Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
Results for Pou2f1
Z-value: 1.00
Transcription factors associated with Pou2f1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Pou2f1
|
ENSRNOG00000003581 | POU class 2 homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Pou2f1 | rn6_v1_chr13_-_84217332_84217366 | 0.48 | 1.5e-19 | Click! |
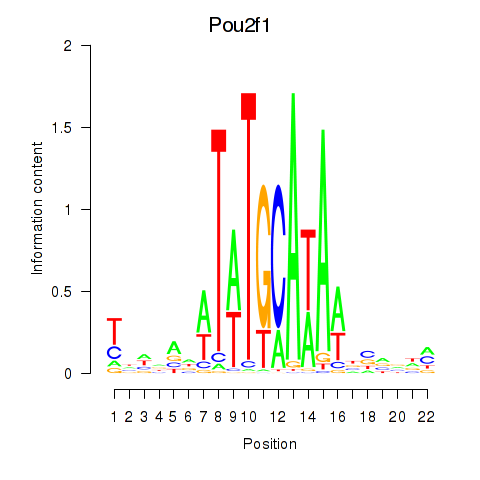
Activity profile of Pou2f1 motif
Sorted Z-values of Pou2f1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_+_57241968 | 30.60 |
ENSRNOT00000082191
|
Lcp1
|
lymphocyte cytosolic protein 1 |
| chr6_-_143065639 | 30.59 |
ENSRNOT00000070923
|
AABR07065827.1
|
|
| chr4_+_106323089 | 29.02 |
ENSRNOT00000091402
|
AABR07061134.1
|
|
| chr2_+_223029559 | 27.55 |
ENSRNOT00000045584
|
AABR07013111.1
|
|
| chr4_+_98370797 | 24.55 |
ENSRNOT00000031991
|
AABR07060872.1
|
|
| chr6_+_139486775 | 23.41 |
ENSRNOT00000077771
|
AABR07065699.3
|
|
| chr3_+_16846412 | 23.06 |
ENSRNOT00000074266
|
AABR07051551.1
|
|
| chr6_-_142676432 | 22.55 |
ENSRNOT00000074947
|
AABR07065815.2
|
|
| chr10_+_45289741 | 21.58 |
ENSRNOT00000066190
|
Hist3h2ba
|
histone cluster 3, H2ba |
| chr6_-_142880382 | 20.83 |
ENSRNOT00000074706
ENSRNOT00000072861 |
AABR07065821.1
|
|
| chr3_+_18315320 | 19.73 |
ENSRNOT00000006954
|
AABR07051626.2
|
|
| chr3_+_20641664 | 19.41 |
ENSRNOT00000044699
|
AABR07051741.1
|
|
| chr6_-_142353308 | 18.85 |
ENSRNOT00000066416
|
AABR07065814.2
|
|
| chr6_-_142585188 | 18.61 |
ENSRNOT00000067437
|
AABR07065815.1
|
|
| chr3_+_19274273 | 18.14 |
ENSRNOT00000040102
|
AABR07051684.1
|
|
| chr4_-_95970666 | 17.80 |
ENSRNOT00000008826
|
Hpgds
|
hematopoietic prostaglandin D synthase |
| chr6_-_141488290 | 17.40 |
ENSRNOT00000067336
|
AABR07065792.1
|
|
| chr4_-_103569159 | 17.12 |
ENSRNOT00000084036
|
AABR07061072.1
|
|
| chr14_-_15258207 | 16.78 |
ENSRNOT00000039383
|
Cxcl13
|
C-X-C motif chemokine ligand 13 |
| chr16_-_75481115 | 16.56 |
ENSRNOT00000035128
|
Defa7
|
defensin alpha 7 |
| chr6_-_143131118 | 16.17 |
ENSRNOT00000074930
|
AABR07065834.1
|
|
| chr3_+_20007192 | 15.90 |
ENSRNOT00000075229
|
AABR07051716.1
|
|
| chr4_+_93959152 | 15.77 |
ENSRNOT00000058437
|
AABR07060795.1
|
|
| chr17_+_44758460 | 15.76 |
ENSRNOT00000089436
|
Hist1h2an
|
histone cluster 1, H2an |
| chr3_+_17889972 | 15.68 |
ENSRNOT00000073021
|
AABR07051611.1
|
|
| chr8_-_114617466 | 15.33 |
ENSRNOT00000082992
ENSRNOT00000038160 |
Col6a5
|
collagen type VI alpha 5 chain |
| chr11_+_85532526 | 15.05 |
ENSRNOT00000036565
|
AABR07034730.2
|
|
| chr10_-_45297385 | 15.05 |
ENSRNOT00000041187
|
Hist3h2bb
|
histone cluster 3 H2B family member b |
| chr7_-_18793289 | 14.88 |
ENSRNOT00000036375
|
AABR07056026.1
|
|
| chr3_-_16999720 | 14.88 |
ENSRNOT00000074382
|
RGD1563231
|
similar to immunoglobulin kappa-chain VK-1 |
| chr3_+_19441604 | 14.81 |
ENSRNOT00000090581
|
AABR07051692.1
|
|
| chr6_-_142933110 | 14.78 |
ENSRNOT00000046885
|
AABR07065823.1
|
|
| chr6_-_143702033 | 14.68 |
ENSRNOT00000051410
|
AABR07065886.1
|
|
| chr3_+_19045214 | 14.56 |
ENSRNOT00000070878
|
AABR07051670.1
|
|
| chr8_+_82038967 | 14.38 |
ENSRNOT00000079535
|
Myo5a
|
myosin VA |
| chr1_+_63734135 | 14.37 |
ENSRNOT00000084439
|
Nilr1
|
neutrophil immunoglobulin-like receptor-1 |
| chr4_+_102665529 | 14.32 |
ENSRNOT00000082333
|
AABR07061005.1
|
|
| chr6_-_131926272 | 13.74 |
ENSRNOT00000084057
ENSRNOT00000088421 |
Bcl11b
|
B-cell CLL/lymphoma 11B |
| chr3_+_19690016 | 13.73 |
ENSRNOT00000085460
|
AABR07051707.1
|
|
| chr4_-_69268336 | 13.31 |
ENSRNOT00000018042
|
Prss3b
|
protease, serine, 3B |
| chr3_+_20303979 | 13.13 |
ENSRNOT00000058331
|
AABR07051730.1
|
|
| chr3_+_17546566 | 12.84 |
ENSRNOT00000050825
|
AABR07051583.1
|
|
| chr5_-_169658875 | 12.72 |
ENSRNOT00000015840
|
Kcnab2
|
potassium voltage-gated channel subfamily A regulatory beta subunit 2 |
| chr17_-_44748188 | 12.60 |
ENSRNOT00000081970
|
LOC103690190
|
histone H2A type 1-E |
| chr2_-_35104963 | 12.44 |
ENSRNOT00000018058
|
Rgs7bp
|
regulator of G-protein signaling 7-binding protein |
| chr11_+_85042348 | 12.39 |
ENSRNOT00000042220
|
AABR07034718.1
|
|
| chr14_+_48768537 | 12.22 |
ENSRNOT00000082599
|
Arap2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr5_-_75319765 | 12.11 |
ENSRNOT00000085698
|
Svep1
|
sushi, von Willebrand factor type A, EGF and pentraxin domain containing 1 |
| chr8_+_5606592 | 11.83 |
ENSRNOT00000011727
|
Mmp12
|
matrix metallopeptidase 12 |
| chr2_+_186703541 | 11.57 |
ENSRNOT00000093342
|
Fcrl1
|
Fc receptor-like 1 |
| chr4_-_102124609 | 11.53 |
ENSRNOT00000048263
|
AABR07060979.1
|
|
| chr4_+_94696965 | 11.44 |
ENSRNOT00000064696
|
Grid2
|
glutamate ionotropic receptor delta type subunit 2 |
| chr8_-_49308806 | 11.37 |
ENSRNOT00000047291
|
Cd3e
|
CD3e molecule |
| chr3_+_17139670 | 11.35 |
ENSRNOT00000073316
|
AABR07051564.1
|
|
| chr4_+_101909389 | 11.29 |
ENSRNOT00000086458
|
AABR07060963.2
|
|
| chr6_-_54059119 | 10.66 |
ENSRNOT00000005521
|
Hdac9
|
histone deacetylase 9 |
| chr4_+_102068556 | 10.57 |
ENSRNOT00000077412
|
AABR07060971.1
|
|
| chr5_+_152721940 | 10.55 |
ENSRNOT00000039322
|
Aunip
|
aurora kinase A and ninein interacting protein |
| chr4_-_103761881 | 10.52 |
ENSRNOT00000084103
|
AABR07061087.1
|
|
| chr11_+_85992696 | 10.46 |
ENSRNOT00000084433
|
AABR07034736.1
|
|
| chr4_+_78496043 | 10.43 |
ENSRNOT00000059136
|
Aoc1
|
amine oxidase, copper containing 1 |
| chr17_+_44738643 | 10.32 |
ENSRNOT00000087643
|
LOC100910554
|
histone H2A type 1-like |
| chr6_-_99783047 | 10.18 |
ENSRNOT00000009028
|
Sptb
|
spectrin, beta, erythrocytic |
| chr1_-_116153722 | 10.14 |
ENSRNOT00000041605
|
Fpr3
|
formyl peptide receptor 3 |
| chr4_-_163762434 | 9.95 |
ENSRNOT00000081854
|
Ly49si1
|
immunoreceptor Ly49si1 |
| chr17_-_44738330 | 9.84 |
ENSRNOT00000072195
|
LOC100364835
|
histone cluster 1, H2bd-like |
| chr11_+_85508300 | 9.69 |
ENSRNOT00000038646
|
AABR07034730.3
|
|
| chr9_+_8052210 | 9.49 |
ENSRNOT00000073659
|
Adgre4
|
adhesion G protein-coupled receptor E4 |
| chr17_+_43632397 | 9.47 |
ENSRNOT00000013790
|
Hist1h2ah
|
histone cluster 1, H2ah |
| chr4_+_101949285 | 9.16 |
ENSRNOT00000058446
|
AABR07060963.3
|
|
| chr9_+_8054466 | 9.04 |
ENSRNOT00000081513
|
Adgre4
|
adhesion G protein-coupled receptor E4 |
| chr4_+_101687327 | 8.95 |
ENSRNOT00000082501
|
AABR07060957.1
|
|
| chr1_-_198486157 | 8.85 |
ENSRNOT00000022758
|
Zg16
|
zymogen granule protein 16 |
| chr4_-_162189994 | 8.71 |
ENSRNOT00000045501
|
LOC689757
|
similar to osteoclast inhibitory lectin |
| chr4_-_163049084 | 8.71 |
ENSRNOT00000091644
|
Cd69
|
Cd69 molecule |
| chr1_+_188420461 | 8.69 |
ENSRNOT00000037870
|
Ccp110
|
centriolar coiled-coil protein 110 |
| chr5_+_10178302 | 8.67 |
ENSRNOT00000009679
|
Sntg1
|
syntrophin, gamma 1 |
| chr17_+_78764506 | 8.40 |
ENSRNOT00000077953
|
Suv39h2
|
suppressor of variegation 3-9 homolog 2 |
| chr6_-_143206772 | 8.38 |
ENSRNOT00000073713
|
AABR07065839.1
|
|
| chr1_+_114046478 | 8.34 |
ENSRNOT00000032254
|
Siglech
|
sialic acid binding Ig-like lectin H |
| chr4_-_163423628 | 8.29 |
ENSRNOT00000079526
|
Klrc3
|
killer cell lectin-like receptor subfamily C, member 3 |
| chr13_+_89386023 | 8.05 |
ENSRNOT00000086223
|
Fcgr3a
|
Fc fragment of IgG receptor IIIa |
| chr18_+_30474947 | 7.89 |
ENSRNOT00000027188
|
Pcdhb9
|
protocadherin beta 9 |
| chr2_+_247299433 | 7.82 |
ENSRNOT00000050330
|
Unc5c
|
unc-5 netrin receptor C |
| chr1_+_227640680 | 7.77 |
ENSRNOT00000033613
|
Ms4a4c
|
membrane-spanning 4-domains, subfamily A, member 4C |
| chr14_-_28856214 | 7.74 |
ENSRNOT00000044659
|
Adgrl3
|
adhesion G protein-coupled receptor L3 |
| chr16_-_31301880 | 7.73 |
ENSRNOT00000084847
ENSRNOT00000083943 |
AABR07025272.1
|
|
| chr4_-_135069970 | 7.70 |
ENSRNOT00000008221
|
Cntn3
|
contactin 3 |
| chr1_-_17378047 | 7.67 |
ENSRNOT00000020102
|
Themis
|
thymocyte selection associated |
| chr4_-_128266082 | 7.51 |
ENSRNOT00000039549
|
Nup50
|
nucleoporin 50 |
| chr17_-_31916553 | 7.43 |
ENSRNOT00000074220
|
LOC100911032
|
uncharacterized LOC100911032 |
| chr7_+_11356118 | 7.38 |
ENSRNOT00000041325
|
Atcay
|
ATCAY, caytaxin |
| chr9_-_119869639 | 7.37 |
ENSRNOT00000080498
|
Ndc80
|
NDC80 kinetochore complex component |
| chr8_-_23148396 | 7.37 |
ENSRNOT00000075237
|
Acp5
|
acid phosphatase 5, tartrate resistant |
| chr8_+_58347736 | 7.32 |
ENSRNOT00000080227
ENSRNOT00000066222 |
Slc35f2
|
solute carrier family 35, member F2 |
| chr4_+_101531378 | 7.32 |
ENSRNOT00000087546
|
AABR07060944.1
|
|
| chr2_-_41871858 | 7.18 |
ENSRNOT00000039720
|
Gapt
|
Grb2-binding adaptor protein, transmembrane |
| chr10_+_42614713 | 7.15 |
ENSRNOT00000081136
ENSRNOT00000073148 |
Gria1
|
glutamate ionotropic receptor AMPA type subunit 1 |
| chr4_-_70628470 | 7.13 |
ENSRNOT00000029319
|
Try5
|
trypsin 5 |
| chr8_+_107875991 | 7.10 |
ENSRNOT00000090299
|
Dzip1l
|
DAZ interacting zinc finger protein 1-like |
| chr6_-_143768985 | 7.00 |
ENSRNOT00000046694
|
Ighv15-2
|
immunoglobulin heavy variable V15-2 |
| chr1_+_156552328 | 7.00 |
ENSRNOT00000055401
|
Dlg2
|
discs large MAGUK scaffold protein 2 |
| chr4_+_69457472 | 6.98 |
ENSRNOT00000067597
|
Trbv19
|
T cell receptor beta, variable 19 |
| chr8_-_13906355 | 6.98 |
ENSRNOT00000029634
|
Cep295
|
centrosomal protein 295 |
| chr16_-_48921242 | 6.83 |
ENSRNOT00000041596
|
Cenpu
|
centromere protein U |
| chr5_+_122019301 | 6.81 |
ENSRNOT00000068158
|
Pde4b
|
phosphodiesterase 4B |
| chr3_+_19366370 | 6.72 |
ENSRNOT00000086557
|
AABR07051689.1
|
|
| chr15_-_34647421 | 6.70 |
ENSRNOT00000072426
|
Mcpt8
|
mast cell protease 8 |
| chr1_-_73226777 | 6.63 |
ENSRNOT00000025080
|
Ncr1
|
natural cytotoxicity triggering receptor 1 |
| chr4_+_69138525 | 6.61 |
ENSRNOT00000073589
|
Trbv1
|
T cell receptor beta, variable 1 |
| chr15_-_34694180 | 6.53 |
ENSRNOT00000079505
ENSRNOT00000027950 ENSRNOT00000079782 ENSRNOT00000080221 |
Mcpt8
|
mast cell protease 8 |
| chr4_-_108717309 | 6.51 |
ENSRNOT00000085062
|
AABR07061178.1
|
|
| chr3_-_7203420 | 6.49 |
ENSRNOT00000015236
|
Gfi1b
|
growth factor independent 1B transcriptional repressor |
| chr15_-_34625121 | 6.48 |
ENSRNOT00000073555
|
Mcpt8l2
|
mast cell protease 8-like 2 |
| chr10_+_61685241 | 6.46 |
ENSRNOT00000092606
|
Mnt
|
MAX network transcriptional repressor |
| chr2_-_250778269 | 6.43 |
ENSRNOT00000085670
ENSRNOT00000083535 ENSRNOT00000017824 |
Clca5
|
chloride channel accessory 5 |
| chr10_-_18574710 | 6.42 |
ENSRNOT00000079031
|
Kcnip1
|
potassium voltage-gated channel interacting protein 1 |
| chrX_+_96863891 | 6.39 |
ENSRNOT00000085665
|
AABR07040284.1
|
|
| chr4_-_164691405 | 6.38 |
ENSRNOT00000090979
ENSRNOT00000091932 ENSRNOT00000078219 |
Ly49s4
Ly49i2
|
Ly49 stimulatory receptor 4 Ly49 inhibitory receptor 2 |
| chr6_-_141198565 | 6.38 |
ENSRNOT00000064361
|
AABR07065782.1
|
|
| chr20_+_4043741 | 6.33 |
ENSRNOT00000000525
|
RT1-Bb
|
RT1 class II, locus Bb |
| chr6_+_111076351 | 6.30 |
ENSRNOT00000089644
|
Tmem63c
|
transmembrane protein 63c |
| chr16_+_39909270 | 6.29 |
ENSRNOT00000081994
|
Wdr17
|
WD repeat domain 17 |
| chr14_-_21252538 | 6.24 |
ENSRNOT00000005003
|
Ambn
|
ameloblastin |
| chr6_-_142060032 | 6.22 |
ENSRNOT00000064717
|
AABR07065811.1
|
|
| chr6_+_45683234 | 6.22 |
ENSRNOT00000052003
|
Cmpk2
|
cytidine/uridine monophosphate kinase 2 |
| chr8_+_39305128 | 6.20 |
ENSRNOT00000008285
|
Fez1
|
fasciculation and elongation protein zeta 1 |
| chr1_+_151439409 | 6.20 |
ENSRNOT00000022060
ENSRNOT00000050639 |
Grm5
|
glutamate metabotropic receptor 5 |
| chr2_-_165600748 | 6.15 |
ENSRNOT00000013216
|
Ift80
|
intraflagellar transport 80 |
| chr4_+_162437089 | 6.10 |
ENSRNOT00000038801
|
Clec2d2
|
C-type lectin domain family 2 member D2 |
| chr9_-_85243001 | 6.05 |
ENSRNOT00000020219
|
Scg2
|
secretogranin II |
| chr9_+_92681078 | 6.01 |
ENSRNOT00000034152
|
Sp100
|
SP100 nuclear antigen |
| chr4_-_165118062 | 5.95 |
ENSRNOT00000087219
|
LOC502908
|
Ly49s8 |
| chr6_-_143537030 | 5.89 |
ENSRNOT00000071876
|
LOC100360610
|
rCG64220-like |
| chr6_+_100337226 | 5.71 |
ENSRNOT00000011220
|
Fut8
|
fucosyltransferase 8 |
| chrX_+_118197217 | 5.62 |
ENSRNOT00000090922
|
Htr2c
|
5-hydroxytryptamine receptor 2C |
| chr18_+_30398113 | 5.59 |
ENSRNOT00000027206
|
Pcdhb5
|
protocadherin beta 5 |
| chr5_-_150723321 | 5.57 |
ENSRNOT00000018038
|
Atpif1
|
ATPase inhibitory factor 1 |
| chr18_+_30387937 | 5.57 |
ENSRNOT00000027210
|
Pcdhb4
|
protocadherin beta 4 |
| chr2_-_184263564 | 5.55 |
ENSRNOT00000015279
|
Fbxw7
|
F-box and WD repeat domain containing 7 |
| chr9_+_91001828 | 5.40 |
ENSRNOT00000080956
|
AABR07068214.1
|
|
| chr15_+_32832035 | 5.35 |
ENSRNOT00000060250
|
AABR07017902.1
|
|
| chr4_+_147333056 | 5.33 |
ENSRNOT00000012137
|
Pparg
|
peroxisome proliferator-activated receptor gamma |
| chr3_+_19205460 | 5.33 |
ENSRNOT00000070869
|
Igkv4-80
|
immunoglobulin kappa variable 4-80 |
| chr11_+_42259761 | 5.30 |
ENSRNOT00000047310
|
Epha6
|
Eph receptor A6 |
| chr15_+_41643541 | 5.29 |
ENSRNOT00000019646
|
Arl11
|
ADP-ribosylation factor like GTPase 11 |
| chr3_-_173944396 | 5.29 |
ENSRNOT00000079019
|
Sycp2
|
synaptonemal complex protein 2 |
| chr4_+_153330069 | 5.29 |
ENSRNOT00000015560
|
Slc25a18
|
solute carrier family 25 member 18 |
| chr1_-_62558033 | 5.27 |
ENSRNOT00000015520
|
Zfp40
|
zinc finger protein 40 |
| chr14_+_48740190 | 5.23 |
ENSRNOT00000031638
|
Arap2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr17_+_44520537 | 5.21 |
ENSRNOT00000077985
|
hist1h2ail2
|
histone cluster 1, H2ai-like2 |
| chr8_-_21481735 | 5.14 |
ENSRNOT00000038069
|
Zfp426
|
zinc finger protein 426 |
| chr13_-_49487892 | 5.11 |
ENSRNOT00000044972
|
Nfasc
|
neurofascin |
| chr2_-_173668555 | 5.10 |
ENSRNOT00000013452
|
Serpini2
|
serpin family I member 2 |
| chr17_-_81002838 | 5.07 |
ENSRNOT00000089807
|
St8sia6
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 6 |
| chr14_+_94590765 | 5.03 |
ENSRNOT00000072533
|
LOC690700
|
similar to similar to RIKEN cDNA 1700001E04 |
| chr2_+_206392200 | 5.03 |
ENSRNOT00000026631
|
Rsbn1
|
round spermatid basic protein 1 |
| chrX_+_120901495 | 5.02 |
ENSRNOT00000040742
|
Wdr44
|
WD repeat domain 44 |
| chr3_-_66335869 | 5.00 |
ENSRNOT00000079781
ENSRNOT00000043238 ENSRNOT00000073412 ENSRNOT00000057878 ENSRNOT00000079212 |
Cerkl
|
ceramide kinase-like |
| chr2_+_86891092 | 4.92 |
ENSRNOT00000077162
ENSRNOT00000083066 |
LOC100360380
|
zinc finger protein 457-like |
| chr13_+_43850751 | 4.88 |
ENSRNOT00000004995
|
Mgat5
|
mannosyl (alpha-1,6-)-glycoprotein beta-1,6-N-acetyl-glucosaminyltransferase |
| chr3_-_48831417 | 4.87 |
ENSRNOT00000009920
ENSRNOT00000085246 |
Kcnh7
|
potassium voltage-gated channel subfamily H member 7 |
| chr4_+_138269142 | 4.77 |
ENSRNOT00000007788
|
Cntn4
|
contactin 4 |
| chrX_+_65566047 | 4.75 |
ENSRNOT00000092103
|
Heph
|
hephaestin |
| chr13_-_79890134 | 4.74 |
ENSRNOT00000083043
|
RGD1309106
|
similar to hypothetical protein |
| chr2_-_140387505 | 4.71 |
ENSRNOT00000047635
|
Elf2
|
E74-like factor 2 |
| chr1_+_55219773 | 4.65 |
ENSRNOT00000041610
|
RGD1561667
|
similar to putative protein kinase |
| chr6_+_22167919 | 4.64 |
ENSRNOT00000007655
|
Nlrc4
|
NLR family, CARD domain containing 4 |
| chr15_+_75067297 | 4.64 |
ENSRNOT00000057931
|
LOC498555
|
similar to 60S acidic ribosomal protein P2 |
| chr12_-_23727535 | 4.57 |
ENSRNOT00000085911
ENSRNOT00000001950 |
Dtx2
|
deltex E3 ubiquitin ligase 2 |
| chr6_-_76535517 | 4.57 |
ENSRNOT00000083857
|
Ralgapa1
|
Ral GTPase activating protein catalytic alpha subunit 1 |
| chr4_+_136676254 | 4.55 |
ENSRNOT00000088070
|
Cntn6
|
contactin 6 |
| chr9_+_110721322 | 4.52 |
ENSRNOT00000079170
|
AABR07068593.1
|
|
| chr19_-_914880 | 4.51 |
ENSRNOT00000017127
|
Cklf
|
chemokine-like factor |
| chrX_+_65563122 | 4.47 |
ENSRNOT00000017312
|
Heph
|
hephaestin |
| chr4_+_31229913 | 4.47 |
ENSRNOT00000077134
ENSRNOT00000087897 |
Samd9l
|
sterile alpha motif domain containing 9-like |
| chr12_-_22021851 | 4.47 |
ENSRNOT00000039280
|
Tsc22d4
|
TSC22 domain family, member 4 |
| chr2_-_173563273 | 4.46 |
ENSRNOT00000081423
|
Zbbx
|
zinc finger, B-box domain containing |
| chr13_+_57243877 | 4.46 |
ENSRNOT00000083693
|
Kcnt2
|
potassium sodium-activated channel subfamily T member 2 |
| chr7_+_78188912 | 4.43 |
ENSRNOT00000043079
|
Rims2
|
regulating synaptic membrane exocytosis 2 |
| chr4_+_136512201 | 4.39 |
ENSRNOT00000051645
|
Cntn6
|
contactin 6 |
| chr15_+_31642169 | 4.34 |
ENSRNOT00000072362
|
AABR07017833.1
|
|
| chr13_+_48455923 | 4.31 |
ENSRNOT00000009280
|
Rab7b
|
Rab7b, member RAS oncogene family |
| chr7_+_44009069 | 4.29 |
ENSRNOT00000005523
|
Mgat4c
|
MGAT4 family, member C |
| chr18_+_30890869 | 4.26 |
ENSRNOT00000060466
|
Pcdhgb6
|
protocadherin gamma subfamily B, 6 |
| chr7_+_48867664 | 4.24 |
ENSRNOT00000005862
|
Ppfia2
|
PTPRF interacting protein alpha 2 |
| chr2_-_170301348 | 4.22 |
ENSRNOT00000088131
|
Si
|
sucrase-isomaltase |
| chr14_-_72025137 | 4.22 |
ENSRNOT00000080604
|
C1qtnf7
|
C1q and tumor necrosis factor related protein 7 |
| chr6_+_97110158 | 4.21 |
ENSRNOT00000088430
|
Syt16
|
synaptotagmin 16 |
| chr4_-_11610518 | 4.16 |
ENSRNOT00000066643
|
Magi2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr4_+_163174487 | 4.15 |
ENSRNOT00000088108
|
Clec9a
|
C-type lectin domain family 9, member A |
| chr1_+_99561523 | 4.13 |
ENSRNOT00000038229
|
Ceacam18
|
carcinoembryonic antigen-related cell adhesion molecule 18 |
| chr9_+_1313209 | 4.09 |
ENSRNOT00000083387
|
Tbc1d5
|
TBC1 domain family, member 5 |
| chr13_+_50873605 | 4.08 |
ENSRNOT00000004382
|
Fmod
|
fibromodulin |
| chr3_-_147188929 | 4.07 |
ENSRNOT00000055455
|
Rad21l1
|
RAD21 cohesin complex component like 1 |
| chr9_-_9142339 | 4.06 |
ENSRNOT00000046852
|
MGC116197
|
similar to RIKEN cDNA 1700001E04 |
| chr4_-_87873247 | 4.05 |
ENSRNOT00000044351
|
Vom1r77
|
vomeronasal 1 receptor 77 |
| chr2_-_157066781 | 4.04 |
ENSRNOT00000001187
|
LOC304239
|
similar to RalA binding protein 1 |
| chr3_+_149466770 | 4.03 |
ENSRNOT00000065104
|
Bpifa6
|
BPI fold containing family A, member 6 |
| chr4_-_164899041 | 3.99 |
ENSRNOT00000090316
|
Ly49i2
|
Ly49 inhibitory receptor 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Pou2f1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.4 | 17.8 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 4.2 | 16.8 | GO:2001027 | negative regulation of endothelial cell chemotaxis(GO:2001027) |
| 3.6 | 14.4 | GO:0051643 | endoplasmic reticulum localization(GO:0051643) |
| 3.0 | 11.8 | GO:0072642 | type I interferon secretion(GO:0072641) interferon-alpha secretion(GO:0072642) regulation of interferon-alpha secretion(GO:1902739) positive regulation of interferon-alpha secretion(GO:1902741) |
| 2.9 | 8.7 | GO:0032053 | ciliary basal body organization(GO:0032053) |
| 2.5 | 7.4 | GO:0045763 | negative regulation of cellular amino acid metabolic process(GO:0045763) |
| 2.3 | 13.7 | GO:0002681 | somatic diversification of T cell receptor genes(GO:0002568) somatic recombination of T cell receptor gene segments(GO:0002681) T cell receptor V(D)J recombination(GO:0033153) |
| 2.1 | 6.2 | GO:0046077 | dUDP biosynthetic process(GO:0006227) pyrimidine nucleoside diphosphate biosynthetic process(GO:0009139) pyrimidine deoxyribonucleoside diphosphate metabolic process(GO:0009196) pyrimidine deoxyribonucleoside diphosphate biosynthetic process(GO:0009197) dUDP metabolic process(GO:0046077) |
| 2.1 | 6.2 | GO:0099552 | trans-synaptic signaling by lipid, modulating synaptic transmission(GO:0099552) trans-synaptic signaling by endocannabinoid, modulating synaptic transmission(GO:0099553) |
| 2.0 | 30.6 | GO:0051764 | actin filament network formation(GO:0051639) actin crosslink formation(GO:0051764) |
| 2.0 | 6.0 | GO:1902044 | regulation of Fas signaling pathway(GO:1902044) |
| 2.0 | 3.9 | GO:0071226 | cellular response to molecule of fungal origin(GO:0071226) |
| 1.9 | 11.4 | GO:0002669 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 1.9 | 5.6 | GO:0031583 | phospholipase C-activating serotonin receptor signaling pathway(GO:0007208) phospholipase D-activating G-protein coupled receptor signaling pathway(GO:0031583) |
| 1.9 | 5.6 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 1.8 | 5.3 | GO:2000230 | negative regulation of pancreatic stellate cell proliferation(GO:2000230) |
| 1.7 | 6.6 | GO:0009597 | detection of virus(GO:0009597) |
| 1.6 | 6.3 | GO:0002343 | peripheral B cell selection(GO:0002343) B cell affinity maturation(GO:0002344) |
| 1.4 | 4.3 | GO:0034499 | late endosome to Golgi transport(GO:0034499) |
| 1.4 | 5.7 | GO:0046368 | GDP-L-fucose metabolic process(GO:0046368) |
| 1.4 | 5.6 | GO:1901526 | positive regulation of macromitophagy(GO:1901526) positive regulation of mitophagy in response to mitochondrial depolarization(GO:1904925) |
| 1.4 | 13.7 | GO:0034983 | peptidyl-lysine deacetylation(GO:0034983) |
| 1.3 | 4.0 | GO:0015688 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 1.3 | 7.8 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 1.3 | 3.9 | GO:0021849 | neuroblast division in subventricular zone(GO:0021849) |
| 1.3 | 12.7 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 1.2 | 7.4 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 1.2 | 3.6 | GO:0009177 | pyrimidine deoxyribonucleoside monophosphate biosynthetic process(GO:0009177) dUMP metabolic process(GO:0046078) |
| 1.2 | 7.2 | GO:0002322 | B cell proliferation involved in immune response(GO:0002322) |
| 1.2 | 7.1 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 1.1 | 6.8 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 1.1 | 5.3 | GO:0032707 | negative regulation of interleukin-23 production(GO:0032707) |
| 1.1 | 8.4 | GO:0036123 | histone H3-K9 dimethylation(GO:0036123) |
| 1.0 | 3.1 | GO:1901740 | negative regulation of myoblast fusion(GO:1901740) |
| 1.0 | 5.1 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 1.0 | 2.9 | GO:0021836 | chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) |
| 1.0 | 3.8 | GO:0006272 | leading strand elongation(GO:0006272) |
| 0.9 | 10.1 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.9 | 7.4 | GO:0090267 | positive regulation of spindle checkpoint(GO:0090232) positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) |
| 0.9 | 2.8 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.9 | 2.7 | GO:0072720 | response to dithiothreitol(GO:0072720) |
| 0.9 | 5.3 | GO:0090598 | male genitalia morphogenesis(GO:0048808) male anatomical structure morphogenesis(GO:0090598) |
| 0.9 | 5.1 | GO:0071205 | protein localization to juxtaparanode region of axon(GO:0071205) |
| 0.8 | 11.4 | GO:0021684 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.8 | 6.5 | GO:0051572 | negative regulation of histone H3-K4 methylation(GO:0051572) |
| 0.8 | 3.0 | GO:1900063 | regulation of peroxisome organization(GO:1900063) |
| 0.7 | 10.4 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.7 | 9.8 | GO:0038095 | Fc-epsilon receptor signaling pathway(GO:0038095) |
| 0.7 | 4.2 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.7 | 6.1 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.7 | 2.7 | GO:0036394 | amylase secretion(GO:0036394) |
| 0.7 | 4.6 | GO:0070269 | pyroptosis(GO:0070269) |
| 0.6 | 1.9 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.6 | 1.9 | GO:0010956 | negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) |
| 0.6 | 7.0 | GO:0046710 | GDP metabolic process(GO:0046710) |
| 0.6 | 8.7 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.6 | 54.7 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.6 | 2.8 | GO:1990918 | meiotic DNA recombinase assembly(GO:0000707) double-strand break repair involved in meiotic recombination(GO:1990918) |
| 0.5 | 2.2 | GO:1903999 | negative regulation of eating behavior(GO:1903999) regulation of locomotor rhythm(GO:1904059) |
| 0.5 | 1.6 | GO:2000567 | defense response to nematode(GO:0002215) memory T cell activation(GO:0035709) positive regulation of memory T cell differentiation(GO:0043382) regulation of memory T cell activation(GO:2000567) positive regulation of memory T cell activation(GO:2000568) |
| 0.5 | 6.2 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.5 | 2.8 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.5 | 7.7 | GO:0031987 | locomotion involved in locomotory behavior(GO:0031987) |
| 0.4 | 4.5 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.4 | 2.6 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.4 | 6.0 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 0.4 | 2.5 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.4 | 9.2 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.4 | 5.5 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.4 | 1.1 | GO:1905218 | cellular response to astaxanthin(GO:1905218) |
| 0.4 | 4.1 | GO:0044821 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.4 | 10.2 | GO:0006779 | porphyrin-containing compound biosynthetic process(GO:0006779) |
| 0.4 | 5.8 | GO:0043383 | negative T cell selection(GO:0043383) |
| 0.4 | 1.8 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.3 | 4.4 | GO:0098814 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.3 | 1.7 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.3 | 1.3 | GO:0051344 | negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.3 | 30.4 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.3 | 1.7 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.3 | 5.9 | GO:0045760 | positive regulation of action potential(GO:0045760) |
| 0.3 | 1.3 | GO:1903896 | positive regulation of IRE1-mediated unfolded protein response(GO:1903896) positive regulation of PERK-mediated unfolded protein response(GO:1903899) |
| 0.3 | 4.3 | GO:0006999 | nuclear pore organization(GO:0006999) |
| 0.2 | 0.7 | GO:1902953 | positive regulation of ER to Golgi vesicle-mediated transport(GO:1902953) |
| 0.2 | 1.4 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.2 | 4.7 | GO:0035404 | histone-serine phosphorylation(GO:0035404) |
| 0.2 | 3.6 | GO:0042136 | neurotransmitter biosynthetic process(GO:0042136) |
| 0.2 | 2.1 | GO:0060693 | regulation of branching involved in salivary gland morphogenesis(GO:0060693) |
| 0.2 | 0.6 | GO:1901295 | arterial endothelial cell fate commitment(GO:0060844) blood vessel endothelial cell fate commitment(GO:0060846) blood vessel lumenization(GO:0072554) blood vessel endothelial cell fate specification(GO:0097101) regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901295) regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) |
| 0.2 | 3.1 | GO:0021860 | pyramidal neuron development(GO:0021860) |
| 0.2 | 1.4 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) positive regulation of energy homeostasis(GO:2000507) |
| 0.2 | 0.8 | GO:0038110 | interleukin-2-mediated signaling pathway(GO:0038110) |
| 0.2 | 1.1 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.2 | 6.2 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.2 | 3.7 | GO:0008340 | determination of adult lifespan(GO:0008340) |
| 0.2 | 3.0 | GO:2000780 | negative regulation of double-strand break repair(GO:2000780) |
| 0.2 | 6.1 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.2 | 2.3 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.1 | 2.5 | GO:0009750 | response to fructose(GO:0009750) |
| 0.1 | 3.3 | GO:0048520 | positive regulation of behavior(GO:0048520) |
| 0.1 | 5.3 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.1 | 1.0 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.1 | 5.8 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.1 | 2.6 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.1 | 2.1 | GO:0006271 | DNA strand elongation involved in DNA replication(GO:0006271) |
| 0.1 | 2.0 | GO:0010763 | positive regulation of fibroblast migration(GO:0010763) |
| 0.1 | 2.6 | GO:0051567 | histone H3-K9 methylation(GO:0051567) |
| 0.1 | 1.7 | GO:0043518 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) |
| 0.1 | 0.9 | GO:0042073 | intraciliary transport(GO:0042073) |
| 0.1 | 2.1 | GO:0051895 | negative regulation of focal adhesion assembly(GO:0051895) |
| 0.1 | 19.5 | GO:0007416 | synapse assembly(GO:0007416) |
| 0.1 | 0.7 | GO:2000381 | negative regulation of mesoderm development(GO:2000381) |
| 0.1 | 7.4 | GO:0030148 | sphingolipid biosynthetic process(GO:0030148) |
| 0.1 | 0.4 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.1 | 1.4 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.1 | 3.1 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.1 | 1.1 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.1 | 9.0 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.1 | 1.7 | GO:0071044 | histone mRNA catabolic process(GO:0071044) |
| 0.1 | 3.0 | GO:0030318 | melanocyte differentiation(GO:0030318) |
| 0.1 | 0.6 | GO:0061304 | retinal blood vessel morphogenesis(GO:0061304) |
| 0.1 | 1.4 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.1 | 4.2 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.1 | 0.7 | GO:0045059 | positive thymic T cell selection(GO:0045059) |
| 0.1 | 1.4 | GO:0001953 | negative regulation of cell-matrix adhesion(GO:0001953) |
| 0.1 | 0.5 | GO:0071233 | cellular response to leucine(GO:0071233) |
| 0.1 | 1.1 | GO:0021702 | cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.1 | 0.4 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.1 | 6.8 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.1 | 1.7 | GO:0038202 | TORC1 signaling(GO:0038202) |
| 0.1 | 0.8 | GO:0014049 | positive regulation of glutamate secretion(GO:0014049) |
| 0.1 | 6.3 | GO:0042246 | tissue regeneration(GO:0042246) |
| 0.1 | 0.7 | GO:0071675 | positive regulation of macrophage chemotaxis(GO:0010759) mononuclear cell migration(GO:0071674) regulation of mononuclear cell migration(GO:0071675) |
| 0.1 | 2.6 | GO:0050732 | negative regulation of peptidyl-tyrosine phosphorylation(GO:0050732) |
| 0.1 | 4.6 | GO:0006414 | translational elongation(GO:0006414) |
| 0.1 | 0.8 | GO:1904153 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.1 | 2.4 | GO:0046426 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.1 | 3.7 | GO:0034394 | protein localization to cell surface(GO:0034394) |
| 0.1 | 6.2 | GO:0042475 | odontogenesis of dentin-containing tooth(GO:0042475) |
| 0.1 | 3.0 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.1 | 0.5 | GO:0032277 | negative regulation of gonadotropin secretion(GO:0032277) |
| 0.1 | 9.4 | GO:0007051 | spindle organization(GO:0007051) |
| 0.1 | 4.1 | GO:1902117 | positive regulation of organelle assembly(GO:1902117) |
| 0.1 | 3.9 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.1 | 1.0 | GO:0051443 | positive regulation of ubiquitin-protein transferase activity(GO:0051443) |
| 0.0 | 0.5 | GO:2000344 | positive regulation of acrosome reaction(GO:2000344) |
| 0.0 | 3.4 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.0 | 5.9 | GO:0007586 | digestion(GO:0007586) |
| 0.0 | 4.2 | GO:0050715 | positive regulation of cytokine secretion(GO:0050715) |
| 0.0 | 1.4 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 0.7 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.0 | 4.9 | GO:0051291 | protein heterooligomerization(GO:0051291) |
| 0.0 | 13.5 | GO:0043547 | positive regulation of GTPase activity(GO:0043547) |
| 0.0 | 0.8 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 0.8 | GO:0051310 | metaphase plate congression(GO:0051310) |
| 0.0 | 4.8 | GO:0045665 | negative regulation of neuron differentiation(GO:0045665) |
| 0.0 | 0.2 | GO:0006353 | DNA-templated transcription, termination(GO:0006353) |
| 0.0 | 1.0 | GO:0090263 | positive regulation of canonical Wnt signaling pathway(GO:0090263) |
| 0.0 | 3.1 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.0 | 1.1 | GO:0008637 | apoptotic mitochondrial changes(GO:0008637) |
| 0.0 | 0.8 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.3 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 1.9 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 4.0 | GO:0060271 | cilium morphogenesis(GO:0060271) |
| 0.0 | 2.6 | GO:2001234 | negative regulation of apoptotic signaling pathway(GO:2001234) |
| 0.0 | 0.6 | GO:0006278 | RNA-dependent DNA biosynthetic process(GO:0006278) telomere maintenance via telomerase(GO:0007004) |
| 0.0 | 2.1 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.8 | 14.4 | GO:0042642 | actomyosin, myosin complex part(GO:0042642) |
| 1.5 | 7.4 | GO:0031262 | condensed nuclear chromosome outer kinetochore(GO:0000942) Ndc80 complex(GO:0031262) |
| 1.4 | 11.4 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 1.4 | 22.7 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 1.3 | 35.9 | GO:0001891 | phagocytic cup(GO:0001891) |
| 1.3 | 8.9 | GO:0060205 | cytoplasmic membrane-bounded vesicle lumen(GO:0060205) |
| 1.1 | 5.6 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 1.0 | 3.1 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 1.0 | 5.1 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 1.0 | 10.2 | GO:0014731 | spectrin(GO:0008091) spectrin-associated cytoskeleton(GO:0014731) |
| 0.9 | 4.6 | GO:0072557 | IPAF inflammasome complex(GO:0072557) |
| 0.9 | 85.8 | GO:0000786 | nucleosome(GO:0000786) |
| 0.9 | 6.0 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.7 | 4.4 | GO:0044308 | axonal spine(GO:0044308) |
| 0.6 | 4.1 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 0.5 | 3.1 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.5 | 4.1 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.5 | 3.8 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.4 | 4.2 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.4 | 2.1 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.4 | 6.3 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.4 | 6.0 | GO:0031045 | dense core granule(GO:0031045) |
| 0.4 | 6.2 | GO:0097449 | astrocyte projection(GO:0097449) |
| 0.4 | 5.3 | GO:0000800 | lateral element(GO:0000800) |
| 0.3 | 6.8 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.3 | 12.9 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.3 | 3.9 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.3 | 4.7 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.3 | 1.3 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.3 | 6.1 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.2 | 2.7 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.2 | 1.7 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.2 | 7.7 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.2 | 5.6 | GO:0005753 | mitochondrial proton-transporting ATP synthase complex(GO:0005753) |
| 0.2 | 22.4 | GO:0005814 | centriole(GO:0005814) |
| 0.2 | 11.4 | GO:0098878 | ionotropic glutamate receptor complex(GO:0008328) neurotransmitter receptor complex(GO:0098878) |
| 0.2 | 1.0 | GO:0002142 | stereocilia ankle link complex(GO:0002142) periciliary membrane compartment(GO:1990075) USH2 complex(GO:1990696) |
| 0.2 | 7.4 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.2 | 1.1 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.2 | 8.4 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.2 | 10.7 | GO:0035097 | histone methyltransferase complex(GO:0035097) |
| 0.2 | 0.6 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.2 | 1.4 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.1 | 2.2 | GO:0030128 | clathrin coat of endocytic vesicle(GO:0030128) |
| 0.1 | 3.0 | GO:0031082 | BLOC complex(GO:0031082) |
| 0.1 | 0.6 | GO:0005606 | laminin-1 complex(GO:0005606) |
| 0.1 | 2.9 | GO:0030673 | axolemma(GO:0030673) |
| 0.1 | 13.3 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.1 | 2.2 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.1 | 6.8 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.1 | 8.7 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.1 | 2.6 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.1 | 2.6 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.1 | 2.3 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.1 | 3.0 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.1 | 10.6 | GO:0000922 | spindle pole(GO:0000922) |
| 0.1 | 4.2 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 3.0 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.1 | 4.4 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.1 | 1.1 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.1 | 1.0 | GO:0005858 | axonemal dynein complex(GO:0005858) |
| 0.1 | 15.0 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.1 | 5.2 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.1 | 7.3 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 4.3 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 24.3 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.1 | 5.7 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.1 | 1.5 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.1 | 0.8 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.1 | 2.6 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 1.0 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 3.5 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 2.6 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 2.8 | GO:0005875 | microtubule associated complex(GO:0005875) |
| 0.0 | 1.4 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 2.1 | GO:0097014 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.0 | 2.7 | GO:0000123 | histone acetyltransferase complex(GO:0000123) |
| 0.0 | 18.4 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 0.7 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 4.1 | GO:0010008 | endosome membrane(GO:0010008) |
| 0.0 | 0.3 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 15.6 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 0.8 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 2.2 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
| 0.0 | 2.3 | GO:0098857 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.6 | 16.8 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 3.3 | 13.3 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 2.1 | 6.2 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 1.9 | 5.6 | GO:0071886 | 1-(4-iodo-2,5-dimethoxyphenyl)propan-2-amine binding(GO:0071886) |
| 1.9 | 3.7 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 1.9 | 5.6 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 1.7 | 10.1 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 1.6 | 8.0 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 1.6 | 7.8 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 1.6 | 6.2 | GO:0030345 | extracellular matrix structural constituent conferring compression resistance(GO:0030021) structural constituent of tooth enamel(GO:0030345) |
| 1.5 | 9.2 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 1.5 | 7.4 | GO:0001729 | ceramide kinase activity(GO:0001729) |
| 1.2 | 3.7 | GO:0031783 | corticotropin hormone receptor binding(GO:0031780) type 5 melanocortin receptor binding(GO:0031783) |
| 1.2 | 11.0 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 1.2 | 4.9 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 1.1 | 5.6 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 1.1 | 5.3 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 1.0 | 4.2 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 1.0 | 3.0 | GO:0042903 | tubulin deacetylase activity(GO:0042903) |
| 0.9 | 10.4 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.8 | 2.5 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.8 | 14.4 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.8 | 9.2 | GO:0019103 | pyrimidine nucleotide binding(GO:0019103) |
| 0.8 | 10.7 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.8 | 3.1 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.8 | 5.3 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.8 | 6.0 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.7 | 7.0 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.7 | 5.3 | GO:0051022 | Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.6 | 12.7 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
| 0.6 | 11.4 | GO:0005234 | extracellular-glutamate-gated ion channel activity(GO:0005234) |
| 0.6 | 5.1 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.5 | 1.6 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.5 | 5.5 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.5 | 6.3 | GO:1990405 | protein antigen binding(GO:1990405) |
| 0.5 | 5.7 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.5 | 2.8 | GO:0008318 | protein prenyltransferase activity(GO:0008318) |
| 0.4 | 7.4 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.4 | 1.3 | GO:0008396 | oxysterol 7-alpha-hydroxylase activity(GO:0008396) |
| 0.4 | 2.9 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.4 | 2.0 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.4 | 3.0 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.4 | 8.9 | GO:0005112 | Notch binding(GO:0005112) |
| 0.4 | 1.8 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.4 | 12.5 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.3 | 17.4 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.3 | 4.1 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.3 | 1.3 | GO:0036004 | GAF domain binding(GO:0036004) |
| 0.3 | 2.3 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.3 | 17.1 | GO:0016860 | intramolecular oxidoreductase activity(GO:0016860) |
| 0.3 | 4.7 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.3 | 1.8 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.3 | 4.2 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.3 | 1.1 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.3 | 5.3 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.2 | 2.7 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.2 | 76.4 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.2 | 6.0 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.2 | 8.2 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.2 | 4.3 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.2 | 10.4 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.2 | 4.9 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.2 | 7.5 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.2 | 2.1 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.2 | 7.4 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.2 | 2.7 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.2 | 1.1 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.2 | 9.1 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.2 | 2.6 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 0.2 | 2.2 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.2 | 2.9 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.2 | 2.0 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.2 | 11.8 | GO:0005518 | collagen binding(GO:0005518) |
| 0.2 | 2.6 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.2 | 1.8 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.2 | 98.9 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.2 | 1.9 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.2 | 1.1 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.2 | 0.8 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.2 | 3.9 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.2 | 2.2 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.1 | 0.4 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.1 | 23.7 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.1 | 0.9 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.1 | 3.0 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.1 | 1.7 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.1 | 1.9 | GO:0008373 | sialyltransferase activity(GO:0008373) |
| 0.1 | 0.3 | GO:0102390 | mycophenolic acid acyl-glucuronide esterase activity(GO:0102390) |
| 0.1 | 9.4 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.1 | 0.4 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.1 | 3.0 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.1 | 3.1 | GO:0015301 | anion:anion antiporter activity(GO:0015301) |
| 0.1 | 1.4 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.1 | 2.1 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.1 | 0.6 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.1 | 0.5 | GO:0070728 | leucine binding(GO:0070728) |
| 0.1 | 1.5 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.1 | 1.3 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.1 | 2.6 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.1 | 3.9 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.1 | 2.1 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 10.6 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 2.5 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 1.6 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 24.3 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 10.3 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.0 | 0.1 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.0 | 0.7 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 2.8 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.0 | 2.6 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 2.5 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 0.6 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.8 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.2 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.9 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 7.8 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.0 | 1.4 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 1.0 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 1.5 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.0 | 0.3 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 0.1 | GO:0015929 | hexosaminidase activity(GO:0015929) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 21.6 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.4 | 8.0 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.4 | 17.9 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.4 | 17.4 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.3 | 11.4 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.3 | 15.9 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.3 | 7.8 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.3 | 20.9 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.2 | 15.1 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.2 | 5.3 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.2 | 8.8 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.2 | 5.4 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.2 | 4.7 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.2 | 3.3 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.1 | 3.4 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.1 | 2.8 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.1 | 23.4 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 0.8 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.1 | 8.3 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.1 | 2.2 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.1 | 2.7 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.1 | 4.0 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 4.1 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.1 | 1.2 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.1 | 3.8 | PID E2F PATHWAY | E2F transcription factor network |
| 0.1 | 0.9 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 2.1 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 5.9 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 2.9 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 11.4 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.8 | 16.8 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.7 | 18.5 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.6 | 14.4 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.4 | 2.5 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.4 | 11.8 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.4 | 5.6 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.4 | 7.1 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.4 | 8.2 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.3 | 9.2 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.3 | 17.6 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.3 | 5.9 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 0.3 | 10.2 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.3 | 5.8 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.3 | 15.8 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.2 | 11.7 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.2 | 4.2 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.2 | 3.1 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.2 | 6.7 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.2 | 6.8 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.2 | 8.0 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.2 | 7.8 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.2 | 2.6 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.2 | 4.6 | REACTOME SIGNALING BY NOTCH1 | Genes involved in Signaling by NOTCH1 |
| 0.2 | 5.5 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.2 | 11.0 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.1 | 1.8 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.1 | 3.6 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 4.0 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.1 | 1.6 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.1 | 1.8 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 3.6 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.1 | 2.0 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.1 | 1.7 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.1 | 14.4 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 0.9 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.1 | 7.4 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.1 | 1.3 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.1 | 2.7 | REACTOME REGULATION OF INSULIN SECRETION BY GLUCAGON LIKE PEPTIDE1 | Genes involved in Regulation of Insulin Secretion by Glucagon-like Peptide-1 |
| 0.1 | 10.1 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.1 | 0.8 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.1 | 1.7 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 5.8 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.0 | 1.7 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 2.3 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 0.8 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.4 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |