Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
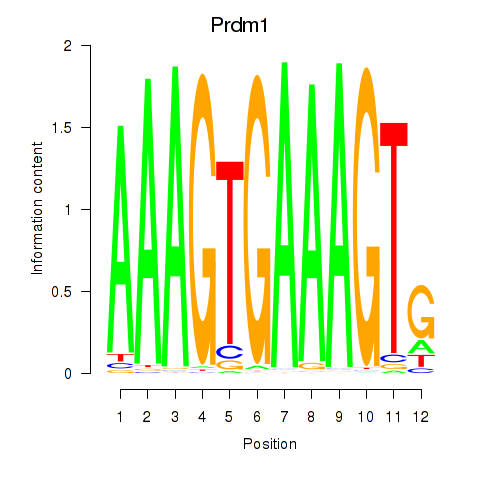
Results for Prdm1
Z-value: 0.84
Transcription factors associated with Prdm1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Prdm1
|
ENSRNOG00000000323 | PR/SET domain 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Prdm1 | rn6_v1_chr20_-_49486550_49486552 | 0.55 | 1.1e-26 | Click! |
Activity profile of Prdm1 motif
Sorted Z-values of Prdm1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_215660492 | 37.04 |
ENSRNOT00000079442
|
AC132720.1
|
|
| chr5_-_58198782 | 34.86 |
ENSRNOT00000023951
|
Ccl21
|
C-C motif chemokine ligand 21 |
| chr10_-_5251809 | 33.83 |
ENSRNOT00000085479
|
Ciita
|
class II, major histocompatibility complex, transactivator |
| chr1_-_237910012 | 26.81 |
ENSRNOT00000023664
|
Anxa1
|
annexin A1 |
| chr4_+_98337367 | 21.89 |
ENSRNOT00000042165
|
AABR07060872.1
|
|
| chr8_+_49713190 | 21.29 |
ENSRNOT00000022074
|
Fxyd2
|
FXYD domain-containing ion transport regulator 2 |
| chr20_-_1878126 | 21.16 |
ENSRNOT00000000995
|
Ubd
|
ubiquitin D |
| chr13_-_109849632 | 20.76 |
ENSRNOT00000005085
|
Atf3
|
activating transcription factor 3 |
| chr13_+_48575091 | 20.69 |
ENSRNOT00000009838
|
Pm20d1
|
peptidase M20 domain containing 1 |
| chr2_+_209097927 | 17.23 |
ENSRNOT00000023807
|
Dennd2d
|
DENN domain containing 2D |
| chr7_+_124460358 | 16.75 |
ENSRNOT00000014089
|
Tspo
|
translocator protein |
| chr4_+_90990088 | 16.66 |
ENSRNOT00000030320
|
Mmrn1
|
multimerin 1 |
| chr6_-_24676524 | 15.38 |
ENSRNOT00000047853
|
Capn13
|
calpain 13 |
| chr3_+_6430201 | 15.08 |
ENSRNOT00000086352
|
Col5a1
|
collagen type V alpha 1 chain |
| chr2_-_238468435 | 14.92 |
ENSRNOT00000055537
|
LOC102556447
|
rho guanine nucleotide exchange factor 38-like |
| chr3_-_160738927 | 14.71 |
ENSRNOT00000043470
|
Slpil2
|
antileukoproteinase-like 2 |
| chr13_+_74410010 | 13.93 |
ENSRNOT00000079152
|
Angptl1
|
angiopoietin-like 1 |
| chr3_+_16413080 | 13.64 |
ENSRNOT00000040386
|
LOC100912707
|
Ig kappa chain V19-17-like |
| chr10_-_87067456 | 13.60 |
ENSRNOT00000014163
|
Ccr7
|
C-C motif chemokine receptor 7 |
| chr15_-_51855325 | 13.38 |
ENSRNOT00000011663
|
Pdlim2
|
PDZ and LIM domain 2 |
| chr17_-_8429338 | 13.19 |
ENSRNOT00000016390
|
Tgfbi
|
transforming growth factor, beta induced |
| chr11_-_37914983 | 12.62 |
ENSRNOT00000039876
|
Mx1
|
myxovirus (influenza virus) resistance 1 |
| chr5_-_58183017 | 12.48 |
ENSRNOT00000020982
|
Ccl19
|
C-C motif chemokine ligand 19 |
| chr3_+_158997834 | 12.33 |
ENSRNOT00000000211
|
LOC108348048
|
von Willebrand factor A domain-containing protein 5A |
| chr4_-_103024841 | 12.17 |
ENSRNOT00000090283
|
AABR07061036.1
|
|
| chrX_+_104684676 | 12.08 |
ENSRNOT00000083229
|
Tnmd
|
tenomodulin |
| chr3_-_7498555 | 12.00 |
ENSRNOT00000017725
|
Barhl1
|
BarH-like homeobox 1 |
| chr7_-_117300662 | 11.97 |
ENSRNOT00000006376
|
Parp10
|
poly (ADP-ribose) polymerase family, member 10 |
| chr1_-_166677026 | 11.88 |
ENSRNOT00000026644
ENSRNOT00000076714 |
Art2b
|
ADP-ribosyltransferase 2b |
| chr3_-_17081510 | 11.83 |
ENSRNOT00000063862
|
AABR07051562.1
|
|
| chr6_-_143195143 | 11.73 |
ENSRNOT00000081337
|
AABR07065837.1
|
|
| chr16_-_85306366 | 11.70 |
ENSRNOT00000089650
|
Tnfsf13b
|
tumor necrosis factor superfamily member 13b |
| chr10_-_57064600 | 11.70 |
ENSRNOT00000032926
|
Cxcl16
|
C-X-C motif chemokine ligand 16 |
| chr2_-_149564113 | 11.55 |
ENSRNOT00000018628
|
Igsf10
|
immunoglobulin superfamily, member 10 |
| chr14_+_6058544 | 11.25 |
ENSRNOT00000061267
|
Abcg3l3
|
ATP-binding cassette, subfamily G (WHITE), member 3-like 3 |
| chr7_+_118685022 | 11.13 |
ENSRNOT00000089135
|
Apol3
|
apolipoprotein L, 3 |
| chr1_-_101236065 | 11.06 |
ENSRNOT00000066834
|
Cd37
|
CD37 molecule |
| chr1_-_88193346 | 10.91 |
ENSRNOT00000078600
|
LOC103690068
|
immortalization up-regulated protein-like |
| chr1_+_226634009 | 10.90 |
ENSRNOT00000028094
|
Cyb561a3
|
cytochrome b561 family, member A3 |
| chr19_-_55367353 | 10.86 |
ENSRNOT00000091139
|
Piezo1
|
piezo-type mechanosensitive ion channel component 1 |
| chr3_-_171286413 | 10.70 |
ENSRNOT00000008365
ENSRNOT00000081036 |
Zbp1
|
Z-DNA binding protein 1 |
| chr5_+_173288447 | 10.69 |
ENSRNOT00000091205
ENSRNOT00000067252 |
Mxra8
|
matrix remodeling associated 8 |
| chr4_+_149261044 | 10.63 |
ENSRNOT00000066670
|
Cxcl12
|
C-X-C motif chemokine ligand 12 |
| chr2_+_248398917 | 10.60 |
ENSRNOT00000045855
|
Gbp1
|
guanylate binding protein 1 |
| chr8_+_42443442 | 10.56 |
ENSRNOT00000015986
|
Vwa5a
|
von Willebrand factor A domain containing 5A |
| chr7_+_107695227 | 10.31 |
ENSRNOT00000009673
|
Wisp1
|
WNT1 inducible signaling pathway protein 1 |
| chr20_+_3995544 | 10.23 |
ENSRNOT00000000527
|
Tap2
|
transporter 2, ATP binding cassette subfamily B member |
| chr1_-_198577226 | 10.20 |
ENSRNOT00000055013
|
Spn
|
sialophorin |
| chr19_+_54060622 | 10.10 |
ENSRNOT00000023607
|
Gse1
|
Gse1 coiled-coil protein |
| chr8_-_128026841 | 9.93 |
ENSRNOT00000018341
|
Myd88
|
myeloid differentiation primary response 88 |
| chr5_-_58950373 | 9.76 |
ENSRNOT00000060442
|
Cd72
|
Cd72 molecule |
| chr13_-_89606326 | 9.76 |
ENSRNOT00000029179
|
Fcer1g
|
Fc fragment of IgE receptor Ig |
| chrX_-_62690806 | 9.74 |
ENSRNOT00000018147
|
Pola1
|
DNA polymerase alpha 1, catalytic subunit |
| chr9_-_76768770 | 9.64 |
ENSRNOT00000087779
ENSRNOT00000057849 |
Ikzf2
|
IKAROS family zinc finger 2 |
| chr9_+_68414339 | 9.61 |
ENSRNOT00000040778
|
Pard3b
|
par-3 family cell polarity regulator beta |
| chr1_-_217844957 | 9.59 |
ENSRNOT00000090653
|
Ano1
|
anoctamin 1 |
| chr20_+_3990820 | 9.30 |
ENSRNOT00000000528
|
Psmb8
|
proteasome subunit beta 8 |
| chr16_-_48692476 | 9.28 |
ENSRNOT00000013118
|
Irf2
|
interferon regulatory factor 2 |
| chr2_+_243656579 | 9.25 |
ENSRNOT00000036993
|
Adh1
|
alcohol dehydrogenase 1 (class I) |
| chr14_-_6541171 | 9.09 |
ENSRNOT00000042532
ENSRNOT00000087758 |
Abcg3l1
|
ATP-binding cassette, subfamily G (WHITE), member 3-like 1 |
| chr8_+_14060394 | 9.03 |
ENSRNOT00000014827
|
Smco4
|
single-pass membrane protein with coiled-coil domains 4 |
| chr8_+_99880073 | 9.02 |
ENSRNOT00000010765
|
Plscr4
|
phospholipid scramblase 4 |
| chr17_-_32661865 | 8.97 |
ENSRNOT00000022194
|
Serpinb9
|
serpin family B member 9 |
| chr9_+_98924134 | 8.97 |
ENSRNOT00000027597
|
Twist2
|
twist family bHLH transcription factor 2 |
| chr4_-_81968832 | 8.90 |
ENSRNOT00000016608
|
Skap2
|
src kinase associated phosphoprotein 2 |
| chr15_+_42489377 | 8.82 |
ENSRNOT00000064306
|
Pbk
|
PDZ binding kinase |
| chrX_-_34794589 | 8.45 |
ENSRNOT00000008703
|
Rai2
|
retinoic acid induced 2 |
| chr4_-_103569159 | 8.28 |
ENSRNOT00000084036
|
AABR07061072.1
|
|
| chr15_+_34256071 | 8.25 |
ENSRNOT00000025887
|
Psme1
|
proteasome activator subunit 1 |
| chr17_-_44840131 | 8.23 |
ENSRNOT00000083417
|
Hist1h3b
|
histone cluster 1, H3b |
| chr3_+_22964230 | 8.07 |
ENSRNOT00000041813
|
LOC100362149
|
ribosomal protein S20-like |
| chr1_+_41323194 | 7.99 |
ENSRNOT00000026350
|
Esr1
|
estrogen receptor 1 |
| chr2_-_235052464 | 7.97 |
ENSRNOT00000093512
|
LOC103691699
|
uncharacterized LOC103691699 |
| chr12_+_2180150 | 7.96 |
ENSRNOT00000001322
|
Stxbp2
|
syntaxin binding protein 2 |
| chr8_-_116469915 | 7.95 |
ENSRNOT00000024193
|
Sema3f
|
semaphorin 3F |
| chr20_+_12225202 | 7.84 |
ENSRNOT00000045315
|
Col18a1
|
collagen type XVIII alpha 1 chain |
| chrX_+_120624518 | 7.81 |
ENSRNOT00000007967
|
Slc6a14
|
solute carrier family 6 member 14 |
| chr9_-_1012450 | 7.81 |
ENSRNOT00000051449
|
LOC100359951
|
ribosomal protein S20-like |
| chr13_+_51795867 | 7.66 |
ENSRNOT00000006747
|
Ube2t
|
ubiquitin-conjugating enzyme E2T |
| chr5_+_161889342 | 7.66 |
ENSRNOT00000040481
|
LOC100362684
|
ribosomal protein S20-like |
| chr16_-_40555576 | 7.47 |
ENSRNOT00000015529
|
Vegfc
|
vascular endothelial growth factor C |
| chr18_+_79773608 | 7.45 |
ENSRNOT00000088484
|
Zfp516
|
zinc finger protein 516 |
| chr5_-_59113773 | 7.43 |
ENSRNOT00000083129
|
AC121204.3
|
|
| chr1_-_101131012 | 7.36 |
ENSRNOT00000082283
ENSRNOT00000093498 ENSRNOT00000093559 |
Flt3lg
Rpl13a
|
fms-related tyrosine kinase 3 ligand ribosomal protein L13A |
| chr20_+_248410 | 7.35 |
ENSRNOT00000049573
|
Tmlhe
|
trimethyllysine hydroxylase, epsilon |
| chr20_-_3978845 | 7.33 |
ENSRNOT00000000532
|
Psmb9
|
proteasome subunit beta 9 |
| chr5_+_157423213 | 7.33 |
ENSRNOT00000023431
|
Tmco4
|
transmembrane and coiled-coil domains 4 |
| chr8_+_62341613 | 7.27 |
ENSRNOT00000066923
|
Scamp2
|
secretory carrier membrane protein 2 |
| chr20_+_8484311 | 7.23 |
ENSRNOT00000050041
|
LOC100364116
|
ribosomal protein S20-like |
| chr7_+_59349745 | 7.15 |
ENSRNOT00000085334
|
Ptprb
|
protein tyrosine phosphatase, receptor type, B |
| chr8_-_107952530 | 7.14 |
ENSRNOT00000052043
|
Cldn18
|
claudin 18 |
| chr2_-_52282548 | 6.96 |
ENSRNOT00000033627
|
Nnt
|
nicotinamide nucleotide transhydrogenase |
| chr17_+_69468427 | 6.93 |
ENSRNOT00000058413
|
RGD1564865
|
similar to 20-alpha-hydroxysteroid dehydrogenase |
| chr4_+_67165883 | 6.80 |
ENSRNOT00000012280
|
Rab19
|
RAB19, member RAS oncogene family |
| chr11_+_60336061 | 6.78 |
ENSRNOT00000084022
|
LOC685767
|
similar to OX-2 membrane glycoprotein precursor (MRC OX-2 antigen) (CD200 antigen) |
| chr17_+_32516679 | 6.64 |
ENSRNOT00000032707
|
Serpinb9d
|
serine (or cysteine) peptidase inhibitor, clade B, member 9d |
| chr4_-_161587222 | 6.56 |
ENSRNOT00000050834
|
Tead4
|
TEA domain transcription factor 4 |
| chr12_-_40686249 | 6.36 |
ENSRNOT00000042264
|
LOC100909911
|
40S ribosomal protein S20-like |
| chr8_+_93439648 | 6.30 |
ENSRNOT00000043008
|
LOC100359563
|
ribosomal protein S20-like |
| chr6_+_135866739 | 6.30 |
ENSRNOT00000013460
|
Exoc3l4
|
exocyst complex component 3-like 4 |
| chr3_-_80842916 | 6.28 |
ENSRNOT00000033978
|
Mdk
|
midkine |
| chr6_-_143702033 | 6.27 |
ENSRNOT00000051410
|
AABR07065886.1
|
|
| chr4_+_179398621 | 6.20 |
ENSRNOT00000049474
ENSRNOT00000067506 |
Lrmp
|
lymphoid-restricted membrane protein |
| chr13_+_99220510 | 6.17 |
ENSRNOT00000004519
|
Tmem63a
|
transmembrane protein 63a |
| chr4_-_163214678 | 6.09 |
ENSRNOT00000091602
|
Clec1a
|
C-type lectin domain family 1, member A |
| chr19_-_25914689 | 6.05 |
ENSRNOT00000091994
ENSRNOT00000039354 |
Nf1x
|
nuclear factor 1 X |
| chr8_-_62616828 | 6.02 |
ENSRNOT00000068340
|
Arid3b
|
AT-rich interaction domain 3B |
| chr5_+_135687538 | 5.97 |
ENSRNOT00000091664
|
AC126292.2
|
|
| chr5_-_136053210 | 5.94 |
ENSRNOT00000025903
|
Kif2c
|
kinesin family member 2C |
| chr11_+_68105369 | 5.89 |
ENSRNOT00000046888
|
Parp14
|
poly (ADP-ribose) polymerase family, member 14 |
| chr3_+_28416954 | 5.75 |
ENSRNOT00000043533
|
Kynu
|
kynureninase |
| chr18_-_30781292 | 5.74 |
ENSRNOT00000027125
|
Taf7
|
TATA-box binding protein associated factor 7 |
| chr8_+_48805684 | 5.63 |
ENSRNOT00000064041
|
Bcl9l
|
B-cell CLL/lymphoma 9-like |
| chr1_+_128637049 | 5.61 |
ENSRNOT00000018639
|
Ttc23
|
tetratricopeptide repeat domain 23 |
| chr6_-_1534488 | 5.52 |
ENSRNOT00000006824
|
Cebpz
|
CCAAT/enhancer binding protein zeta |
| chr13_-_102643223 | 5.51 |
ENSRNOT00000003155
|
Hlx
|
H2.0-like homeobox |
| chr2_-_185444897 | 5.45 |
ENSRNOT00000016329
|
Rps3a
|
ribosomal protein S3a |
| chr3_-_114235933 | 5.44 |
ENSRNOT00000023939
|
Duox2
|
dual oxidase 2 |
| chr1_-_98521551 | 5.39 |
ENSRNOT00000081922
|
Siglec10
|
sialic acid binding Ig-like lectin 10 |
| chr10_-_94652658 | 5.39 |
ENSRNOT00000017371
|
Ern1
|
endoplasmic reticulum to nucleus signaling 1 |
| chr20_+_8486265 | 5.38 |
ENSRNOT00000072213
|
LOC100909911
|
40S ribosomal protein S20-like |
| chr3_-_37480984 | 5.30 |
ENSRNOT00000030373
|
Nmi
|
N-myc (and STAT) interactor |
| chr8_-_115179191 | 5.29 |
ENSRNOT00000017224
|
Parp3
|
poly (ADP-ribose) polymerase family, member 3 |
| chr2_-_88763733 | 5.21 |
ENSRNOT00000059424
|
LOC688389
|
similar to solute carrier family 7 (cationic amino acid transporter, y+ system), member 12 |
| chr10_+_57040267 | 5.11 |
ENSRNOT00000026207
|
Arrb2
|
arrestin, beta 2 |
| chr16_+_75744786 | 5.07 |
ENSRNOT00000032368
|
Xkr5
|
XK related 5 |
| chr4_-_21920651 | 5.06 |
ENSRNOT00000066211
|
Tmem243
|
transmembrane protein 243 |
| chr7_+_144064931 | 5.04 |
ENSRNOT00000048504
|
Prr13
|
proline rich 13 |
| chr6_-_77421286 | 5.04 |
ENSRNOT00000011453
|
Nkx2-1
|
NK2 homeobox 1 |
| chr1_+_201117630 | 5.04 |
ENSRNOT00000077908
|
Tacc2
|
transforming, acidic coiled-coil containing protein 2 |
| chr1_-_87191588 | 5.04 |
ENSRNOT00000027998
|
LOC100361913
|
rCG54286-like |
| chr1_-_217902473 | 5.02 |
ENSRNOT00000028329
|
Ano1
|
anoctamin 1 |
| chr1_-_91855983 | 4.98 |
ENSRNOT00000017172
|
Rgs9bp
|
regulator of G protein signaling 9 binding protein |
| chr2_-_198412350 | 4.93 |
ENSRNOT00000040210
|
LOC100912489
|
histone H4-like |
| chr16_-_85305782 | 4.87 |
ENSRNOT00000067511
ENSRNOT00000076737 |
Tnfsf13b
|
tumor necrosis factor superfamily member 13b |
| chr2_-_257376756 | 4.87 |
ENSRNOT00000065811
|
Gipc2
|
GIPC PDZ domain containing family, member 2 |
| chr3_+_177397892 | 4.85 |
ENSRNOT00000078261
|
AABR07054983.1
|
|
| chr17_+_76079720 | 4.84 |
ENSRNOT00000073933
|
Proser2
|
proline and serine rich 2 |
| chr11_-_71695000 | 4.82 |
ENSRNOT00000073550
|
Ubxn7
|
UBX domain protein 7 |
| chr13_+_91334004 | 4.69 |
ENSRNOT00000041384
|
LOC108352650
|
40S ribosomal protein S29 |
| chr2_-_235852708 | 4.69 |
ENSRNOT00000009046
ENSRNOT00000005772 |
Rpl34
|
ribosomal protein L34 |
| chrX_+_159158194 | 4.68 |
ENSRNOT00000043820
ENSRNOT00000001169 ENSRNOT00000083502 |
Fhl1
|
four and a half LIM domains 1 |
| chr1_+_101517714 | 4.67 |
ENSRNOT00000028423
|
Plekha4
|
pleckstrin homology domain containing A4 |
| chr10_+_57064482 | 4.63 |
ENSRNOT00000046807
|
Zmynd15
|
zinc finger, MYND-type containing 15 |
| chr4_+_56549739 | 4.62 |
ENSRNOT00000084597
|
Hilpda
|
hypoxia inducible lipid droplet-associated |
| chr8_+_116754178 | 4.49 |
ENSRNOT00000068295
ENSRNOT00000084429 |
Uba7
|
ubiquitin-like modifier activating enzyme 7 |
| chrX_-_19386541 | 4.43 |
ENSRNOT00000072722
|
AABR07037356.1
|
|
| chr5_+_104394119 | 4.43 |
ENSRNOT00000093557
ENSRNOT00000093462 |
Adamtsl1
|
ADAMTS-like 1 |
| chr8_-_49573742 | 4.38 |
ENSRNOT00000021926
|
Il10ra
|
interleukin 10 receptor subunit alpha |
| chr6_-_125590049 | 4.36 |
ENSRNOT00000006371
|
Tc2n
|
tandem C2 domains, nuclear |
| chr11_+_64882288 | 4.29 |
ENSRNOT00000077727
|
Pla1a
|
phospholipase A1 member A |
| chr10_-_66482150 | 4.23 |
ENSRNOT00000048407
|
Ksr1
|
kinase suppressor of ras 1 |
| chr17_+_4212890 | 4.23 |
ENSRNOT00000024467
|
Testin
|
testin gene |
| chr3_-_171957996 | 4.12 |
ENSRNOT00000036546
|
Apcdd1l
|
APC down-regulated 1 like |
| chr1_-_167708685 | 4.12 |
ENSRNOT00000092857
ENSRNOT00000024992 |
Trim21
|
tripartite motif-containing 21 |
| chr3_+_91191837 | 4.11 |
ENSRNOT00000006097
|
Rag2
|
recombination activating 2 |
| chr10_-_19652898 | 4.11 |
ENSRNOT00000009648
|
Spdl1
|
spindle apparatus coiled-coil protein 1 |
| chr17_-_81002838 | 4.09 |
ENSRNOT00000089807
|
St8sia6
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 6 |
| chr3_-_95418679 | 4.08 |
ENSRNOT00000018074
|
Rcn1
|
reticulocalbin 1 |
| chr4_-_113957774 | 4.01 |
ENSRNOT00000011921
|
Ino80b
|
INO80 complex subunit B |
| chr12_-_2786628 | 4.00 |
ENSRNOT00000001348
|
RGD1564571
|
similar to DC-SIGN |
| chr4_-_114823500 | 3.99 |
ENSRNOT00000089870
|
NEWGENE_1562258
|
INO80 complex subunit B |
| chr4_+_28989115 | 3.97 |
ENSRNOT00000075326
|
Gng11
|
G protein subunit gamma 11 |
| chr2_-_149563905 | 3.97 |
ENSRNOT00000086186
|
Igsf10
|
immunoglobulin superfamily, member 10 |
| chr1_-_234704792 | 3.96 |
ENSRNOT00000017405
|
Carnmt1
|
carnosine N-methyltransferase 1 |
| chr2_+_208738132 | 3.95 |
ENSRNOT00000023972
|
AABR07012795.1
|
|
| chrX_-_123600890 | 3.95 |
ENSRNOT00000067942
|
Sept6
|
septin 6 |
| chr11_+_38035611 | 3.92 |
ENSRNOT00000087603
ENSRNOT00000002695 |
Mx2
|
MX dynamin like GTPase 2 |
| chr1_+_15062432 | 3.92 |
ENSRNOT00000016286
|
Ifngr1
|
interferon gamma receptor 1 |
| chr1_+_86972244 | 3.92 |
ENSRNOT00000036907
|
Rinl
|
Ras and Rab interactor-like |
| chr13_+_52588917 | 3.92 |
ENSRNOT00000011999
|
Phlda3
|
pleckstrin homology-like domain, family A, member 3 |
| chr2_+_60920257 | 3.92 |
ENSRNOT00000025170
|
C1qtnf3
|
C1q and tumor necrosis factor related protein 3 |
| chr16_+_41079444 | 3.90 |
ENSRNOT00000015623
|
Neil3
|
nei-like DNA glycosylase 3 |
| chr19_+_60017746 | 3.88 |
ENSRNOT00000042623
|
Pard3
|
par-3 family cell polarity regulator |
| chr7_+_144065650 | 3.87 |
ENSRNOT00000090695
|
Prr13
|
proline rich 13 |
| chr8_-_66863476 | 3.81 |
ENSRNOT00000018820
|
Rplp1
|
ribosomal protein lateral stalk subunit P1 |
| chr1_-_167347490 | 3.79 |
ENSRNOT00000076499
|
Rhog
|
ras homolog family member G |
| chr3_-_2123820 | 3.76 |
ENSRNOT00000086843
|
Ehmt1
|
euchromatic histone lysine methyltransferase 1 |
| chr15_-_12513931 | 3.72 |
ENSRNOT00000010103
|
Atxn7
|
ataxin 7 |
| chr17_-_66295014 | 3.71 |
ENSRNOT00000023974
|
Mtr
|
5-methyltetrahydrofolate-homocysteine methyltransferase |
| chr16_+_50016857 | 3.71 |
ENSRNOT00000035781
|
Tlr3
|
toll-like receptor 3 |
| chr7_-_23953002 | 3.69 |
ENSRNOT00000047587
|
RGD1563124
|
similar to 40S ribosomal protein S20 |
| chr7_+_31784438 | 3.65 |
ENSRNOT00000010914
ENSRNOT00000010929 |
Ikbip
|
IKBKB interacting protein |
| chr20_-_5476193 | 3.64 |
ENSRNOT00000044975
|
Tapbp
|
TAP binding protein |
| chrX_-_159891326 | 3.63 |
ENSRNOT00000001154
|
Rbmx
|
RNA binding motif protein, X-linked |
| chr1_-_214159157 | 3.59 |
ENSRNOT00000091292
ENSRNOT00000022241 |
Rnh1
|
ribonuclease/angiogenin inhibitor 1 |
| chr3_+_114236718 | 3.58 |
ENSRNOT00000024201
|
Duoxa2
|
dual oxidase maturation factor 2 |
| chr5_-_150653172 | 3.56 |
ENSRNOT00000072028
|
Med18
|
mediator complex subunit 18 |
| chr5_+_135351816 | 3.55 |
ENSRNOT00000082602
|
Ipp
|
intracisternal A particle-promoted polypeptide |
| chr17_-_66288202 | 3.51 |
ENSRNOT00000083172
|
Mtr
|
5-methyltetrahydrofolate-homocysteine methyltransferase |
| chr10_-_5344121 | 3.45 |
ENSRNOT00000003479
|
Nubp1
|
nucleotide binding protein 1 |
| chr8_-_126495347 | 3.42 |
ENSRNOT00000013467
|
Cmc1
|
C-x(9)-C motif containing 1 |
| chr2_+_231941794 | 3.40 |
ENSRNOT00000091475
|
AABR07013288.4
|
|
| chr18_+_68408890 | 3.32 |
ENSRNOT00000039702
|
Ccdc68
|
coiled-coil domain containing 68 |
| chr4_+_87943152 | 3.32 |
ENSRNOT00000080536
|
Vom1r78
|
vomeronasal 1 receptor 78 |
| chr4_-_97784842 | 3.31 |
ENSRNOT00000007698
|
Gadd45a
|
growth arrest and DNA-damage-inducible, alpha |
| chr9_-_54351339 | 3.30 |
ENSRNOT00000080522
|
Stat1
|
signal transducer and activator of transcription 1 |
| chr3_+_72191533 | 3.30 |
ENSRNOT00000044319
|
Ube2l6
|
ubiquitin-conjugating enzyme E2L 6 |
| chr9_-_65329066 | 3.28 |
ENSRNOT00000018284
|
Ppil3
|
peptidylprolyl isomerase like 3 |
| chr16_-_60427474 | 3.26 |
ENSRNOT00000051720
|
Ppp1r3b
|
protein phosphatase 1, regulatory subunit 3B |
Network of associatons between targets according to the STRING database.
First level regulatory network of Prdm1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 12.2 | 60.9 | GO:0002408 | myeloid dendritic cell chemotaxis(GO:0002408) |
| 8.1 | 32.3 | GO:0045627 | positive regulation of T-helper 1 cell differentiation(GO:0045627) |
| 6.9 | 20.7 | GO:0043605 | cellular amide catabolic process(GO:0043605) |
| 5.2 | 20.8 | GO:1903984 | positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 4.8 | 33.8 | GO:0045343 | MHC class I biosynthetic process(GO:0045341) regulation of MHC class I biosynthetic process(GO:0045343) positive regulation of MHC class I biosynthetic process(GO:0045345) |
| 4.0 | 12.0 | GO:0010847 | regulation of chromatin assembly(GO:0010847) protein poly-ADP-ribosylation(GO:0070212) |
| 4.0 | 11.9 | GO:0019677 | NAD catabolic process(GO:0019677) |
| 3.8 | 15.1 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 3.4 | 10.2 | GO:0002481 | antigen processing and presentation of exogenous peptide antigen via MHC class Ib(GO:0002477) antigen processing and presentation of exogenous protein antigen via MHC class Ib, TAP-dependent(GO:0002481) antigen processing and presentation of endogenous peptide antigen via MHC class Ib via ER pathway(GO:0002488) antigen processing and presentation of endogenous peptide antigen via MHC class Ib via ER pathway, TAP-dependent(GO:0002489) positive regulation of antigen processing and presentation of peptide antigen(GO:0002585) cytosol to ER transport(GO:0046967) peptide antigen transport(GO:0046968) |
| 3.4 | 10.2 | GO:0001807 | regulation of type IV hypersensitivity(GO:0001807) |
| 3.3 | 16.6 | GO:0002636 | positive regulation of germinal center formation(GO:0002636) B cell costimulation(GO:0031296) |
| 3.3 | 9.9 | GO:0072566 | chemokine (C-X-C motif) ligand 1 production(GO:0072566) regulation of chemokine (C-X-C motif) ligand 1 production(GO:2000338) |
| 3.3 | 9.8 | GO:0002554 | serotonin secretion by platelet(GO:0002554) |
| 3.1 | 9.3 | GO:0006117 | acetaldehyde metabolic process(GO:0006117) |
| 2.8 | 16.7 | GO:0010266 | response to vitamin B1(GO:0010266) maintenance of protein location in mitochondrion(GO:0072656) |
| 2.8 | 11.1 | GO:0030886 | negative regulation of myeloid dendritic cell activation(GO:0030886) |
| 2.7 | 10.7 | GO:0060857 | establishment of glial blood-brain barrier(GO:0060857) |
| 2.7 | 10.6 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 2.6 | 21.2 | GO:0070842 | aggresome assembly(GO:0070842) |
| 2.5 | 7.5 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 2.4 | 9.7 | GO:0006272 | leading strand elongation(GO:0006272) |
| 2.4 | 7.1 | GO:2001205 | negative regulation of osteoclast development(GO:2001205) |
| 2.0 | 12.1 | GO:0035990 | tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) |
| 2.0 | 8.0 | GO:1990375 | baculum development(GO:1990375) |
| 2.0 | 17.6 | GO:0003374 | dynamin polymerization involved in membrane fission(GO:0003373) dynamin polymerization involved in mitochondrial fission(GO:0003374) |
| 1.9 | 7.7 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 1.8 | 7.4 | GO:0090290 | positive regulation of osteoclast proliferation(GO:0090290) |
| 1.8 | 7.3 | GO:1901423 | response to benzene(GO:1901423) |
| 1.8 | 5.4 | GO:0042335 | cuticle development(GO:0042335) hypophysis morphogenesis(GO:0048850) |
| 1.8 | 9.0 | GO:0042270 | protection from natural killer cell mediated cytotoxicity(GO:0042270) |
| 1.8 | 10.6 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) |
| 1.8 | 5.3 | GO:0032687 | negative regulation of interferon-alpha production(GO:0032687) |
| 1.8 | 5.3 | GO:1990166 | protein localization to site of double-strand break(GO:1990166) |
| 1.6 | 14.6 | GO:0015705 | iodide transport(GO:0015705) |
| 1.6 | 7.8 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 1.6 | 10.9 | GO:0033634 | positive regulation of integrin activation(GO:0033625) positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 1.5 | 4.6 | GO:0035425 | autocrine signaling(GO:0035425) |
| 1.4 | 7.2 | GO:1904640 | response to methionine(GO:1904640) |
| 1.4 | 5.8 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 1.3 | 8.0 | GO:0097490 | trunk segmentation(GO:0035290) trunk neural crest cell migration(GO:0036484) ventral trunk neural crest cell migration(GO:0036486) sympathetic neuron projection extension(GO:0097490) sympathetic neuron projection guidance(GO:0097491) |
| 1.3 | 5.1 | GO:0002032 | desensitization of G-protein coupled receptor protein signaling pathway by arrestin(GO:0002032) |
| 1.3 | 6.3 | GO:0030421 | defecation(GO:0030421) |
| 1.3 | 21.3 | GO:0010248 | establishment or maintenance of transmembrane electrochemical gradient(GO:0010248) |
| 1.2 | 7.4 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 1.2 | 3.6 | GO:0002397 | MHC class I protein complex assembly(GO:0002397) |
| 1.2 | 3.5 | GO:0072697 | protein localization to cell cortex(GO:0072697) |
| 1.1 | 6.5 | GO:0045080 | positive regulation of chemokine biosynthetic process(GO:0045080) |
| 1.1 | 3.2 | GO:1903972 | regulation of macrophage colony-stimulating factor signaling pathway(GO:1902226) regulation of response to macrophage colony-stimulating factor(GO:1903969) regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903972) microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 1.1 | 10.7 | GO:0060340 | positive regulation of type I interferon-mediated signaling pathway(GO:0060340) |
| 1.0 | 4.1 | GO:0002331 | pre-B cell allelic exclusion(GO:0002331) |
| 1.0 | 9.0 | GO:0061303 | cornea development in camera-type eye(GO:0061303) |
| 1.0 | 5.9 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.9 | 5.4 | GO:0036289 | peptidyl-serine autophosphorylation(GO:0036289) |
| 0.8 | 5.0 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.8 | 3.3 | GO:0034240 | negative regulation of macrophage fusion(GO:0034240) |
| 0.8 | 4.1 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.8 | 3.2 | GO:1902623 | leukocyte chemotaxis involved in inflammatory response(GO:0002232) negative regulation of neutrophil migration(GO:1902623) |
| 0.8 | 11.7 | GO:0010818 | T cell chemotaxis(GO:0010818) |
| 0.8 | 4.7 | GO:0046292 | formaldehyde metabolic process(GO:0046292) |
| 0.8 | 3.8 | GO:1904401 | cellular response to Thyroid stimulating hormone(GO:1904401) |
| 0.8 | 2.3 | GO:1901740 | negative regulation of myoblast fusion(GO:1901740) |
| 0.8 | 20.3 | GO:0006855 | drug transmembrane transport(GO:0006855) |
| 0.7 | 2.2 | GO:0045716 | positive regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045716) |
| 0.7 | 7.8 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.7 | 3.9 | GO:0071638 | negative regulation of monocyte chemotactic protein-1 production(GO:0071638) |
| 0.6 | 3.9 | GO:0003383 | apical constriction(GO:0003383) |
| 0.6 | 1.9 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.6 | 10.3 | GO:0061051 | positive regulation of cell growth involved in cardiac muscle cell development(GO:0061051) |
| 0.6 | 2.5 | GO:0060474 | positive regulation of sperm motility involved in capacitation(GO:0060474) |
| 0.5 | 1.6 | GO:1990091 | sodium-dependent self proteolysis(GO:1990091) |
| 0.5 | 8.0 | GO:0043312 | neutrophil degranulation(GO:0043312) |
| 0.5 | 4.1 | GO:0090086 | negative regulation of protein deubiquitination(GO:0090086) |
| 0.5 | 3.6 | GO:2000609 | regulation of thyroid hormone generation(GO:2000609) |
| 0.5 | 9.0 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.5 | 1.5 | GO:1905006 | positive regulation of activation-induced cell death of T cells(GO:0070237) negative regulation of epithelial to mesenchymal transition involved in endocardial cushion formation(GO:1905006) |
| 0.4 | 6.3 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.4 | 6.6 | GO:1902459 | positive regulation of stem cell population maintenance(GO:1902459) |
| 0.4 | 6.1 | GO:0021684 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.4 | 2.6 | GO:0060723 | regulation of spongiotrophoblast cell proliferation(GO:0060721) regulation of cell proliferation involved in embryonic placenta development(GO:0060723) |
| 0.4 | 2.1 | GO:1900425 | negative regulation of defense response to bacterium(GO:1900425) |
| 0.4 | 5.0 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.4 | 2.3 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.4 | 3.8 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.4 | 1.9 | GO:0016557 | peroxisome membrane biogenesis(GO:0016557) |
| 0.4 | 1.1 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 0.3 | 5.5 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.3 | 2.3 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.3 | 8.3 | GO:0019884 | antigen processing and presentation of exogenous antigen(GO:0019884) |
| 0.3 | 1.6 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.3 | 1.8 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.3 | 4.9 | GO:0045653 | negative regulation of megakaryocyte differentiation(GO:0045653) |
| 0.3 | 4.1 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.3 | 1.1 | GO:1902994 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) regulation of phospholipid efflux(GO:1902994) positive regulation of phospholipid efflux(GO:1902995) |
| 0.3 | 1.4 | GO:0036123 | histone H3-K9 dimethylation(GO:0036123) |
| 0.3 | 3.3 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.3 | 2.6 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.3 | 1.3 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.2 | 5.0 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.2 | 0.7 | GO:0002462 | tolerance induction to nonself antigen(GO:0002462) |
| 0.2 | 5.7 | GO:0035067 | negative regulation of histone acetylation(GO:0035067) |
| 0.2 | 1.6 | GO:0030397 | membrane disassembly(GO:0030397) nuclear envelope disassembly(GO:0051081) |
| 0.2 | 6.5 | GO:1900027 | regulation of ruffle assembly(GO:1900027) |
| 0.2 | 0.6 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.2 | 2.1 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.2 | 3.0 | GO:1902358 | oxalate transport(GO:0019532) sulfate transmembrane transport(GO:1902358) |
| 0.2 | 15.5 | GO:0042246 | tissue regeneration(GO:0042246) |
| 0.2 | 0.5 | GO:2000567 | memory T cell activation(GO:0035709) regulation of memory T cell activation(GO:2000567) positive regulation of memory T cell activation(GO:2000568) |
| 0.2 | 0.5 | GO:1905243 | response to 3,3',5-triiodo-L-thyronine(GO:1905242) cellular response to 3,3',5-triiodo-L-thyronine(GO:1905243) |
| 0.2 | 3.7 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.2 | 2.2 | GO:0034285 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.2 | 5.5 | GO:0010718 | positive regulation of epithelial to mesenchymal transition(GO:0010718) |
| 0.1 | 7.4 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.1 | 5.2 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.1 | 0.7 | GO:0010694 | positive regulation of alkaline phosphatase activity(GO:0010694) |
| 0.1 | 1.2 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.1 | 5.2 | GO:1902475 | L-alpha-amino acid transmembrane transport(GO:1902475) |
| 0.1 | 6.1 | GO:0032435 | negative regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032435) |
| 0.1 | 4.7 | GO:0010972 | negative regulation of G2/M transition of mitotic cell cycle(GO:0010972) |
| 0.1 | 12.5 | GO:0030901 | midbrain development(GO:0030901) |
| 0.1 | 0.8 | GO:0034201 | response to oleic acid(GO:0034201) |
| 0.1 | 0.9 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.1 | 14.0 | GO:0042157 | lipoprotein metabolic process(GO:0042157) |
| 0.1 | 14.9 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.1 | 1.7 | GO:0043552 | positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
| 0.1 | 7.6 | GO:0019882 | antigen processing and presentation(GO:0019882) |
| 0.1 | 3.9 | GO:0051898 | negative regulation of protein kinase B signaling(GO:0051898) |
| 0.1 | 1.1 | GO:0002031 | G-protein coupled receptor internalization(GO:0002031) |
| 0.1 | 1.6 | GO:0010758 | regulation of macrophage chemotaxis(GO:0010758) |
| 0.1 | 1.7 | GO:0030449 | regulation of complement activation(GO:0030449) |
| 0.1 | 14.3 | GO:0007596 | blood coagulation(GO:0007596) |
| 0.1 | 3.9 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.1 | 1.6 | GO:0097286 | iron ion import(GO:0097286) |
| 0.1 | 3.3 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 0.5 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.1 | 16.3 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.1 | 0.7 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 0.1 | 2.4 | GO:0006182 | cGMP biosynthetic process(GO:0006182) |
| 0.0 | 1.7 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 0.4 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.0 | 30.8 | GO:0006412 | translation(GO:0006412) |
| 0.0 | 8.0 | GO:0030198 | extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |
| 0.0 | 2.3 | GO:0008542 | visual learning(GO:0008542) |
| 0.0 | 0.6 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.0 | 4.0 | GO:0006338 | chromatin remodeling(GO:0006338) |
| 0.0 | 0.2 | GO:0071000 | response to magnetism(GO:0071000) |
| 0.0 | 4.0 | GO:0006518 | peptide metabolic process(GO:0006518) |
| 0.0 | 3.5 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 0.9 | GO:0031648 | protein destabilization(GO:0031648) |
| 0.0 | 0.1 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.0 | 1.0 | GO:0019098 | reproductive behavior(GO:0019098) |
| 0.0 | 0.9 | GO:0060976 | coronary vasculature development(GO:0060976) |
| 0.0 | 3.6 | GO:0007286 | spermatid development(GO:0007286) |
| 0.0 | 1.2 | GO:0030500 | regulation of bone mineralization(GO:0030500) |
| 0.0 | 1.8 | GO:0051209 | release of sequestered calcium ion into cytosol(GO:0051209) negative regulation of sequestering of calcium ion(GO:0051283) |
| 0.0 | 3.2 | GO:0015992 | proton transport(GO:0015992) |
| 0.0 | 0.2 | GO:0030643 | cellular phosphate ion homeostasis(GO:0030643) cellular divalent inorganic anion homeostasis(GO:0072501) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.0 | 0.0 | GO:0032741 | positive regulation of interleukin-18 production(GO:0032741) |
| 0.0 | 4.0 | GO:0008380 | RNA splicing(GO:0008380) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.9 | 20.8 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 3.8 | 15.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 3.0 | 26.8 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 2.8 | 8.3 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 2.7 | 10.6 | GO:0020005 | symbiont-containing vacuole membrane(GO:0020005) |
| 2.6 | 15.8 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 2.0 | 13.9 | GO:0042825 | TAP complex(GO:0042825) |
| 1.8 | 21.3 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 1.3 | 5.4 | GO:1990630 | IRE1-RACK1-PP2A complex(GO:1990630) |
| 1.2 | 9.7 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 1.1 | 3.2 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 1.0 | 8.0 | GO:0044194 | cytolytic granule(GO:0044194) tertiary granule(GO:0070820) |
| 0.9 | 7.4 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.9 | 7.8 | GO:0098651 | basement membrane collagen trimer(GO:0098651) |
| 0.8 | 8.0 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.8 | 3.0 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.7 | 3.6 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.7 | 4.3 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.7 | 10.2 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.6 | 1.9 | GO:0046691 | intracellular canaliculus(GO:0046691) |
| 0.6 | 5.7 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.6 | 1.8 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.6 | 10.9 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.6 | 10.3 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.5 | 4.8 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.4 | 6.2 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.4 | 21.2 | GO:0016235 | aggresome(GO:0016235) |
| 0.4 | 2.1 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.3 | 11.1 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.3 | 3.3 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.3 | 3.9 | GO:0043219 | lateral loop(GO:0043219) |
| 0.3 | 32.7 | GO:0016605 | PML body(GO:0016605) |
| 0.3 | 7.3 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.3 | 6.3 | GO:0000145 | exocyst(GO:0000145) |
| 0.2 | 4.0 | GO:0031011 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.2 | 2.3 | GO:0042555 | MCM complex(GO:0042555) |
| 0.2 | 15.8 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.2 | 5.3 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.2 | 13.4 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.2 | 5.9 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.2 | 4.9 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.2 | 4.3 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.2 | 6.5 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.2 | 52.8 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.2 | 17.2 | GO:0005604 | basement membrane(GO:0005604) |
| 0.2 | 1.3 | GO:0000796 | condensin complex(GO:0000796) |
| 0.2 | 2.5 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.2 | 3.2 | GO:0019013 | viral nucleocapsid(GO:0019013) |
| 0.2 | 0.8 | GO:0030689 | Noc complex(GO:0030689) |
| 0.1 | 36.1 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.1 | 3.9 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 1.9 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.1 | 17.6 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.1 | 16.7 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.1 | 2.4 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.1 | 8.5 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 1.6 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 4.1 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.1 | 8.8 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.1 | 5.3 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.1 | 4.6 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 0.7 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.1 | 1.1 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.1 | 2.1 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.1 | 18.0 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.1 | 8.4 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.1 | 1.2 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.1 | 14.4 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 3.5 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 3.3 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 2.9 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.5 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.0 | 0.5 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 1.4 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.0 | 3.9 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 1.8 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 33.6 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 8.2 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 5.6 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 15.6 | GO:0031966 | mitochondrial membrane(GO:0031966) |
| 0.0 | 3.5 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 1.2 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 1.5 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 1.5 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 6.3 | GO:0000785 | chromatin(GO:0000785) |
| 0.0 | 3.3 | GO:0005635 | nuclear envelope(GO:0005635) |
| 0.0 | 10.0 | GO:0005615 | extracellular space(GO:0005615) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.4 | 26.8 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 4.2 | 12.5 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 3.7 | 14.6 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 3.4 | 10.2 | GO:0015433 | peptide antigen-transporting ATPase activity(GO:0015433) tapasin binding(GO:0046980) |
| 3.3 | 9.9 | GO:0070976 | TIR domain binding(GO:0070976) |
| 2.7 | 10.7 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 2.7 | 10.6 | GO:0019002 | GMP binding(GO:0019002) |
| 1.9 | 5.8 | GO:0016822 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 1.9 | 5.7 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 1.7 | 5.1 | GO:0031762 | follicle-stimulating hormone receptor binding(GO:0031762) type 2A serotonin receptor binding(GO:0031826) |
| 1.7 | 8.3 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 1.6 | 57.2 | GO:0008009 | chemokine activity(GO:0008009) |
| 1.6 | 9.8 | GO:0019767 | IgE receptor activity(GO:0019767) |
| 1.6 | 8.0 | GO:0031798 | type 1 metabotropic glutamate receptor binding(GO:0031798) |
| 1.6 | 8.0 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 1.5 | 9.3 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 1.5 | 21.3 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 1.5 | 11.9 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 1.4 | 9.7 | GO:0003896 | DNA primase activity(GO:0003896) |
| 1.4 | 13.6 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 1.4 | 25.8 | GO:0070628 | proteasome binding(GO:0070628) |
| 1.2 | 13.4 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 1.2 | 3.6 | GO:0046979 | TAP binding(GO:0046977) TAP1 binding(GO:0046978) TAP2 binding(GO:0046979) |
| 1.2 | 10.9 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 1.1 | 23.1 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 1.1 | 3.3 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 1.1 | 15.1 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 1.1 | 3.2 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.9 | 7.2 | GO:0031419 | S-methyltransferase activity(GO:0008172) cobalamin binding(GO:0031419) |
| 0.9 | 20.3 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 0.9 | 4.4 | GO:0004920 | interleukin-10 receptor activity(GO:0004920) |
| 0.7 | 4.3 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.7 | 17.0 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.7 | 2.0 | GO:0071208 | histone pre-mRNA DCP binding(GO:0071208) U7 snRNA binding(GO:0071209) |
| 0.6 | 9.0 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.6 | 7.5 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.6 | 8.0 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.6 | 4.2 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.6 | 5.4 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.6 | 3.6 | GO:0008428 | ribonuclease inhibitor activity(GO:0008428) |
| 0.6 | 2.3 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.6 | 5.1 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.6 | 3.9 | GO:0000405 | bubble DNA binding(GO:0000405) |
| 0.5 | 1.6 | GO:0046592 | polyamine oxidase activity(GO:0046592) |
| 0.5 | 2.0 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.5 | 41.3 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.4 | 5.0 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) |
| 0.4 | 4.5 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 0.4 | 16.8 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.4 | 10.3 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.4 | 2.6 | GO:0015119 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.4 | 3.3 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.4 | 3.3 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.4 | 5.4 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.4 | 1.8 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.3 | 8.9 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.3 | 8.0 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.3 | 7.4 | GO:0031418 | L-ascorbic acid binding(GO:0031418) |
| 0.3 | 2.3 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.3 | 5.9 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.3 | 9.0 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.3 | 1.5 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.3 | 20.7 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.3 | 1.7 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.3 | 9.7 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.2 | 7.7 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.2 | 4.7 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.2 | 0.7 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.2 | 13.2 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.2 | 60.0 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.2 | 1.1 | GO:0090554 | phosphatidylcholine-translocating ATPase activity(GO:0090554) |
| 0.2 | 3.0 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.2 | 1.7 | GO:0001846 | opsonin binding(GO:0001846) |
| 0.2 | 7.9 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.2 | 0.8 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.2 | 2.4 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.2 | 3.9 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.2 | 16.7 | GO:0005496 | steroid binding(GO:0005496) |
| 0.2 | 0.5 | GO:0023029 | MHC class Ib protein binding(GO:0023029) |
| 0.2 | 4.1 | GO:0008373 | sialyltransferase activity(GO:0008373) |
| 0.2 | 14.9 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.2 | 20.8 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.1 | 1.9 | GO:0015250 | water channel activity(GO:0015250) |
| 0.1 | 1.1 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.1 | 14.5 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.1 | 0.5 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 0.1 | 4.3 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.1 | 3.4 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 2.3 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.1 | 7.0 | GO:0050661 | NADP binding(GO:0050661) |
| 0.1 | 3.5 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.1 | 40.5 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.1 | 30.9 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.1 | 18.4 | GO:0061135 | endopeptidase inhibitor activity(GO:0004866) endopeptidase regulator activity(GO:0061135) |
| 0.1 | 2.4 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.1 | 0.7 | GO:0001848 | complement binding(GO:0001848) |
| 0.1 | 1.6 | GO:0005521 | lamin binding(GO:0005521) |
| 0.1 | 3.3 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.1 | 5.2 | GO:0015297 | antiporter activity(GO:0015297) |
| 0.1 | 4.4 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.1 | 2.1 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.1 | 2.9 | GO:0016409 | palmitoyltransferase activity(GO:0016409) |
| 0.1 | 2.2 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.1 | 3.7 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.1 | 0.9 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.1 | 0.9 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.1 | 4.2 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.1 | 4.8 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.1 | 6.0 | GO:0001076 | transcription factor activity, RNA polymerase II transcription factor binding(GO:0001076) |
| 0.0 | 3.6 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 2.6 | GO:0048029 | monosaccharide binding(GO:0048029) |
| 0.0 | 1.1 | GO:0016755 | transferase activity, transferring amino-acyl groups(GO:0016755) |
| 0.0 | 0.6 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.0 | 3.7 | GO:0019955 | cytokine binding(GO:0019955) |
| 0.0 | 6.7 | GO:0004721 | phosphoprotein phosphatase activity(GO:0004721) |
| 0.0 | 0.2 | GO:0071253 | connexin binding(GO:0071253) |
| 0.0 | 1.6 | GO:0038024 | cargo receptor activity(GO:0038024) |
| 0.0 | 11.7 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.0 | 0.9 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 5.2 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 2.5 | GO:0004540 | ribonuclease activity(GO:0004540) |
| 0.0 | 0.5 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 9.3 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 1.3 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.8 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.1 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 14.9 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.7 | 10.0 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.7 | 7.5 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.6 | 13.6 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.5 | 22.9 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.4 | 16.9 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.4 | 11.4 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.4 | 8.0 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.4 | 24.5 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.3 | 44.8 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.2 | 48.1 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.2 | 5.0 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.2 | 52.7 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.2 | 7.2 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.2 | 9.8 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.2 | 7.7 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.1 | 5.5 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.1 | 7.3 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.1 | 5.9 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.1 | 9.7 | PID E2F PATHWAY | E2F transcription factor network |
| 0.1 | 4.2 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.1 | 5.0 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 6.5 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.1 | 1.6 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.1 | 2.6 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.1 | 7.6 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.1 | 1.2 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.1 | 1.7 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 1.2 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 4.9 | NABA MATRISOME ASSOCIATED | Ensemble of genes encoding ECM-associated proteins including ECM-affilaited proteins, ECM regulators and secreted factors |
| 0.0 | 0.8 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.2 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 83.3 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 1.1 | 10.0 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 1.1 | 63.7 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.8 | 14.4 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.8 | 26.8 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.7 | 20.8 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.7 | 11.8 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.7 | 8.9 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.7 | 9.8 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.6 | 5.8 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.6 | 7.5 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.6 | 9.7 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.5 | 20.1 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.4 | 16.2 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.4 | 7.0 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.4 | 10.2 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.3 | 11.0 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.3 | 12.9 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.3 | 7.7 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.3 | 9.1 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.3 | 7.8 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.3 | 7.8 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.2 | 24.4 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.2 | 6.6 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.2 | 4.1 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.2 | 2.3 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.2 | 5.9 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.2 | 2.0 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.2 | 26.8 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |
| 0.2 | 8.0 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.2 | 1.9 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.1 | 1.7 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 1.2 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.1 | 5.6 | REACTOME VIF MEDIATED DEGRADATION OF APOBEC3G | Genes involved in Vif-mediated degradation of APOBEC3G |
| 0.1 | 5.4 | REACTOME UNFOLDED PROTEIN RESPONSE | Genes involved in Unfolded Protein Response |
| 0.1 | 1.6 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.1 | 1.2 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 0.8 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.1 | 6.9 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 1.7 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.1 | 2.3 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.1 | 1.1 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 3.0 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.6 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.1 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.0 | 0.2 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 1.4 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 1.2 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.5 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |