Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
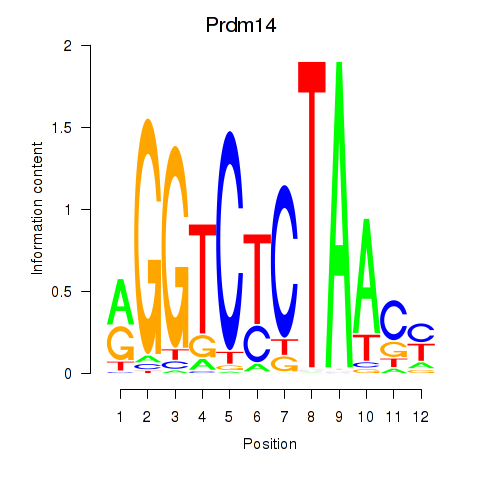
Results for Prdm14
Z-value: 0.66
Transcription factors associated with Prdm14
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Prdm14
|
ENSRNOG00000021792 | PR/SET domain 14 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Prdm14 | rn6_v1_chr5_+_5710392_5710392 | 0.06 | 2.9e-01 | Click! |
Activity profile of Prdm14 motif
Sorted Z-values of Prdm14 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_148119857 | 48.07 |
ENSRNOT00000040325
|
LOC100361547
|
Cytochrome P450, family 2, subfamily c, polypeptide 7-like |
| chr20_-_4542073 | 35.00 |
ENSRNOT00000000477
|
Cfb
|
complement factor B |
| chr13_+_78805347 | 32.70 |
ENSRNOT00000003748
|
Serpinc1
|
serpin family C member 1 |
| chr9_+_95241609 | 26.88 |
ENSRNOT00000032634
ENSRNOT00000077416 |
Ugt1a5
|
UDP glucuronosyltransferase family 1 member A5 |
| chr2_-_210809306 | 19.68 |
ENSRNOT00000047139
|
Gstm1
|
glutathione S-transferase mu 1 |
| chr1_-_102826965 | 16.69 |
ENSRNOT00000078692
|
Saa4
|
serum amyloid A4 |
| chr3_+_92640752 | 16.07 |
ENSRNOT00000007604
|
Slc1a2
|
solute carrier family 1 member 2 |
| chr3_-_138116664 | 12.78 |
ENSRNOT00000055602
|
Rrbp1
|
ribosome binding protein 1 |
| chrX_-_138435391 | 12.06 |
ENSRNOT00000043258
|
Mbnl3
|
muscleblind-like splicing regulator 3 |
| chr1_+_184301493 | 11.87 |
ENSRNOT00000078096
|
Insc
|
inscuteable homolog (Drosophila) |
| chr7_-_132343169 | 10.09 |
ENSRNOT00000021064
|
Abcd2
|
ATP binding cassette subfamily D member 2 |
| chr4_-_50860756 | 8.96 |
ENSRNOT00000068404
|
Cadps2
|
calcium dependent secretion activator 2 |
| chr13_+_100980574 | 8.87 |
ENSRNOT00000067005
ENSRNOT00000004649 |
Capn8
|
calpain 8 |
| chr19_+_15339152 | 7.80 |
ENSRNOT00000060929
|
Ces1a
|
carboxylesterase 1A |
| chr4_+_100218661 | 5.83 |
ENSRNOT00000079415
|
Tmem150a
|
transmembrane protein 150A |
| chrX_-_14972675 | 5.11 |
ENSRNOT00000079664
|
Slc38a5
|
solute carrier family 38, member 5 |
| chrX_-_14972389 | 4.86 |
ENSRNOT00000038068
|
Slc38a5
|
solute carrier family 38, member 5 |
| chr8_-_53816447 | 4.73 |
ENSRNOT00000011454
|
Ttc12
|
tetratricopeptide repeat domain 12 |
| chr15_-_45927804 | 4.33 |
ENSRNOT00000086271
|
Ints6
|
integrator complex subunit 6 |
| chr20_-_28814636 | 4.28 |
ENSRNOT00000086030
|
Sept10
|
septin 10 |
| chr10_-_65963932 | 3.56 |
ENSRNOT00000011726
|
Nlk
|
nemo like kinase |
| chr4_-_123713319 | 3.03 |
ENSRNOT00000012875
ENSRNOT00000086982 |
Slc6a6
|
solute carrier family 6 member 6 |
| chr1_+_225129097 | 2.86 |
ENSRNOT00000026938
|
Eml3
|
echinoderm microtubule associated protein like 3 |
| chr2_-_188645196 | 2.85 |
ENSRNOT00000083793
|
Efna3
|
ephrin A3 |
| chr8_+_55202725 | 2.58 |
ENSRNOT00000029166
|
Alg9
|
ALG9, alpha-1,2-mannosyltransferase |
| chr1_+_79894450 | 1.82 |
ENSRNOT00000057963
|
Nanos2
|
nanos C2HC-type zinc finger 2 |
| chr1_-_88111293 | 1.33 |
ENSRNOT00000077195
|
Spred3
|
sprouty-related, EVH1 domain containing 3 |
| chr2_-_189573280 | 1.32 |
ENSRNOT00000022897
|
Rps27
|
ribosomal protein S27 |
| chr2_-_35104963 | 1.28 |
ENSRNOT00000018058
|
Rgs7bp
|
regulator of G-protein signaling 7-binding protein |
| chr2_-_78427751 | 1.00 |
ENSRNOT00000077768
|
AC113901.1
|
|
| chr4_+_78496043 | 0.97 |
ENSRNOT00000059136
|
Aoc1
|
amine oxidase, copper containing 1 |
| chr2_+_1410934 | 0.97 |
ENSRNOT00000013625
ENSRNOT00000080222 |
Erap1
|
endoplasmic reticulum aminopeptidase 1 |
| chr3_-_147481073 | 0.91 |
ENSRNOT00000084722
|
Fam110a
|
family with sequence similarity 110, member A |
| chr2_-_179704629 | 0.89 |
ENSRNOT00000083361
ENSRNOT00000077941 |
Gria2
|
glutamate ionotropic receptor AMPA type subunit 2 |
| chr14_-_12995584 | 0.75 |
ENSRNOT00000035446
ENSRNOT00000077861 |
Fgf5
|
fibroblast growth factor 5 |
| chr6_+_137164535 | 0.61 |
ENSRNOT00000018225
|
Inf2
|
inverted formin, FH2 and WH2 domain containing |
| chr7_-_116920507 | 0.58 |
ENSRNOT00000048363
|
Mroh6
|
maestro heat-like repeat family member 6 |
| chr20_+_3582743 | 0.52 |
ENSRNOT00000001103
|
Gtf2h4
|
general transcription factor 2H subunit 4 |
| chr11_-_43617633 | 0.49 |
ENSRNOT00000060874
|
Olr1557
|
olfactory receptor 1557 |
| chr12_+_36871999 | 0.44 |
ENSRNOT00000001334
ENSRNOT00000084236 |
Ncor2
|
nuclear receptor co-repressor 2 |
| chr18_-_69780922 | 0.37 |
ENSRNOT00000079093
|
Me2
|
malic enzyme 2 |
| chr1_-_89509343 | 0.32 |
ENSRNOT00000028637
|
Fxyd3
|
FXYD domain-containing ion transport regulator 3 |
| chr1_-_225128740 | 0.27 |
ENSRNOT00000026897
|
Rom1
|
retinal outer segment membrane protein 1 |
| chr3_-_10694649 | 0.23 |
ENSRNOT00000037742
|
Gpr107
|
G protein-coupled receptor 107 |
| chr12_+_7081895 | 0.19 |
ENSRNOT00000047163
|
Hmgb1
|
high mobility group box 1 |
| chr3_-_2938663 | 0.09 |
ENSRNOT00000050294
|
LOC108348105
|
allergen Fel d 4-like |
Network of associatons between targets according to the STRING database.
First level regulatory network of Prdm14
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.2 | 32.7 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 6.6 | 19.7 | GO:0018916 | nitrobenzene metabolic process(GO:0018916) |
| 3.2 | 16.1 | GO:0070777 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 3.2 | 35.0 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 3.0 | 26.9 | GO:0052697 | xenobiotic glucuronidation(GO:0052697) |
| 1.8 | 48.1 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 1.3 | 10.1 | GO:0042760 | very long-chain fatty acid catabolic process(GO:0042760) |
| 1.1 | 9.0 | GO:1990504 | dense core granule exocytosis(GO:1990504) |
| 0.7 | 10.0 | GO:0015816 | glycine transport(GO:0015816) |
| 0.4 | 11.9 | GO:0000132 | establishment of mitotic spindle orientation(GO:0000132) |
| 0.4 | 12.1 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.3 | 1.8 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.3 | 16.7 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.2 | 4.3 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.1 | 5.8 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.1 | 1.0 | GO:0019885 | antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.1 | 3.0 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.1 | 2.6 | GO:0097502 | mannosylation(GO:0097502) |
| 0.1 | 0.4 | GO:1903799 | negative regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903799) |
| 0.1 | 1.0 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.1 | 0.2 | GO:0035711 | T cell mediated immune response to tumor cell(GO:0002424) regulation of T cell mediated immune response to tumor cell(GO:0002840) negative regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0017055) regulation of mismatch repair(GO:0032423) positive regulation of toll-like receptor 9 signaling pathway(GO:0034165) T-helper 1 cell activation(GO:0035711) positive regulation of glycogen catabolic process(GO:0045819) negative regulation of apoptotic cell clearance(GO:2000426) |
| 0.1 | 2.9 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.1 | 8.9 | GO:0007586 | digestion(GO:0007586) |
| 0.0 | 1.3 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 12.8 | GO:0001649 | osteoblast differentiation(GO:0001649) |
| 0.0 | 1.3 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.9 | GO:0060992 | response to fungicide(GO:0060992) |
| 0.0 | 0.7 | GO:0023019 | signal transduction involved in regulation of gene expression(GO:0023019) |
| 0.0 | 0.4 | GO:1902031 | malate metabolic process(GO:0006108) regulation of NADP metabolic process(GO:1902031) |
| 0.0 | 3.6 | GO:0030178 | negative regulation of Wnt signaling pathway(GO:0030178) |
| 0.0 | 0.5 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.0 | 0.6 | GO:0090140 | regulation of mitochondrial fission(GO:0090140) |
| 0.0 | 0.3 | GO:0061298 | retina vasculature development in camera-type eye(GO:0061298) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 16.7 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.6 | 67.7 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.5 | 16.1 | GO:0030673 | axolemma(GO:0030673) |
| 0.3 | 4.3 | GO:0032039 | integrator complex(GO:0032039) |
| 0.2 | 0.5 | GO:0000438 | core TFIIH complex portion of holo TFIIH complex(GO:0000438) |
| 0.1 | 10.1 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.1 | 9.9 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.1 | 12.8 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.1 | 19.7 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.3 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 11.9 | GO:0045177 | apical part of cell(GO:0045177) |
| 0.0 | 1.3 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 1.8 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 1.0 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 5.8 | GO:0005764 | lytic vacuole(GO:0000323) lysosome(GO:0005764) |
| 0.0 | 1.0 | GO:0005923 | bicellular tight junction(GO:0005923) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.2 | 16.1 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 2.5 | 19.7 | GO:0016151 | nickel cation binding(GO:0016151) |
| 1.6 | 48.1 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 1.0 | 3.0 | GO:0030977 | taurine binding(GO:0030977) |
| 0.9 | 26.9 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.8 | 10.0 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.4 | 8.9 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.3 | 32.7 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.2 | 1.3 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.2 | 0.7 | GO:0005163 | nerve growth factor receptor binding(GO:0005163) |
| 0.2 | 0.9 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.2 | 35.0 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 1.0 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.1 | 3.6 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.1 | 2.6 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.1 | 1.0 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.1 | 2.9 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 10.1 | GO:0042626 | ATPase activity, coupled to transmembrane movement of substances(GO:0042626) |
| 0.1 | 0.4 | GO:0004473 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.0 | 7.8 | GO:0052689 | carboxylic ester hydrolase activity(GO:0052689) |
| 0.0 | 0.2 | GO:0010858 | calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.0 | 0.5 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.2 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.0 | 0.4 | GO:0016922 | Notch binding(GO:0005112) ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 25.6 | GO:0003723 | RNA binding(GO:0003723) |
| 0.0 | 4.3 | GO:0005525 | GTP binding(GO:0005525) |
| 0.0 | 2.9 | GO:0008017 | microtubule binding(GO:0008017) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 35.0 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.8 | 32.7 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.2 | 23.2 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.1 | 2.9 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.9 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.4 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 35.0 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 2.2 | 32.7 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 1.4 | 26.9 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.7 | 19.7 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.5 | 10.1 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.5 | 29.1 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.1 | 2.6 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.1 | 0.7 | REACTOME FGFR2C LIGAND BINDING AND ACTIVATION | Genes involved in FGFR2c ligand binding and activation |
| 0.1 | 1.0 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.9 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.0 | 1.3 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.5 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 0.4 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.2 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |