Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
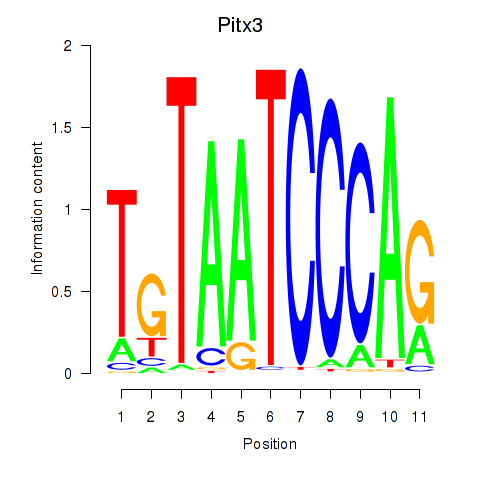
Results for Pitx3
Z-value: 1.07
Transcription factors associated with Pitx3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Pitx3
|
ENSRNOG00000019194 | paired-like homeodomain 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Pitx3 | rn6_v1_chr1_-_265899958_265899958 | 0.27 | 1.0e-06 | Click! |
Activity profile of Pitx3 motif
Sorted Z-values of Pitx3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_29152442 | 42.59 |
ENSRNOT00000079774
|
Mybpc1
|
myosin binding protein C, slow type |
| chr12_+_23752844 | 31.57 |
ENSRNOT00000001953
|
Ssc4d
|
scavenger receptor cysteine rich family member with 4 domains |
| chr18_+_63098144 | 29.10 |
ENSRNOT00000024968
|
Cidea
|
cell death-inducing DFFA-like effector a |
| chr1_+_91716383 | 28.08 |
ENSRNOT00000016919
|
Slc7a9
|
solute carrier family 7 member 9 |
| chr7_+_11769400 | 26.36 |
ENSRNOT00000044417
|
Jsrp1
|
junctional sarcoplasmic reticulum protein 1 |
| chr9_-_93404883 | 24.54 |
ENSRNOT00000025024
|
Nmur1
|
neuromedin U receptor 1 |
| chr12_-_48048540 | 23.17 |
ENSRNOT00000071335
ENSRNOT00000078807 |
Myo1h
|
myosin 1H |
| chr7_-_145174771 | 23.17 |
ENSRNOT00000055270
|
Glycam1
|
glycosylation dependent cell adhesion molecule 1 |
| chr1_-_102255459 | 22.56 |
ENSRNOT00000067392
|
Ush1c
|
USH1 protein network component harmonin |
| chr1_+_215666628 | 22.43 |
ENSRNOT00000040598
ENSRNOT00000066135 ENSRNOT00000051425 ENSRNOT00000080339 ENSRNOT00000066896 ENSRNOT00000063918 |
Tnnt3
|
troponin T3, fast skeletal type |
| chr10_-_88050622 | 22.41 |
ENSRNOT00000019037
|
Krt15
|
keratin 15 |
| chr10_-_40525008 | 22.12 |
ENSRNOT00000016440
|
Slc36a2
|
solute carrier family 36 member 2 |
| chr12_+_22153983 | 21.79 |
ENSRNOT00000080775
ENSRNOT00000001894 |
Pcolce
|
procollagen C-endopeptidase enhancer |
| chr2_-_187706300 | 21.71 |
ENSRNOT00000092349
ENSRNOT00000026414 |
Tmem79
|
transmembrane protein 79 |
| chr1_+_48273611 | 21.56 |
ENSRNOT00000022254
ENSRNOT00000022068 |
Slc22a1
|
solute carrier family 22 member 1 |
| chr1_-_216663720 | 21.30 |
ENSRNOT00000078944
ENSRNOT00000077409 |
Cdkn1c
|
cyclin-dependent kinase inhibitor 1C |
| chr18_+_35574002 | 20.27 |
ENSRNOT00000093442
ENSRNOT00000070817 ENSRNOT00000093356 |
Myot
|
myotilin |
| chr1_+_263554453 | 20.24 |
ENSRNOT00000070861
|
Abcc2
|
ATP binding cassette subfamily C member 2 |
| chr9_+_37727942 | 19.98 |
ENSRNOT00000016511
ENSRNOT00000074276 |
LOC100912306
|
myotilin-like |
| chr3_-_10371240 | 19.88 |
ENSRNOT00000012075
|
Ass1
|
argininosuccinate synthase 1 |
| chr10_+_40247436 | 19.74 |
ENSRNOT00000079830
|
Gpx3
|
glutathione peroxidase 3 |
| chr1_-_80689171 | 18.89 |
ENSRNOT00000045574
|
Bcam
|
basal cell adhesion molecule (Lutheran blood group) |
| chr2_+_189423559 | 18.24 |
ENSRNOT00000029076
|
Tpm3
|
tropomyosin 3 |
| chr19_-_24875137 | 17.52 |
ENSRNOT00000006152
|
Adgre5
|
adhesion G protein-coupled receptor E5 |
| chr2_+_256964860 | 17.46 |
ENSRNOT00000073547
|
Ifi44l
|
interferon-induced protein 44-like |
| chr3_-_80012750 | 16.68 |
ENSRNOT00000018154
|
Nr1h3
|
nuclear receptor subfamily 1, group H, member 3 |
| chr10_-_94576512 | 16.65 |
ENSRNOT00000035474
|
Icam2
|
intercellular adhesion molecule 2 |
| chrX_-_32095867 | 16.56 |
ENSRNOT00000049947
ENSRNOT00000080730 |
Ace2
|
angiotensin I converting enzyme 2 |
| chr12_+_38828444 | 16.15 |
ENSRNOT00000001809
|
Hpd
|
4-hydroxyphenylpyruvate dioxygenase |
| chr13_+_78805347 | 15.81 |
ENSRNOT00000003748
|
Serpinc1
|
serpin family C member 1 |
| chr2_-_200762492 | 15.73 |
ENSRNOT00000056172
|
Hsd3b2
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 2 |
| chr1_-_80331626 | 15.60 |
ENSRNOT00000022577
|
AABR07002677.1
|
|
| chr1_+_189359853 | 15.08 |
ENSRNOT00000055083
|
Acsm1
|
acyl-CoA synthetase medium-chain family member 1 |
| chrM_+_10160 | 14.75 |
ENSRNOT00000042928
|
Mt-nd4
|
mitochondrially encoded NADH dehydrogenase 4 |
| chr8_+_115151627 | 14.60 |
ENSRNOT00000064252
|
Abhd14b
|
abhydrolase domain containing 14b |
| chr12_+_25264400 | 14.31 |
ENSRNOT00000087656
|
Gtf2ird1
|
GTF2I repeat domain containing 1 |
| chr10_+_55626741 | 14.30 |
ENSRNOT00000008492
|
Aurkb
|
aurora kinase B |
| chr10_-_70337532 | 14.18 |
ENSRNOT00000055963
|
Slfn13
|
schlafen family member 13 |
| chr4_-_10517832 | 14.08 |
ENSRNOT00000039953
ENSRNOT00000083964 |
Gsap
|
gamma-secretase activating protein |
| chr5_-_137321121 | 13.83 |
ENSRNOT00000027414
|
Tie1
|
tyrosine kinase with immunoglobulin-like and EGF-like domains 1 |
| chr12_+_25264192 | 13.65 |
ENSRNOT00000079392
|
Gtf2ird1
|
GTF2I repeat domain containing 1 |
| chrX_+_77230180 | 13.31 |
ENSRNOT00000085879
|
Tlr13
|
toll-like receptor 13 |
| chr9_-_113598477 | 13.27 |
ENSRNOT00000035606
ENSRNOT00000084884 |
Ralbp1
|
ralA binding protein 1 |
| chr4_-_120840111 | 12.67 |
ENSRNOT00000022231
|
Mcm2
|
minichromosome maintenance complex component 2 |
| chr17_-_10004321 | 12.64 |
ENSRNOT00000042394
|
Fgfr4
|
fibroblast growth factor receptor 4 |
| chr14_+_84447885 | 12.60 |
ENSRNOT00000009150
|
Gatsl3
|
GATS protein-like 3 |
| chr8_+_128044084 | 12.47 |
ENSRNOT00000019106
|
Xylb
|
xylulokinase |
| chr16_+_20109200 | 12.31 |
ENSRNOT00000079784
|
Jak3
|
Janus kinase 3 |
| chr10_-_40419054 | 12.18 |
ENSRNOT00000039040
|
Ccdc69
|
coiled-coil domain containing 69 |
| chr2_+_125752130 | 11.98 |
ENSRNOT00000038703
|
Fat4
|
FAT atypical cadherin 4 |
| chr8_-_21831668 | 11.96 |
ENSRNOT00000027897
|
Col5a3
|
collagen type V alpha 3 chain |
| chr10_-_47775055 | 11.95 |
ENSRNOT00000057864
|
LOC100912585
|
mitogen-activated protein kinase 7-like |
| chr11_+_83855753 | 11.63 |
ENSRNOT00000030686
|
RGD1563956
|
similar to 60S ribosomal protein L12 |
| chr13_-_91981432 | 10.96 |
ENSRNOT00000004637
|
AABR07021804.1
|
|
| chr1_+_214005791 | 10.86 |
ENSRNOT00000071831
|
LOC100911692
|
EF-hand calcium-binding domain-containing protein 4A-like |
| chr16_+_83358116 | 10.61 |
ENSRNOT00000031109
|
Rab20
|
RAB20, member RAS oncogene family |
| chr1_+_141821916 | 10.55 |
ENSRNOT00000071574
|
Echs1
|
enoyl-CoA hydratase, short chain 1 |
| chr2_+_260884337 | 10.54 |
ENSRNOT00000032808
|
Cryz
|
crystallin zeta |
| chr4_-_157433467 | 10.51 |
ENSRNOT00000028965
|
Lag3
|
lymphocyte activating 3 |
| chr17_+_44556039 | 10.50 |
ENSRNOT00000086540
|
Prss16
|
protease, serine 16 |
| chrX_+_71174699 | 10.47 |
ENSRNOT00000076957
ENSRNOT00000090192 ENSRNOT00000040334 |
Med12
|
mediator complex subunit 12 |
| chr10_-_49007089 | 10.45 |
ENSRNOT00000073706
|
Mapk7
|
mitogen-activated protein kinase 7 |
| chr7_-_80796670 | 10.06 |
ENSRNOT00000010539
|
Abra
|
actin-binding Rho activating protein |
| chr1_+_101554642 | 10.00 |
ENSRNOT00000028474
|
Bcat2
|
branched chain amino acid transaminase 2 |
| chr10_-_109909646 | 9.98 |
ENSRNOT00000074362
ENSRNOT00000088907 |
Dcxr
|
dicarbonyl and L-xylulose reductase |
| chr11_+_84399417 | 9.96 |
ENSRNOT00000048306
ENSRNOT00000091595 |
Abcc5
|
ATP binding cassette subfamily C member 5 |
| chr4_+_179481263 | 9.93 |
ENSRNOT00000021284
|
Etfrf1
|
electron transfer flavoprotein regulatory factor 1 |
| chr2_-_100249811 | 9.91 |
ENSRNOT00000086760
|
Hnf4g
|
hepatocyte nuclear factor 4, gamma |
| chr1_+_81499821 | 9.88 |
ENSRNOT00000027100
|
Lypd3
|
Ly6/Plaur domain containing 3 |
| chr7_+_120067379 | 9.87 |
ENSRNOT00000011270
|
Cdc42ep1
|
CDC42 effector protein 1 |
| chr3_-_12502859 | 9.77 |
ENSRNOT00000022814
|
Zbtb43
|
zinc finger and BTB domain containing 43 |
| chr1_-_80609836 | 9.67 |
ENSRNOT00000091647
ENSRNOT00000046169 |
Apoc1
|
apolipoprotein C1 |
| chr9_+_16529579 | 9.67 |
ENSRNOT00000067894
|
Ptcra
|
pre T-cell antigen receptor alpha |
| chr8_+_76754492 | 9.60 |
ENSRNOT00000085727
|
Myo1e
|
myosin IE |
| chr3_-_57957346 | 9.53 |
ENSRNOT00000036728
|
Slc25a12
|
solute carrier family 25 member 12 |
| chr16_-_47535358 | 9.43 |
ENSRNOT00000040731
|
Cldn22
|
claudin 22 |
| chr8_+_103459161 | 9.43 |
ENSRNOT00000074047
|
PCOLCE2
|
procollagen C-endopeptidase enhancer 2 |
| chr10_-_86930947 | 9.42 |
ENSRNOT00000081440
|
Top2a
|
topoisomerase (DNA) II alpha |
| chr1_-_212579057 | 9.40 |
ENSRNOT00000025446
|
LOC100911186
|
enoyl-CoA hydratase, mitochondrial-like |
| chr5_+_128190708 | 9.37 |
ENSRNOT00000012492
|
Orc1
|
origin recognition complex, subunit 1 |
| chr15_-_27819376 | 9.30 |
ENSRNOT00000067400
|
A930018M24Rik
|
RIKEN cDNA A930018M24 gene |
| chr19_+_46455761 | 9.30 |
ENSRNOT00000034012
ENSRNOT00000093392 |
Nudt7
|
nudix hydrolase 7 |
| chr4_-_98995590 | 9.25 |
ENSRNOT00000008698
|
Thnsl2
|
threonine synthase-like 2 |
| chr16_-_21017163 | 9.20 |
ENSRNOT00000027661
|
Mef2b
|
myocyte enhancer factor 2B |
| chr7_+_11490852 | 9.14 |
ENSRNOT00000044484
|
Creb3l3
|
cAMP responsive element binding protein 3-like 3 |
| chr7_+_141666111 | 9.11 |
ENSRNOT00000083250
|
AABR07058886.1
|
|
| chr16_+_20020444 | 8.99 |
ENSRNOT00000024827
|
Pgls
|
6-phosphogluconolactonase |
| chr7_-_120077612 | 8.87 |
ENSRNOT00000011750
|
Lgals2
|
galectin 2 |
| chr4_-_151428894 | 8.79 |
ENSRNOT00000010556
|
Adipor2
|
adiponectin receptor 2 |
| chr15_-_29548400 | 8.52 |
ENSRNOT00000078176
|
AABR07017639.2
|
|
| chr1_+_189514553 | 8.47 |
ENSRNOT00000020039
|
Acsm3
|
acyl-CoA synthetase medium-chain family member 3 |
| chr1_+_221735517 | 8.46 |
ENSRNOT00000028628
ENSRNOT00000044866 |
Sf1
|
splicing factor 1 |
| chr19_-_10884047 | 8.39 |
ENSRNOT00000088196
ENSRNOT00000063791 |
Cpne2
|
copine 2 |
| chr7_-_130151880 | 8.32 |
ENSRNOT00000088623
|
Plxnb2
|
plexin B2 |
| chr20_-_4935887 | 8.30 |
ENSRNOT00000064734
|
RT1-CE4
|
RT1 class I, locus CE4 |
| chr1_-_77893509 | 7.97 |
ENSRNOT00000015059
|
Gltscr1
|
glioma tumor suppressor candidate region gene 1 |
| chr7_+_120176530 | 7.94 |
ENSRNOT00000087548
|
Triobp
|
TRIO and F-actin binding protein |
| chr1_-_166939541 | 7.92 |
ENSRNOT00000079675
|
Folr1
|
folate receptor 1 |
| chr12_-_19294888 | 7.92 |
ENSRNOT00000001815
|
Zfp113
|
zinc finger protein 3 |
| chr13_-_86451002 | 7.85 |
ENSRNOT00000043004
ENSRNOT00000027996 |
Pbx1
|
PBX homeobox 1 |
| chr8_+_71914867 | 7.67 |
ENSRNOT00000023372
|
Dapk2
|
death-associated protein kinase 2 |
| chr1_-_263885169 | 7.60 |
ENSRNOT00000030782
|
Chuk
|
conserved helix-loop-helix ubiquitous kinase |
| chr5_+_107712579 | 7.59 |
ENSRNOT00000090739
|
Mtap
|
methylthioadenosine phosphorylase |
| chr20_-_3813673 | 7.59 |
ENSRNOT00000081085
|
Ring1
|
ring finger protein 1 |
| chr1_-_216920625 | 7.53 |
ENSRNOT00000028119
|
Osbpl5
|
oxysterol binding protein-like 5 |
| chr3_+_132052612 | 7.53 |
ENSRNOT00000030148
|
AABR07053879.1
|
|
| chr9_-_53118613 | 7.50 |
ENSRNOT00000065581
|
Ormdl1
|
ORMDL sphingolipid biosynthesis regulator 1 |
| chr1_-_85317968 | 7.48 |
ENSRNOT00000026891
|
Gmfg
|
glia maturation factor, gamma |
| chr1_+_88078350 | 7.46 |
ENSRNOT00000048677
|
Rasgrp4
|
RAS guanyl releasing protein 4 |
| chr12_-_48365784 | 7.43 |
ENSRNOT00000077317
|
Dao
|
D-amino-acid oxidase |
| chr3_-_110556808 | 7.41 |
ENSRNOT00000092158
ENSRNOT00000045362 |
RGD1565536
|
similar to hypothetical protein |
| chr1_+_99599620 | 7.26 |
ENSRNOT00000024690
|
Ctu1
|
cytosolic thiouridylase subunit 1 |
| chr5_-_173611202 | 7.24 |
ENSRNOT00000047854
|
Agrn
|
agrin |
| chr6_+_26566494 | 7.24 |
ENSRNOT00000079292
|
Gtf3c2
|
general transcription factor IIIC subunit 2 |
| chr5_+_151343493 | 7.01 |
ENSRNOT00000077807
|
Wasf2
|
WAS protein family, member 2 |
| chr12_+_47239663 | 6.96 |
ENSRNOT00000077871
|
Unc119b
|
unc-119 lipid binding chaperone B |
| chr11_-_65845418 | 6.96 |
ENSRNOT00000091057
|
Fstl1
|
follistatin-like 1 |
| chr15_-_24199341 | 6.92 |
ENSRNOT00000015553
|
Dlgap5
|
DLG associated protein 5 |
| chr19_-_43848937 | 6.85 |
ENSRNOT00000044844
|
Ldhd
|
lactate dehydrogenase D |
| chr16_+_69046514 | 6.84 |
ENSRNOT00000092344
|
Rab11fip1
|
RAB11 family interacting protein 1 |
| chr7_+_117763783 | 6.78 |
ENSRNOT00000021208
|
Mfsd3
|
major facilitator superfamily domain containing 3 |
| chr5_+_159735008 | 6.63 |
ENSRNOT00000064310
|
Rsg1
|
REM2 and RAB-like small GTPase 1 |
| chr5_+_169506138 | 6.62 |
ENSRNOT00000014904
|
Rpl22
|
ribosomal protein L22 |
| chr10_+_37215937 | 6.60 |
ENSRNOT00000006567
|
Sar1b
|
secretion associated, Ras related GTPase 1B |
| chr16_+_7758996 | 6.60 |
ENSRNOT00000061063
|
Btd
|
biotinidase |
| chr8_-_21453190 | 6.58 |
ENSRNOT00000078192
|
Zfp26
|
zinc finger protein 26 |
| chr20_+_2057878 | 6.50 |
ENSRNOT00000051480
|
RT1-M6-1
|
RT1 class I, locus M6, gene 1 |
| chr8_-_61919874 | 6.41 |
ENSRNOT00000064830
|
RGD1305464
|
similar to human chromosome 15 open reading frame 39 |
| chr20_+_31096214 | 6.40 |
ENSRNOT00000000676
|
Lrrc20
|
leucine rich repeat containing 20 |
| chr10_-_45514878 | 6.34 |
ENSRNOT00000036940
|
Iba57
|
IBA57 homolog, iron-sulfur cluster assembly |
| chr1_+_101397828 | 6.32 |
ENSRNOT00000028189
|
Kcna7
|
potassium voltage-gated channel subfamily A member 7 |
| chr3_+_55910177 | 6.32 |
ENSRNOT00000009969
|
Klhl41
|
kelch-like family member 41 |
| chr3_+_170996193 | 6.28 |
ENSRNOT00000008959
|
Spo11
|
SPO11, initiator of meiotic double stranded breaks |
| chr12_+_29012852 | 6.28 |
ENSRNOT00000002006
|
LOC100912262
|
general transcription factor II-I repeat domain-containing protein 1-like |
| chr17_+_57040023 | 6.22 |
ENSRNOT00000020204
|
Crem
|
cAMP responsive element modulator |
| chr4_+_57378069 | 6.15 |
ENSRNOT00000080173
ENSRNOT00000011851 |
Nrf1
|
nuclear respiratory factor 1 |
| chr3_-_153321352 | 6.14 |
ENSRNOT00000064824
|
Rbl1
|
RB transcriptional corepressor like 1 |
| chr6_+_6908684 | 6.07 |
ENSRNOT00000079095
|
Mta3
|
metastasis associated 1 family, member 3 |
| chr20_+_49319307 | 6.03 |
ENSRNOT00000057078
|
Atg5
|
autophagy related 5 |
| chr8_+_75625174 | 6.02 |
ENSRNOT00000013742
|
Ice2
|
interactor of little elongation complex ELL subunit 2 |
| chr10_+_89352835 | 5.99 |
ENSRNOT00000028060
|
Rpl27
|
ribosomal protein L27 |
| chr4_+_180062799 | 5.99 |
ENSRNOT00000021623
|
Rassf8
|
Ras association domain family member 8 |
| chr12_+_29012695 | 5.97 |
ENSRNOT00000085696
|
LOC100912262
|
general transcription factor II-I repeat domain-containing protein 1-like |
| chr5_+_159734838 | 5.95 |
ENSRNOT00000079905
|
Rsg1
|
REM2 and RAB-like small GTPase 1 |
| chr10_-_67478848 | 5.94 |
ENSRNOT00000005325
|
Tefm
|
transcription elongation factor, mitochondrial |
| chr3_-_119405453 | 5.92 |
ENSRNOT00000090355
|
Sppl2a
|
signal peptide peptidase-like 2A |
| chr7_+_38819771 | 5.90 |
ENSRNOT00000006109
|
Lum
|
lumican |
| chr16_-_47537476 | 5.87 |
ENSRNOT00000050279
|
Cldn24
|
claudin 24 |
| chr14_+_84282073 | 5.79 |
ENSRNOT00000078271
|
Sec14l4
|
SEC14-like lipid binding 4 |
| chr4_+_100277391 | 5.79 |
ENSRNOT00000086820
ENSRNOT00000084281 ENSRNOT00000017927 |
Ggcx
|
gamma-glutamyl carboxylase |
| chr6_-_91456696 | 5.68 |
ENSRNOT00000005577
|
Rps29
|
ribosomal protein S29 |
| chr16_+_70778913 | 5.63 |
ENSRNOT00000019281
|
Hook3
|
hook microtubule-tethering protein 3 |
| chr20_-_27308069 | 5.59 |
ENSRNOT00000056047
|
Slc25a16
|
solute carrier family 25 member 16 |
| chr12_+_25305823 | 5.54 |
ENSRNOT00000002010
|
Gtf2ird1
|
GTF2I repeat domain containing 1 |
| chr5_+_152613255 | 5.52 |
ENSRNOT00000037097
|
Pafah2
|
platelet-activating factor acetylhydrolase 2 |
| chr10_-_59883839 | 5.43 |
ENSRNOT00000093579
|
Aspa
|
aspartoacylase |
| chr19_-_26243223 | 5.42 |
ENSRNOT00000037766
|
Zfp791
|
zinc finger protein 791 |
| chr5_-_136023511 | 5.38 |
ENSRNOT00000081747
|
Rps8
|
ribosomal protein S8 |
| chr6_+_123361864 | 5.37 |
ENSRNOT00000057577
|
AABR07065353.1
|
|
| chr19_+_52664322 | 5.36 |
ENSRNOT00000082754
|
Crispld2
|
cysteine-rich secretory protein LCCL domain containing 2 |
| chr7_+_23854846 | 5.33 |
ENSRNOT00000037290
|
Bpifc
|
BPI fold containing family C |
| chr4_+_147692028 | 5.32 |
ENSRNOT00000014207
|
Cand2
|
cullin-associated and neddylation-dissociated 2 (putative) |
| chr11_-_87924816 | 5.32 |
ENSRNOT00000031819
|
Serpind1
|
serpin family D member 1 |
| chr17_-_78812111 | 5.30 |
ENSRNOT00000021506
|
Dclre1c
|
DNA cross-link repair 1C |
| chr3_+_66673071 | 5.30 |
ENSRNOT00000034769
|
Ppp1r1c
|
protein phosphatase 1, regulatory (inhibitor) subunit 1C |
| chrX_-_123486814 | 5.28 |
ENSRNOT00000016993
|
RGD1564541
|
similar to hypothetical protein FLJ22965 |
| chr10_-_56506446 | 5.22 |
ENSRNOT00000021357
|
Acap1
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 1 |
| chr1_-_87174530 | 5.22 |
ENSRNOT00000089578
|
Kcnk6
|
potassium two pore domain channel subfamily K member 6 |
| chr5_+_58144705 | 5.18 |
ENSRNOT00000019886
|
Galt
|
galactose-1-phosphate uridylyltransferase |
| chr7_+_121361415 | 5.13 |
ENSRNOT00000067904
|
Tab1
|
TGF-beta activated kinase 1/MAP3K7 binding protein 1 |
| chr1_+_190555177 | 5.07 |
ENSRNOT00000021514
|
Uqcrc2
|
ubiquinol cytochrome c reductase core protein 2 |
| chr3_+_145764932 | 5.05 |
ENSRNOT00000075257
|
AABR07054264.1
|
|
| chr5_+_173256834 | 5.03 |
ENSRNOT00000089936
|
Ccnl2
|
cyclin L2 |
| chr13_+_50196042 | 5.03 |
ENSRNOT00000090858
|
Zc3h11a
|
zinc finger CCCH-type containing 11A |
| chr5_-_61130026 | 5.00 |
ENSRNOT00000060149
|
Slc25a51
|
solute carrier family 25, member 51 |
| chr8_-_116993193 | 5.00 |
ENSRNOT00000026327
|
Dag1
|
dystroglycan 1 |
| chr9_-_4945352 | 4.85 |
ENSRNOT00000082530
|
Sult1c3
|
sulfotransferase family 1C member 3 |
| chr16_-_20562399 | 4.82 |
ENSRNOT00000031692
|
Lrrc25
|
leucine rich repeat containing 25 |
| chr20_+_2057689 | 4.82 |
ENSRNOT00000086007
|
RT1-M6-1
|
RT1 class I, locus M6, gene 1 |
| chr14_+_44524753 | 4.78 |
ENSRNOT00000076245
|
Rpl9
|
ribosomal protein L9 |
| chr19_+_42066351 | 4.75 |
ENSRNOT00000066733
|
Dhodh
|
dihydroorotate dehydrogenase (quinone) |
| chr2_+_52397175 | 4.72 |
ENSRNOT00000082614
|
Ccl28
|
C-C motif chemokine ligand 28 |
| chr5_+_173256637 | 4.71 |
ENSRNOT00000025531
|
Ccnl2
|
cyclin L2 |
| chr1_-_101514974 | 4.68 |
ENSRNOT00000044788
|
Ppp1r15a
|
protein phosphatase 1, regulatory subunit 15A |
| chr2_+_93656946 | 4.66 |
ENSRNOT00000013905
|
Zfand1
|
zinc finger AN1-type containing 1 |
| chr9_+_95274707 | 4.63 |
ENSRNOT00000045163
|
Ugt1a5
|
UDP glucuronosyltransferase family 1 member A5 |
| chr8_+_118926478 | 4.63 |
ENSRNOT00000028426
|
Ccdc12
|
coiled-coil domain containing 12 |
| chr7_+_144899534 | 4.63 |
ENSRNOT00000065270
|
Copz1
|
coatomer protein complex, subunit zeta 1 |
| chr12_+_30165694 | 4.62 |
ENSRNOT00000001211
|
Asl
|
argininosuccinate lyase |
| chr10_-_37215899 | 4.59 |
ENSRNOT00000006356
|
Sec24a
|
SEC24 homolog A, COPII coat complex component |
| chr17_+_45188248 | 4.56 |
ENSRNOT00000091609
|
Zscan26
|
zinc finger and SCAN domain containing 26 |
| chr8_+_129240528 | 4.55 |
ENSRNOT00000083427
|
Rpl14
|
ribosomal protein L14 |
| chr3_-_112320762 | 4.51 |
ENSRNOT00000081074
ENSRNOT00000078893 ENSRNOT00000089857 |
Zfp106
|
zinc finger protein 106 |
| chr3_+_66673246 | 4.47 |
ENSRNOT00000081338
|
Ppp1r1c
|
protein phosphatase 1, regulatory (inhibitor) subunit 1C |
| chr11_-_68197772 | 4.45 |
ENSRNOT00000003060
ENSRNOT00000081875 |
Hspbap1
|
Hspb associated protein 1 |
| chr19_-_19727081 | 4.43 |
ENSRNOT00000020700
|
Adcy7
|
adenylate cyclase 7 |
| chr14_-_114649173 | 4.41 |
ENSRNOT00000083528
|
Sptbn1
|
spectrin, beta, non-erythrocytic 1 |
| chr4_-_163528220 | 4.39 |
ENSRNOT00000090813
|
Klri1
|
killer cell lectin-like receptor family I member 1 |
| chr10_-_56300077 | 4.38 |
ENSRNOT00000064401
|
Tnfsf12
|
tumor necrosis factor superfamily member 12 |
| chr10_-_110431792 | 4.36 |
ENSRNOT00000054922
|
LOC619574
|
hypothetical protein LOC619574 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Pitx3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.2 | 24.5 | GO:0000053 | argininosuccinate metabolic process(GO:0000053) |
| 7.5 | 22.5 | GO:0042732 | D-xylose metabolic process(GO:0042732) |
| 7.2 | 21.7 | GO:0070268 | cuticle development(GO:0042335) cornification(GO:0070268) |
| 7.0 | 28.1 | GO:0015811 | L-cystine transport(GO:0015811) |
| 5.5 | 22.1 | GO:0010958 | regulation of amino acid import(GO:0010958) |
| 5.5 | 16.6 | GO:0003051 | angiotensin-mediated drinking behavior(GO:0003051) tryptophan transport(GO:0015827) positive regulation of gap junction assembly(GO:1903598) |
| 5.1 | 20.2 | GO:0050787 | detoxification of mercury ion(GO:0050787) |
| 4.8 | 33.5 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 4.5 | 22.6 | GO:1904970 | brush border assembly(GO:1904970) |
| 4.3 | 42.6 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 4.2 | 16.7 | GO:0048550 | negative regulation of pinocytosis(GO:0048550) |
| 4.2 | 29.1 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 4.1 | 12.3 | GO:0045210 | negative regulation of dendritic cell cytokine production(GO:0002731) FasL biosynthetic process(GO:0045210) |
| 4.0 | 12.0 | GO:0072137 | condensed mesenchymal cell proliferation(GO:0072137) |
| 4.0 | 15.8 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 3.6 | 14.3 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 3.5 | 21.3 | GO:1902746 | regulation of lens fiber cell differentiation(GO:1902746) |
| 3.3 | 10.0 | GO:0009099 | branched-chain amino acid biosynthetic process(GO:0009082) leucine biosynthetic process(GO:0009098) valine biosynthetic process(GO:0009099) |
| 3.2 | 12.6 | GO:0007037 | vacuolar phosphate transport(GO:0007037) positive regulation of parathyroid hormone secretion(GO:2000830) |
| 3.1 | 9.3 | GO:0034034 | coenzyme A catabolic process(GO:0015938) nucleoside bisphosphate catabolic process(GO:0033869) ribonucleoside bisphosphate catabolic process(GO:0034031) purine nucleoside bisphosphate catabolic process(GO:0034034) acetyl-CoA catabolic process(GO:0046356) |
| 2.8 | 19.7 | GO:0002238 | response to molecule of fungal origin(GO:0002238) |
| 2.7 | 16.2 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 2.6 | 7.9 | GO:0060974 | cell migration involved in heart formation(GO:0060974) |
| 2.6 | 15.7 | GO:0034757 | negative regulation of iron ion transport(GO:0034757) |
| 2.6 | 7.7 | GO:2000424 | regulation of eosinophil chemotaxis(GO:2000422) positive regulation of eosinophil chemotaxis(GO:2000424) |
| 2.5 | 7.6 | GO:0010034 | response to acetate(GO:0010034) |
| 2.5 | 7.6 | GO:0070638 | nicotinamide riboside catabolic process(GO:0006738) nicotinamide riboside metabolic process(GO:0046495) pyridine nucleoside metabolic process(GO:0070637) pyridine nucleoside catabolic process(GO:0070638) |
| 2.5 | 7.5 | GO:1900060 | negative regulation of ceramide biosynthetic process(GO:1900060) |
| 2.5 | 7.4 | GO:0046416 | D-amino acid metabolic process(GO:0046416) |
| 2.4 | 9.7 | GO:0010916 | regulation of phosphatidylcholine catabolic process(GO:0010899) regulation of very-low-density lipoprotein particle clearance(GO:0010915) negative regulation of very-low-density lipoprotein particle clearance(GO:0010916) |
| 2.4 | 7.2 | GO:0045887 | positive regulation of synaptic growth at neuromuscular junction(GO:0045887) regulation of sodium:potassium-exchanging ATPase activity(GO:1903406) regulation of protein geranylgeranylation(GO:2000539) positive regulation of protein geranylgeranylation(GO:2000541) |
| 2.4 | 9.5 | GO:0089712 | malate-aspartate shuttle(GO:0043490) L-aspartate transport(GO:0070778) L-aspartate transmembrane transport(GO:0089712) |
| 2.2 | 11.1 | GO:1902990 | mitotic telomere maintenance via semi-conservative replication(GO:1902990) |
| 2.2 | 21.6 | GO:0015697 | quaternary ammonium group transport(GO:0015697) |
| 2.1 | 10.5 | GO:0045085 | negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 1.9 | 9.4 | GO:0070318 | positive regulation of G0 to G1 transition(GO:0070318) |
| 1.9 | 9.3 | GO:0006566 | threonine metabolic process(GO:0006566) short-chain fatty acid biosynthetic process(GO:0051790) |
| 1.8 | 5.5 | GO:0009644 | response to high light intensity(GO:0009644) |
| 1.8 | 12.7 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 1.8 | 5.3 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 1.8 | 10.6 | GO:0090383 | phagosome acidification(GO:0090383) |
| 1.7 | 10.5 | GO:0071499 | cellular response to laminar fluid shear stress(GO:0071499) |
| 1.7 | 5.2 | GO:0009227 | nucleotide-sugar catabolic process(GO:0009227) |
| 1.6 | 9.6 | GO:0035166 | post-embryonic hemopoiesis(GO:0035166) |
| 1.6 | 9.4 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 1.6 | 4.7 | GO:0060734 | regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:0060734) response to methyl methanesulfonate(GO:0072702) cellular response to methyl methanesulfonate(GO:0072703) response to environmental enrichment(GO:0090648) positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 1.5 | 5.9 | GO:0032914 | positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 1.5 | 7.3 | GO:0034227 | tRNA thio-modification(GO:0034227) |
| 1.4 | 5.8 | GO:0018214 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 1.4 | 4.3 | GO:0072429 | response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 1.4 | 4.1 | GO:0046078 | pyrimidine deoxyribonucleoside monophosphate biosynthetic process(GO:0009177) dUMP metabolic process(GO:0046078) |
| 1.3 | 10.5 | GO:0048702 | embryonic neurocranium morphogenesis(GO:0048702) axis elongation involved in somitogenesis(GO:0090245) |
| 1.3 | 5.2 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 1.3 | 6.3 | GO:2000619 | negative regulation of histone H4-K16 acetylation(GO:2000619) |
| 1.3 | 6.3 | GO:1990918 | meiotic DNA double-strand break processing(GO:0000706) double-strand break repair involved in meiotic recombination(GO:1990918) |
| 1.2 | 7.5 | GO:0006041 | glucosamine metabolic process(GO:0006041) |
| 1.2 | 6.0 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 1.2 | 4.7 | GO:1903576 | response to L-arginine(GO:1903576) |
| 1.2 | 4.7 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) negative regulation of leukocyte adhesion to vascular endothelial cell(GO:1904995) |
| 1.2 | 3.5 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 1.2 | 14.1 | GO:1902993 | positive regulation of beta-amyloid formation(GO:1902004) positive regulation of amyloid precursor protein catabolic process(GO:1902993) |
| 1.2 | 16.2 | GO:0031272 | regulation of pseudopodium assembly(GO:0031272) |
| 1.1 | 6.8 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 1.1 | 3.4 | GO:0002752 | cell surface pattern recognition receptor signaling pathway(GO:0002752) |
| 1.1 | 26.4 | GO:0060314 | regulation of ryanodine-sensitive calcium-release channel activity(GO:0060314) |
| 1.1 | 4.4 | GO:1902994 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) regulation of phospholipid efflux(GO:1902994) positive regulation of phospholipid efflux(GO:1902995) |
| 1.1 | 11.8 | GO:0070244 | negative regulation of thymocyte apoptotic process(GO:0070244) |
| 1.1 | 2.1 | GO:0052564 | response to immune response of other organism involved in symbiotic interaction(GO:0052564) response to host immune response(GO:0052572) |
| 1.1 | 3.2 | GO:1901143 | insulin catabolic process(GO:1901143) |
| 1.1 | 4.2 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 1.1 | 10.5 | GO:0042178 | xenobiotic catabolic process(GO:0042178) |
| 1.0 | 4.1 | GO:0046061 | dATP catabolic process(GO:0046061) |
| 1.0 | 6.0 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 1.0 | 1.9 | GO:0070340 | detection of diacyl bacterial lipopeptide(GO:0042496) detection of bacterial lipopeptide(GO:0070340) |
| 0.9 | 2.8 | GO:0070245 | positive regulation of thymocyte apoptotic process(GO:0070245) |
| 0.9 | 3.6 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.9 | 8.8 | GO:0061042 | adiponectin-activated signaling pathway(GO:0033211) vascular wound healing(GO:0061042) |
| 0.8 | 6.6 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.8 | 4.9 | GO:1902416 | positive regulation of mRNA binding(GO:1902416) |
| 0.8 | 9.7 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.7 | 6.0 | GO:1904044 | response to aldosterone(GO:1904044) |
| 0.7 | 5.2 | GO:0060075 | regulation of resting membrane potential(GO:0060075) |
| 0.7 | 24.5 | GO:0007202 | activation of phospholipase C activity(GO:0007202) |
| 0.7 | 14.8 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.7 | 5.1 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.7 | 2.1 | GO:0097167 | circadian regulation of translation(GO:0097167) response to sodium phosphate(GO:1904383) response to putrescine(GO:1904585) cellular response to putrescine(GO:1904586) |
| 0.7 | 9.6 | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.7 | 3.4 | GO:0032707 | negative regulation of interleukin-23 production(GO:0032707) |
| 0.7 | 2.7 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.7 | 5.3 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.7 | 5.9 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.6 | 3.2 | GO:0071955 | recycling endosome to Golgi transport(GO:0071955) |
| 0.6 | 13.9 | GO:0010971 | positive regulation of G2/M transition of mitotic cell cycle(GO:0010971) |
| 0.6 | 7.6 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.6 | 3.8 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.6 | 4.4 | GO:0007182 | common-partner SMAD protein phosphorylation(GO:0007182) |
| 0.6 | 5.6 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.6 | 3.1 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
| 0.6 | 3.7 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.6 | 4.2 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 0.6 | 1.8 | GO:0008592 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) regulation of Toll signaling pathway(GO:0008592) negative regulation of Toll signaling pathway(GO:0045751) |
| 0.6 | 2.4 | GO:0019046 | viral latency(GO:0019042) release from viral latency(GO:0019046) |
| 0.6 | 2.4 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.6 | 8.5 | GO:0030575 | male sex determination(GO:0030238) nuclear body organization(GO:0030575) |
| 0.6 | 1.7 | GO:0033986 | response to methanol(GO:0033986) |
| 0.5 | 7.5 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.5 | 4.6 | GO:0052697 | xenobiotic glucuronidation(GO:0052697) |
| 0.5 | 7.6 | GO:0035518 | histone H2A monoubiquitination(GO:0035518) |
| 0.5 | 3.3 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.5 | 5.0 | GO:0071679 | cytoskeletal anchoring at plasma membrane(GO:0007016) commissural neuron axon guidance(GO:0071679) |
| 0.4 | 4.0 | GO:0016559 | peroxisome fission(GO:0016559) vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.4 | 3.6 | GO:0060613 | fat pad development(GO:0060613) |
| 0.4 | 2.1 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.4 | 19.6 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.4 | 13.1 | GO:0071526 | semaphorin-plexin signaling pathway(GO:0071526) |
| 0.4 | 9.4 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.4 | 9.0 | GO:0019682 | pentose-phosphate shunt(GO:0006098) glyceraldehyde-3-phosphate metabolic process(GO:0019682) |
| 0.4 | 2.0 | GO:0070827 | chromatin maintenance(GO:0070827) |
| 0.4 | 1.9 | GO:1904798 | regulation of core promoter binding(GO:1904796) positive regulation of core promoter binding(GO:1904798) |
| 0.4 | 4.1 | GO:0060693 | regulation of branching involved in salivary gland morphogenesis(GO:0060693) |
| 0.4 | 13.8 | GO:0045026 | plasma membrane fusion(GO:0045026) |
| 0.4 | 5.0 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.4 | 1.1 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) negative regulation of adipose tissue development(GO:1904178) |
| 0.4 | 1.4 | GO:0008611 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.4 | 7.0 | GO:0072673 | lamellipodium morphogenesis(GO:0072673) |
| 0.3 | 22.4 | GO:0043462 | regulation of ATPase activity(GO:0043462) |
| 0.3 | 2.4 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.3 | 3.4 | GO:0090070 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.3 | 6.1 | GO:0051593 | response to folic acid(GO:0051593) |
| 0.3 | 2.0 | GO:0030222 | eosinophil differentiation(GO:0030222) |
| 0.3 | 3.7 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.3 | 10.1 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.3 | 19.9 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.3 | 4.3 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.3 | 6.9 | GO:0031163 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.3 | 5.1 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.3 | 5.4 | GO:0032291 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.3 | 12.3 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.3 | 1.6 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.3 | 24.4 | GO:0035383 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.3 | 3.0 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.3 | 7.5 | GO:0046579 | positive regulation of Ras protein signal transduction(GO:0046579) |
| 0.3 | 1.2 | GO:0002101 | tRNA wobble cytosine modification(GO:0002101) |
| 0.3 | 1.7 | GO:0030046 | parallel actin filament bundle assembly(GO:0030046) |
| 0.3 | 1.2 | GO:0075525 | viral translational termination-reinitiation(GO:0075525) |
| 0.3 | 4.3 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.3 | 9.8 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.3 | 3.9 | GO:0032025 | response to cobalt ion(GO:0032025) |
| 0.3 | 13.3 | GO:0048662 | negative regulation of smooth muscle cell proliferation(GO:0048662) |
| 0.3 | 0.8 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.3 | 2.4 | GO:0007512 | adult heart development(GO:0007512) |
| 0.3 | 7.9 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.3 | 0.8 | GO:0021699 | cerebellar cortex maturation(GO:0021699) |
| 0.2 | 1.0 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) |
| 0.2 | 16.6 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.2 | 4.8 | GO:0030889 | negative regulation of B cell proliferation(GO:0030889) |
| 0.2 | 0.9 | GO:1990928 | response to amino acid starvation(GO:1990928) |
| 0.2 | 7.0 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.2 | 3.0 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.2 | 9.9 | GO:0022904 | respiratory electron transport chain(GO:0022904) |
| 0.2 | 0.6 | GO:0015965 | diadenosine polyphosphate biosynthetic process(GO:0015960) diadenosine tetraphosphate metabolic process(GO:0015965) diadenosine tetraphosphate biosynthetic process(GO:0015966) |
| 0.2 | 3.4 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.2 | 0.8 | GO:0045650 | negative regulation of macrophage differentiation(GO:0045650) negative regulation of dendritic cell differentiation(GO:2001199) |
| 0.2 | 31.2 | GO:0010952 | positive regulation of peptidase activity(GO:0010952) |
| 0.2 | 6.6 | GO:0006767 | water-soluble vitamin metabolic process(GO:0006767) |
| 0.2 | 11.2 | GO:2000134 | negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.2 | 4.4 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.2 | 2.0 | GO:0061088 | sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.2 | 6.2 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.2 | 1.8 | GO:0072502 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.2 | 1.8 | GO:0001973 | adenosine receptor signaling pathway(GO:0001973) |
| 0.2 | 3.7 | GO:0032098 | regulation of appetite(GO:0032098) |
| 0.2 | 7.0 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) |
| 0.2 | 6.2 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.2 | 4.4 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.2 | 5.4 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.2 | 0.9 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.1 | 3.4 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.1 | 0.3 | GO:0010796 | regulation of multivesicular body size(GO:0010796) |
| 0.1 | 25.0 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
| 0.1 | 0.4 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.1 | 1.3 | GO:0034244 | negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.1 | 7.1 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.1 | 3.3 | GO:0071173 | mitotic spindle assembly checkpoint(GO:0007094) spindle checkpoint(GO:0031577) spindle assembly checkpoint(GO:0071173) mitotic spindle checkpoint(GO:0071174) |
| 0.1 | 4.6 | GO:1901998 | toxin transport(GO:1901998) |
| 0.1 | 2.0 | GO:0014003 | oligodendrocyte development(GO:0014003) |
| 0.1 | 26.7 | GO:0006898 | receptor-mediated endocytosis(GO:0006898) |
| 0.1 | 6.3 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.1 | 4.4 | GO:2001238 | positive regulation of extrinsic apoptotic signaling pathway(GO:2001238) |
| 0.1 | 1.9 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.1 | 2.9 | GO:0050482 | arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.1 | 1.5 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.1 | 1.2 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 0.1 | 1.6 | GO:0010800 | positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.1 | 2.1 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.1 | 1.5 | GO:0042274 | ribosomal small subunit biogenesis(GO:0042274) |
| 0.1 | 0.9 | GO:1902916 | positive regulation of protein polyubiquitination(GO:1902916) |
| 0.1 | 1.7 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.1 | 1.1 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.1 | 2.9 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.1 | 0.4 | GO:0042427 | serotonin biosynthetic process(GO:0042427) primary amino compound biosynthetic process(GO:1901162) |
| 0.1 | 1.9 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.1 | 0.4 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.1 | 0.9 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.1 | 1.3 | GO:0070303 | negative regulation of stress-activated MAPK cascade(GO:0032873) negative regulation of stress-activated protein kinase signaling cascade(GO:0070303) |
| 0.1 | 1.8 | GO:0006379 | mRNA cleavage(GO:0006379) |
| 0.1 | 1.4 | GO:0001522 | pseudouridine synthesis(GO:0001522) |
| 0.1 | 9.9 | GO:0030522 | intracellular receptor signaling pathway(GO:0030522) |
| 0.1 | 3.9 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 2.9 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.0 | 0.4 | GO:2000059 | negative regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000059) |
| 0.0 | 1.4 | GO:0014068 | positive regulation of phosphatidylinositol 3-kinase signaling(GO:0014068) |
| 0.0 | 1.2 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 3.9 | GO:0030100 | regulation of endocytosis(GO:0030100) |
| 0.0 | 2.1 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.0 | 5.4 | GO:0042594 | response to starvation(GO:0042594) |
| 0.0 | 0.6 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 7.8 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 1.4 | GO:0032411 | positive regulation of transporter activity(GO:0032411) |
| 0.0 | 6.3 | GO:0071805 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.0 | 3.5 | GO:0015992 | proton transport(GO:0015992) |
| 0.0 | 2.7 | GO:0042254 | ribosome biogenesis(GO:0042254) |
| 0.0 | 6.2 | GO:0000398 | RNA splicing, via transesterification reactions with bulged adenosine as nucleophile(GO:0000377) mRNA splicing, via spliceosome(GO:0000398) |
| 0.0 | 4.2 | GO:0006790 | sulfur compound metabolic process(GO:0006790) |
| 0.0 | 0.1 | GO:0033522 | histone H2A ubiquitination(GO:0033522) |
| 0.0 | 0.8 | GO:0002279 | mast cell activation involved in immune response(GO:0002279) mast cell degranulation(GO:0043303) |
| 0.0 | 0.3 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 3.7 | GO:0016042 | lipid catabolic process(GO:0016042) |
| 0.0 | 1.5 | GO:0008203 | cholesterol metabolic process(GO:0008203) |
| 0.0 | 7.6 | GO:0006412 | translation(GO:0006412) |
| 0.0 | 0.2 | GO:0006999 | nuclear pore organization(GO:0006999) |
| 0.0 | 0.9 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.0 | 0.6 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.0 | 0.9 | GO:0034968 | histone lysine methylation(GO:0034968) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.5 | 42.6 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 3.0 | 12.0 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 2.6 | 18.2 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 2.5 | 7.6 | GO:0005760 | gamma DNA polymerase complex(GO:0005760) |
| 2.4 | 22.0 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 2.4 | 14.3 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 2.4 | 9.4 | GO:0009330 | DNA topoisomerase complex (ATP-hydrolyzing)(GO:0009330) |
| 1.9 | 7.5 | GO:0035339 | SPOTS complex(GO:0035339) |
| 1.9 | 22.4 | GO:0005861 | troponin complex(GO:0005861) |
| 1.6 | 20.2 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 1.5 | 24.5 | GO:0070852 | cell body fiber(GO:0070852) |
| 1.5 | 6.0 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 1.5 | 6.0 | GO:0035363 | histone locus body(GO:0035363) |
| 1.4 | 14.0 | GO:0032437 | cuticular plate(GO:0032437) |
| 1.3 | 9.4 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 1.3 | 3.9 | GO:1990590 | ATF1-ATF4 transcription factor complex(GO:1990590) |
| 1.2 | 3.7 | GO:0046691 | intracellular canaliculus(GO:0046691) |
| 1.1 | 5.6 | GO:0070695 | FHF complex(GO:0070695) |
| 1.1 | 16.6 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 1.1 | 7.6 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.9 | 4.3 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.8 | 5.9 | GO:0098553 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.8 | 2.5 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) |
| 0.8 | 7.0 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.8 | 21.7 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.7 | 5.9 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.7 | 5.0 | GO:0016011 | dystroglycan complex(GO:0016011) |
| 0.7 | 6.9 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.6 | 1.9 | GO:0035355 | Toll-like receptor 2-Toll-like receptor 6 protein complex(GO:0035355) |
| 0.6 | 1.9 | GO:0035101 | FACT complex(GO:0035101) |
| 0.6 | 7.6 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.6 | 5.3 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 0.6 | 6.3 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.5 | 4.3 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.5 | 8.3 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.5 | 9.7 | GO:0034361 | very-low-density lipoprotein particle(GO:0034361) triglyceride-rich lipoprotein particle(GO:0034385) |
| 0.5 | 7.2 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.5 | 2.9 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.5 | 1.0 | GO:0005668 | RNA polymerase transcription factor SL1 complex(GO:0005668) |
| 0.5 | 5.1 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) |
| 0.4 | 7.9 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.4 | 8.8 | GO:0000346 | transcription export complex(GO:0000346) |
| 0.4 | 6.1 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.4 | 22.6 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.4 | 26.4 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.4 | 4.5 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.4 | 29.1 | GO:0005811 | lipid particle(GO:0005811) |
| 0.4 | 3.4 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.4 | 7.4 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.4 | 18.4 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.4 | 4.3 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.3 | 1.4 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.3 | 5.4 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.3 | 4.6 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.3 | 3.2 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.3 | 7.8 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.3 | 13.9 | GO:0016592 | mediator complex(GO:0016592) |
| 0.3 | 22.4 | GO:0045095 | keratin filament(GO:0045095) |
| 0.3 | 3.9 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.3 | 9.1 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.3 | 1.8 | GO:0000243 | commitment complex(GO:0000243) |
| 0.3 | 30.9 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.3 | 13.3 | GO:0016235 | aggresome(GO:0016235) |
| 0.3 | 6.8 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.3 | 25.5 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.3 | 3.0 | GO:0016342 | catenin complex(GO:0016342) |
| 0.2 | 10.0 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.2 | 2.9 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.2 | 1.2 | GO:0030663 | COPI-coated vesicle membrane(GO:0030663) |
| 0.2 | 3.3 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.2 | 1.8 | GO:0032806 | holo TFIIH complex(GO:0005675) carboxy-terminal domain protein kinase complex(GO:0032806) |
| 0.2 | 2.2 | GO:0030891 | VCB complex(GO:0030891) |
| 0.2 | 9.7 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.2 | 3.2 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.2 | 62.2 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.2 | 3.8 | GO:0019013 | viral nucleocapsid(GO:0019013) |
| 0.2 | 1.6 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.2 | 1.9 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.2 | 22.9 | GO:0030018 | Z disc(GO:0030018) |
| 0.2 | 3.0 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.2 | 3.3 | GO:0045120 | pronucleus(GO:0045120) |
| 0.2 | 3.5 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.2 | 2.4 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.2 | 3.6 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.2 | 4.4 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.2 | 7.4 | GO:0044438 | microbody part(GO:0044438) peroxisomal part(GO:0044439) |
| 0.2 | 11.5 | GO:0022627 | small ribosomal subunit(GO:0015935) cytosolic small ribosomal subunit(GO:0022627) |
| 0.2 | 1.4 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.2 | 1.9 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.2 | 0.9 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.1 | 3.4 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.1 | 21.6 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.1 | 6.6 | GO:0030017 | sarcomere(GO:0030017) |
| 0.1 | 1.5 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.1 | 1.8 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.1 | 9.9 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.1 | 1.1 | GO:0005638 | lamin filament(GO:0005638) |
| 0.1 | 11.7 | GO:0005902 | microvillus(GO:0005902) |
| 0.1 | 1.2 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.1 | 11.2 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 8.0 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.1 | 9.9 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 44.8 | GO:0045177 | apical part of cell(GO:0045177) |
| 0.1 | 1.9 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.1 | 6.6 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.1 | 4.2 | GO:0005844 | polysome(GO:0005844) |
| 0.1 | 6.7 | GO:0005776 | autophagosome(GO:0005776) |
| 0.1 | 10.7 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.1 | 13.0 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.1 | 11.0 | GO:0000781 | chromosome, telomeric region(GO:0000781) |
| 0.1 | 0.3 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.1 | 9.5 | GO:0000922 | spindle pole(GO:0000922) |
| 0.1 | 0.3 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.1 | 208.1 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.1 | 4.3 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 0.6 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.1 | 1.4 | GO:0005753 | mitochondrial proton-transporting ATP synthase complex(GO:0005753) |
| 0.1 | 3.2 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.1 | 8.1 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.1 | 6.3 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.1 | 1.2 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.1 | 5.1 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.1 | 0.9 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 8.0 | GO:0005911 | cell-cell junction(GO:0005911) |
| 0.0 | 6.6 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.2 | GO:0044613 | nuclear pore central transport channel(GO:0044613) nuclear pore nuclear basket(GO:0044615) |
| 0.0 | 1.4 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.1 | GO:0033503 | ubiquitin conjugating enzyme complex(GO:0031371) HULC complex(GO:0033503) |
| 0.0 | 4.2 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 5.8 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.6 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 1.8 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 1.1 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 8.9 | GO:0031966 | mitochondrial membrane(GO:0031966) |
| 0.0 | 1.0 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 1.1 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 1.1 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.4 | 22.1 | GO:0005302 | L-tyrosine transmembrane transporter activity(GO:0005302) |
| 7.2 | 21.6 | GO:1901375 | acetylcholine transmembrane transporter activity(GO:0005277) acetate ester transmembrane transporter activity(GO:1901375) |
| 7.0 | 28.1 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 5.6 | 22.4 | GO:0030899 | troponin C binding(GO:0030172) calcium-dependent ATPase activity(GO:0030899) |
| 5.6 | 16.7 | GO:0032810 | sterol response element binding(GO:0032810) |
| 5.2 | 15.7 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 4.1 | 16.6 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 3.9 | 15.7 | GO:0003918 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 3.3 | 10.0 | GO:0052655 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 3.3 | 42.6 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 3.1 | 9.3 | GO:0070905 | serine binding(GO:0070905) |
| 2.9 | 23.5 | GO:0003996 | acyl-CoA ligase activity(GO:0003996) |
| 2.9 | 23.2 | GO:0043199 | sulfate binding(GO:0043199) |
| 2.5 | 7.6 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 2.5 | 7.6 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 2.5 | 19.7 | GO:0008430 | selenium binding(GO:0008430) |
| 2.3 | 9.3 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 2.3 | 22.6 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 2.2 | 6.6 | GO:0047708 | biotinidase activity(GO:0047708) |
| 2.2 | 8.8 | GO:0055100 | adiponectin binding(GO:0055100) |
| 2.1 | 10.5 | GO:0070404 | NADH binding(GO:0070404) |
| 2.0 | 7.9 | GO:0051870 | methotrexate binding(GO:0051870) |
| 2.0 | 5.9 | GO:0030337 | DNA polymerase processivity factor activity(GO:0030337) |
| 1.9 | 9.5 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 1.9 | 7.6 | GO:0016890 | site-specific endodeoxyribonuclease activity, specific for altered base(GO:0016890) |
| 1.8 | 5.4 | GO:0019807 | aspartoacylase activity(GO:0019807) |
| 1.8 | 10.5 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 1.6 | 12.6 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 1.5 | 21.3 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 1.5 | 6.0 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 1.4 | 4.1 | GO:0008413 | 8-oxo-7,8-dihydroguanosine triphosphate pyrophosphatase activity(GO:0008413) 8-oxo-7,8-dihydrodeoxyguanosine triphosphate pyrophosphatase activity(GO:0035539) |
| 1.3 | 10.6 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 1.3 | 5.2 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 1.2 | 4.8 | GO:0004027 | alcohol sulfotransferase activity(GO:0004027) |
| 1.2 | 4.7 | GO:0016635 | oxidoreductase activity, acting on the CH-CH group of donors, quinone or related compound as acceptor(GO:0016635) |
| 1.2 | 4.6 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 1.1 | 3.4 | GO:0097363 | protein O-GlcNAc transferase activity(GO:0097363) |
| 1.1 | 8.9 | GO:0016936 | galactoside binding(GO:0016936) |
| 1.1 | 9.7 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 1.1 | 4.3 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 1.1 | 41.0 | GO:0043236 | laminin binding(GO:0043236) |
| 1.0 | 19.9 | GO:0015643 | toxic substance binding(GO:0015643) |
| 1.0 | 5.2 | GO:0005534 | galactose binding(GO:0005534) |
| 1.0 | 5.9 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.9 | 7.6 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.9 | 31.2 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.9 | 13.1 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.9 | 16.8 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.9 | 5.5 | GO:0003847 | 1-alkyl-2-acetylglycerophosphocholine esterase activity(GO:0003847) |
| 0.9 | 12.7 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.9 | 14.3 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.8 | 3.2 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.8 | 5.3 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.8 | 4.5 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.7 | 4.4 | GO:0090554 | phosphatidylcholine-translocating ATPase activity(GO:0090554) |
| 0.7 | 16.6 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.7 | 2.0 | GO:0000991 | transcription factor activity, core RNA polymerase II binding(GO:0000991) |
| 0.7 | 20.3 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.6 | 5.1 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.6 | 1.9 | GO:0042498 | Toll-like receptor 2 binding(GO:0035663) diacyl lipopeptide binding(GO:0042498) |
| 0.6 | 12.1 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.6 | 31.4 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.6 | 24.7 | GO:0016655 | oxidoreductase activity, acting on NAD(P)H, quinone or similar compound as acceptor(GO:0016655) |
| 0.6 | 4.2 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.6 | 9.6 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.6 | 11.4 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.6 | 1.8 | GO:2001070 | starch binding(GO:2001070) |
| 0.6 | 16.2 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.6 | 7.3 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.6 | 3.3 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 0.6 | 3.9 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.5 | 3.1 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.5 | 18.2 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.5 | 24.5 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.5 | 3.4 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.5 | 4.3 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.5 | 14.2 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.4 | 2.2 | GO:0005550 | pheromone binding(GO:0005550) |
| 0.4 | 12.0 | GO:0043394 | proteoglycan binding(GO:0043394) |
| 0.4 | 14.6 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.4 | 3.4 | GO:0051022 | Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.4 | 22.6 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.4 | 3.2 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.4 | 12.5 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.4 | 7.4 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.4 | 3.6 | GO:0019808 | polyamine binding(GO:0019808) |
| 0.3 | 1.0 | GO:0070698 | nodal binding(GO:0038100) type I activin receptor binding(GO:0070698) |
| 0.3 | 2.9 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.3 | 41.3 | GO:0008201 | heparin binding(GO:0008201) |
| 0.3 | 0.9 | GO:0036435 | K48-linked polyubiquitin binding(GO:0036435) |
| 0.3 | 26.8 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.3 | 11.3 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.3 | 3.5 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.3 | 6.8 | GO:0015295 | solute:proton symporter activity(GO:0015295) |
| 0.3 | 3.7 | GO:0015250 | water channel activity(GO:0015250) |
| 0.3 | 8.5 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.3 | 1.8 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.3 | 7.5 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.3 | 3.0 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.2 | 2.4 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.2 | 9.2 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.2 | 1.2 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.2 | 2.1 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.2 | 13.1 | GO:0048029 | monosaccharide binding(GO:0048029) |
| 0.2 | 5.8 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.2 | 12.3 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.2 | 3.4 | GO:0015288 | porin activity(GO:0015288) |
| 0.2 | 6.1 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.2 | 3.4 | GO:0038187 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.2 | 4.4 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.2 | 9.9 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.2 | 1.3 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.2 | 3.6 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.2 | 12.4 | GO:0005178 | integrin binding(GO:0005178) |
| 0.2 | 21.9 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.1 | 4.6 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 20.0 | GO:0042626 | ATPase activity, coupled to transmembrane movement of substances(GO:0042626) |
| 0.1 | 2.0 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.1 | 3.3 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.1 | 5.8 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.1 | 51.1 | GO:0005525 | GTP binding(GO:0005525) |
| 0.1 | 2.1 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.1 | 5.9 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 33.6 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.1 | 4.4 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 0.4 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.1 | 1.4 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.1 | 12.5 | GO:0001046 | core promoter sequence-specific DNA binding(GO:0001046) |
| 0.1 | 0.5 | GO:0070137 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.1 | 2.8 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.1 | 3.7 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.1 | 0.9 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.1 | 2.9 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.1 | 1.4 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.1 | 1.4 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.1 | 4.7 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.1 | 6.6 | GO:0004713 | protein tyrosine kinase activity(GO:0004713) |
| 0.1 | 1.6 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.1 | 11.5 | GO:0005267 | potassium channel activity(GO:0005267) |
| 0.1 | 2.6 | GO:0099589 | G-protein coupled serotonin receptor activity(GO:0004993) serotonin receptor activity(GO:0099589) |
| 0.1 | 6.4 | GO:0050660 | flavin adenine dinucleotide binding(GO:0050660) |
| 0.1 | 0.6 | GO:0015266 | protein channel activity(GO:0015266) |
| 0.1 | 2.0 | GO:0016876 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.1 | 3.9 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.1 | 1.4 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.1 | 10.0 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.1 | 1.8 | GO:0017069 | snRNA binding(GO:0017069) |
| 0.1 | 0.4 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.1 | 4.3 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
| 0.1 | 0.2 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 5.1 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 4.2 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 2.3 | GO:0003724 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) RNA-dependent ATPase activity(GO:0008186) |
| 0.0 | 7.1 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.8 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.6 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 20.2 | GO:0001071 | nucleic acid binding transcription factor activity(GO:0001071) transcription factor activity, sequence-specific DNA binding(GO:0003700) |
| 0.0 | 0.3 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.4 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.6 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 2.0 | GO:0070851 | growth factor receptor binding(GO:0070851) |
| 0.0 | 0.2 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 1.5 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.0 | 2.3 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 11.4 | GO:0003723 | RNA binding(GO:0003723) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 12.3 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.7 | 5.0 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.5 | 21.2 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.4 | 13.8 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.4 | 7.5 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.4 | 16.8 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.4 | 9.5 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.4 | 13.3 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.4 | 16.7 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.3 | 2.0 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.3 | 10.5 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.3 | 10.9 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.3 | 8.6 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.3 | 5.3 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.3 | 11.9 | PID ATR PATHWAY | ATR signaling pathway |
| 0.3 | 19.6 | PID E2F PATHWAY | E2F transcription factor network |
| 0.2 | 12.0 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.2 | 7.0 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.2 | 4.4 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.2 | 4.1 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.2 | 7.2 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.2 | 34.4 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.2 | 12.6 | PID FGF PATHWAY | FGF signaling pathway |
| 0.2 | 2.4 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.2 | 1.8 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.2 | 6.2 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.2 | 10.0 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.2 | 3.9 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.2 | 23.4 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.2 | 7.8 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.2 | 10.5 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.1 | 10.5 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.1 | 5.9 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 1.9 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.1 | 2.2 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.1 | 5.2 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.1 | 2.0 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.1 | 2.7 | SIG CHEMOTAXIS | Genes related to chemotaxis |
| 0.0 | 1.7 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.8 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 1.8 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.7 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 4.1 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 21.6 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 1.6 | 60.8 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 1.3 | 15.7 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 1.3 | 50.2 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 1.2 | 12.7 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 1.0 | 15.8 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 1.0 | 5.8 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.9 | 34.5 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.9 | 12.6 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.9 | 9.4 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.8 | 7.6 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.8 | 12.3 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.8 | 10.6 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.7 | 10.5 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.7 | 43.2 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.7 | 14.3 | REACTOME APC C CDH1 MEDIATED DEGRADATION OF CDC20 AND OTHER APC C CDH1 TARGETED PROTEINS IN LATE MITOSIS EARLY G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |
| 0.6 | 7.6 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.6 | 10.0 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.6 | 10.0 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.5 | 5.1 | REACTOME TRAF6 MEDIATED INDUCTION OF TAK1 COMPLEX | Genes involved in TRAF6 mediated induction of TAK1 complex |
| 0.5 | 13.8 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.5 | 5.4 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.4 | 6.6 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.4 | 7.2 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.4 | 5.9 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.4 | 5.2 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.4 | 10.1 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.4 | 37.0 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.4 | 7.0 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.3 | 16.9 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.3 | 6.2 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.3 | 8.9 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.3 | 3.7 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.3 | 15.2 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.3 | 9.4 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.3 | 7.9 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.3 | 4.1 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.3 | 18.9 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.3 | 2.1 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.3 | 6.1 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.2 | 4.7 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.2 | 3.4 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.2 | 4.6 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.2 | 3.9 | REACTOME NUCLEAR EVENTS KINASE AND TRANSCRIPTION FACTOR ACTIVATION | Genes involved in Nuclear Events (kinase and transcription factor activation) |
| 0.2 | 37.9 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.2 | 2.4 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.2 | 14.4 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.2 | 3.2 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.2 | 4.1 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.2 | 20.3 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.2 | 4.4 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.2 | 2.2 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.2 | 2.9 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.2 | 6.4 | REACTOME TRANSPORT OF MATURE MRNA DERIVED FROM AN INTRONLESS TRANSCRIPT | Genes involved in Transport of Mature mRNA Derived from an Intronless Transcript |
| 0.2 | 4.9 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.2 | 3.6 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.2 | 4.3 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.1 | 1.9 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.1 | 2.9 | REACTOME KERATAN SULFATE KERATIN METABOLISM | Genes involved in Keratan sulfate/keratin metabolism |
| 0.1 | 6.3 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 3.9 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.1 | 21.3 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |
| 0.1 | 12.1 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.1 | 1.6 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.1 | 1.0 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 1.5 | REACTOME TRANSLATION | Genes involved in Translation |
| 0.1 | 14.6 | REACTOME METABOLISM OF CARBOHYDRATES | Genes involved in Metabolism of carbohydrates |
| 0.1 | 2.0 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.1 | 4.2 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 3.1 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 1.9 | REACTOME MYD88 MAL CASCADE INITIATED ON PLASMA MEMBRANE | Genes involved in MyD88:Mal cascade initiated on plasma membrane |
| 0.0 | 0.7 | REACTOME CTLA4 INHIBITORY SIGNALING | Genes involved in CTLA4 inhibitory signaling |
| 0.0 | 5.1 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.8 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.6 | REACTOME TRNA AMINOACYLATION | Genes involved in tRNA Aminoacylation |
| 0.0 | 1.8 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.0 | 0.3 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |