1 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 2.1 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 2.2 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.1 | 1.9 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.1 | 5.5 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.1 | 6.3 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.1 | 1.9 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.1 | 2.0 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.9 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.3 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 1.5 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.3 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 1.9 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |