Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
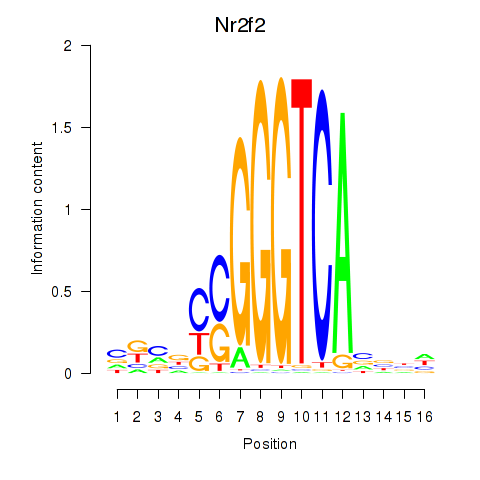
Results for Nr2f2
Z-value: 1.76
Transcription factors associated with Nr2f2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nr2f2
|
ENSRNOG00000010308 | nuclear receptor subfamily 2, group F, member 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nr2f2 | rn6_v1_chr1_-_131460473_131460473 | 0.22 | 5.2e-05 | Click! |
Activity profile of Nr2f2 motif
Sorted Z-values of Nr2f2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_-_32868680 | 58.55 |
ENSRNOT00000015974
ENSRNOT00000082392 |
Aadat
|
aminoadipate aminotransferase |
| chr12_+_943006 | 57.36 |
ENSRNOT00000001449
|
Kl
|
Klotho |
| chr5_-_137372993 | 48.33 |
ENSRNOT00000092823
|
Tmem125
|
transmembrane protein 125 |
| chr9_-_88357182 | 47.70 |
ENSRNOT00000041176
|
Col4a4
|
collagen type IV alpha 4 chain |
| chrX_-_142131545 | 40.73 |
ENSRNOT00000077402
|
Fgf13
|
fibroblast growth factor 13 |
| chr16_+_20521956 | 40.68 |
ENSRNOT00000026597
|
Pgpep1
|
pyroglutamyl-peptidase I |
| chr3_-_2689084 | 39.83 |
ENSRNOT00000020926
|
Ptgds
|
prostaglandin D2 synthase |
| chr10_+_92337879 | 38.88 |
ENSRNOT00000042984
|
Mapt
|
microtubule-associated protein tau |
| chr14_-_115052450 | 38.30 |
ENSRNOT00000067998
|
Acyp2
|
acylphosphatase 2 |
| chr9_-_88356716 | 38.11 |
ENSRNOT00000077503
|
Col4a4
|
collagen type IV alpha 4 chain |
| chr2_-_23289266 | 38.04 |
ENSRNOT00000061708
|
Bhmt2
|
betaine-homocysteine S-methyltransferase 2 |
| chr1_+_100131449 | 36.71 |
ENSRNOT00000091964
ENSRNOT00000071416 |
Klk1
|
kallikrein 1 |
| chr14_-_106393670 | 36.49 |
ENSRNOT00000011429
|
Mdh1
|
malate dehydrogenase 1 |
| chr19_+_24701049 | 35.00 |
ENSRNOT00000035890
|
Ndufb7
|
NADH:ubiquinone oxidoreductase subunit B7 |
| chr5_-_137372524 | 33.65 |
ENSRNOT00000009061
|
Tmem125
|
transmembrane protein 125 |
| chr17_-_417480 | 31.93 |
ENSRNOT00000023685
|
Fbp1
|
fructose-bisphosphatase 1 |
| chr3_-_9326993 | 31.67 |
ENSRNOT00000090137
|
Lamc3
|
laminin subunit gamma 3 |
| chr1_-_70235091 | 31.63 |
ENSRNOT00000075745
|
Apeg3
|
antisense paternally expressed gene 3 |
| chr10_+_15088935 | 31.41 |
ENSRNOT00000030273
|
Gng13
|
G protein subunit gamma 13 |
| chr14_+_39154529 | 30.52 |
ENSRNOT00000003191
|
Gabra4
|
gamma-aminobutyric acid type A receptor alpha4 subunit |
| chr1_-_81881549 | 29.34 |
ENSRNOT00000027497
|
Atp1a3
|
ATPase Na+/K+ transporting subunit alpha 3 |
| chr1_+_80321585 | 28.26 |
ENSRNOT00000022895
|
Ckm
|
creatine kinase, M-type |
| chr8_+_59507990 | 28.00 |
ENSRNOT00000018003
|
Hykk
|
hydroxylysine kinase |
| chr10_-_103848035 | 27.01 |
ENSRNOT00000029001
|
Fads6
|
fatty acid desaturase 6 |
| chr14_+_16491573 | 26.86 |
ENSRNOT00000002995
|
Sowahb
|
sosondowah ankyrin repeat domain family member B |
| chr9_+_16647922 | 26.85 |
ENSRNOT00000031625
|
Klc4
|
kinesin light chain 4 |
| chr10_-_109909646 | 26.39 |
ENSRNOT00000074362
ENSRNOT00000088907 |
Dcxr
|
dicarbonyl and L-xylulose reductase |
| chr6_+_105364668 | 25.78 |
ENSRNOT00000009513
ENSRNOT00000087090 |
Ttc9
|
tetratricopeptide repeat domain 9 |
| chr1_+_80195532 | 25.73 |
ENSRNOT00000022528
|
Rtn2
|
reticulon 2 |
| chr1_+_250426158 | 25.38 |
ENSRNOT00000067643
|
A1cf
|
APOBEC1 complementation factor |
| chr9_-_55673704 | 25.27 |
ENSRNOT00000066231
ENSRNOT00000081677 |
Tmeff2
|
transmembrane protein with EGF-like and two follistatin-like domains 2 |
| chr8_-_73164620 | 25.18 |
ENSRNOT00000031988
|
Tln2
|
talin 2 |
| chr7_-_12518684 | 25.02 |
ENSRNOT00000018691
ENSRNOT00000093426 |
Gpx4
|
glutathione peroxidase 4 |
| chrX_-_54303729 | 24.99 |
ENSRNOT00000087919
ENSRNOT00000064340 ENSRNOT00000051249 ENSRNOT00000087547 |
Gk
|
glycerol kinase |
| chr9_+_16647598 | 24.88 |
ENSRNOT00000087413
|
Klc4
|
kinesin light chain 4 |
| chr8_+_82257849 | 24.44 |
ENSRNOT00000011963
ENSRNOT00000075708 |
Gnb5
|
G protein subunit beta 5 |
| chr1_+_191704311 | 24.10 |
ENSRNOT00000024057
|
Scnn1g
|
sodium channel epithelial 1 gamma subunit |
| chr1_+_15620653 | 23.80 |
ENSRNOT00000017401
|
Map7
|
microtubule-associated protein 7 |
| chr5_+_147288723 | 23.75 |
ENSRNOT00000033285
|
Tmem54
|
transmembrane protein 54 |
| chr2_+_17616401 | 23.33 |
ENSRNOT00000064991
|
Edil3
|
EGF like repeats and discoidin domains 3 |
| chr10_+_39435227 | 23.24 |
ENSRNOT00000042144
ENSRNOT00000077185 |
P4ha2
|
prolyl 4-hydroxylase subunit alpha 2 |
| chr1_-_222167447 | 23.13 |
ENSRNOT00000028687
|
Prdx5
|
peroxiredoxin 5 |
| chr3_+_119014620 | 22.80 |
ENSRNOT00000067700
|
Slc27a2
|
solute carrier family 27 member 2 |
| chr8_+_111209908 | 22.75 |
ENSRNOT00000090221
|
Amotl2
|
angiomotin like 2 |
| chr5_+_158090173 | 22.53 |
ENSRNOT00000088766
ENSRNOT00000079516 ENSRNOT00000092026 |
Tas1r2
|
taste 1 receptor member 2 |
| chr13_+_75111778 | 22.16 |
ENSRNOT00000006924
|
Tp53i3
|
tumor protein p53 inducible protein 3 |
| chr6_-_133716847 | 21.85 |
ENSRNOT00000072399
|
Rtl1
|
retrotransposon-like 1 |
| chr20_+_6869767 | 21.81 |
ENSRNOT00000000631
ENSRNOT00000086330 ENSRNOT00000093188 ENSRNOT00000093736 |
RGD735065
|
similar to GI:13385412-like protein splice form I |
| chr15_+_33632416 | 21.41 |
ENSRNOT00000068212
|
AC130940.1
|
|
| chr12_-_46920952 | 21.35 |
ENSRNOT00000001532
|
Msi1
|
musashi RNA-binding protein 1 |
| chr11_-_25350974 | 21.26 |
ENSRNOT00000002187
|
Adamts1
|
ADAM metallopeptidase with thrombospondin type 1 motif, 1 |
| chr1_+_29413365 | 21.04 |
ENSRNOT00000019047
ENSRNOT00000088885 |
Hint3
|
histidine triad nucleotide binding protein 3 |
| chr1_-_265560386 | 20.97 |
ENSRNOT00000048592
|
LOC100911951
|
Kv channel-interacting protein 2-like |
| chr12_+_48316143 | 20.91 |
ENSRNOT00000084511
|
Svop
|
SV2 related protein |
| chr10_+_65448950 | 20.83 |
ENSRNOT00000082348
ENSRNOT00000037016 |
Rab34
|
RAB34, member RAS oncogene family |
| chr6_-_105261549 | 20.75 |
ENSRNOT00000009122
|
Synj2bp
|
synaptojanin 2 binding protein |
| chr3_-_55822551 | 20.58 |
ENSRNOT00000082624
|
Lrp2
|
LDL receptor related protein 2 |
| chr5_+_4982348 | 20.50 |
ENSRNOT00000010369
|
Lactb2
|
lactamase, beta 2 |
| chr1_+_262905570 | 20.18 |
ENSRNOT00000090765
|
Kcnip2
|
potassium voltage-gated channel interacting protein 2 |
| chr1_+_141821916 | 20.13 |
ENSRNOT00000071574
|
Echs1
|
enoyl-CoA hydratase, short chain 1 |
| chr1_+_219833299 | 20.11 |
ENSRNOT00000087432
|
Pc
|
pyruvate carboxylase |
| chr3_+_145032200 | 19.52 |
ENSRNOT00000068210
|
Syndig1
|
synapse differentiation inducing 1 |
| chr18_+_30574627 | 19.38 |
ENSRNOT00000060484
|
Pcdhb19
|
protocadherin beta 19 |
| chr1_-_216663720 | 19.23 |
ENSRNOT00000078944
ENSRNOT00000077409 |
Cdkn1c
|
cyclin-dependent kinase inhibitor 1C |
| chr2_+_207271853 | 18.77 |
ENSRNOT00000085355
ENSRNOT00000017254 |
Rhoc
|
ras homolog family member C |
| chr17_-_21705773 | 18.71 |
ENSRNOT00000078010
|
Gcnt2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme |
| chr1_-_89539210 | 18.67 |
ENSRNOT00000077462
ENSRNOT00000028644 |
Hpn
|
hepsin |
| chr2_+_23385183 | 18.64 |
ENSRNOT00000014860
|
Arsb
|
arylsulfatase B |
| chr1_-_222189604 | 18.55 |
ENSRNOT00000028704
|
Kcnk4
|
potassium two pore domain channel subfamily K member 4 |
| chr16_+_20691978 | 18.38 |
ENSRNOT00000038139
ENSRNOT00000082319 |
Tmem59l
|
transmembrane protein 59-like |
| chr2_+_242882306 | 18.24 |
ENSRNOT00000013661
|
Ddit4l
|
DNA-damage-inducible transcript 4-like |
| chr2_-_144323254 | 18.21 |
ENSRNOT00000018780
|
Sertm1
|
serine-rich and transmembrane domain containing 1 |
| chr17_-_16695126 | 17.86 |
ENSRNOT00000021550
|
Id4
|
inhibitor of DNA binding 4, HLH protein |
| chr5_-_99566317 | 17.62 |
ENSRNOT00000051028
ENSRNOT00000089235 |
Mpdz
|
multiple PDZ domain crumbs cell polarity complex component |
| chr8_+_48422036 | 17.36 |
ENSRNOT00000036051
|
Usp2
|
ubiquitin specific peptidase 2 |
| chr2_+_115678344 | 17.14 |
ENSRNOT00000036717
|
Slc7a14
|
solute carrier family 7, member 14 |
| chr18_-_28444880 | 17.08 |
ENSRNOT00000060696
|
Prob1
|
proline rich basic protein 1 |
| chr1_-_220467159 | 17.07 |
ENSRNOT00000075365
|
Tmem151a
|
transmembrane protein 151A |
| chr15_+_34167945 | 17.05 |
ENSRNOT00000032252
|
Carmil3
|
capping protein regulator and myosin 1 linker 3 |
| chr17_-_69862110 | 17.04 |
ENSRNOT00000058312
|
Akr1cl
|
aldo-keto reductase family 1, member C-like |
| chr8_-_49106177 | 17.03 |
ENSRNOT00000067558
|
Tmem25
|
transmembrane protein 25 |
| chrX_+_26294066 | 17.03 |
ENSRNOT00000037862
|
Hccs
|
holocytochrome c synthase |
| chr2_+_187344056 | 17.00 |
ENSRNOT00000025314
|
Nes
|
nestin |
| chr17_+_47397558 | 16.66 |
ENSRNOT00000085923
|
Epdr1
|
ependymin related 1 |
| chr1_+_266530477 | 16.61 |
ENSRNOT00000054699
|
Cnnm2
|
cyclin and CBS domain divalent metal cation transport mediator 2 |
| chr1_+_177048655 | 16.61 |
ENSRNOT00000081595
|
Mical2
|
microtubule associated monooxygenase, calponin and LIM domain containing 2 |
| chr7_-_126465723 | 16.44 |
ENSRNOT00000021196
|
Wnt7b
|
wingless-type MMTV integration site family, member 7B |
| chr19_+_25526751 | 16.41 |
ENSRNOT00000083448
|
Cacna1a
|
calcium voltage-gated channel subunit alpha1 A |
| chr1_-_42587666 | 16.38 |
ENSRNOT00000083225
ENSRNOT00000025355 |
Rgs17
|
regulator of G-protein signaling 17 |
| chr5_+_2813131 | 16.13 |
ENSRNOT00000009530
|
Sbspon
|
somatomedin B and thrombospondin, type 1 domain containing |
| chr6_+_92229686 | 15.99 |
ENSRNOT00000046085
|
Atl1
|
atlastin GTPase 1 |
| chr2_+_266317745 | 15.66 |
ENSRNOT00000080305
|
Wls
|
wntless Wnt ligand secretion mediator |
| chr5_-_105582375 | 15.58 |
ENSRNOT00000083373
|
Slc24a2
|
solute carrier family 24 member 2 |
| chr2_-_208420163 | 15.26 |
ENSRNOT00000021920
|
LOC100911417
|
ATP synthase subunit b, mitochondrial-like |
| chrX_+_5825711 | 15.25 |
ENSRNOT00000037360
|
Efhc2
|
EF-hand domain containing 2 |
| chr3_-_8981362 | 15.17 |
ENSRNOT00000086717
|
Crat
|
carnitine O-acetyltransferase |
| chr1_-_53038229 | 15.17 |
ENSRNOT00000017282
|
Mpc1
|
mitochondrial pyruvate carrier 1 |
| chr3_+_160047296 | 15.01 |
ENSRNOT00000051590
|
Pkig
|
cAMP-dependent protein kinase inhibitor gamma |
| chr12_+_40417944 | 14.98 |
ENSRNOT00000073125
|
Acad10
|
acyl-CoA dehydrogenase family, member 10 |
| chr13_+_113373578 | 14.98 |
ENSRNOT00000009900
|
Plxna2
|
plexin A2 |
| chr12_+_47920743 | 14.80 |
ENSRNOT00000072511
|
Mmab
|
methylmalonic aciduria (cobalamin deficiency) cblB type |
| chr2_+_198321142 | 14.73 |
ENSRNOT00000028760
|
Sv2a
|
synaptic vesicle glycoprotein 2a |
| chr4_-_6330699 | 14.71 |
ENSRNOT00000078077
ENSRNOT00000011814 |
Galnt11
|
polypeptide N-acetylgalactosaminyltransferase 11 |
| chr7_+_34533543 | 14.65 |
ENSRNOT00000007412
ENSRNOT00000084185 |
Ntn4
|
netrin 4 |
| chr18_-_56325095 | 14.64 |
ENSRNOT00000025209
|
Slc6a7
|
solute carrier family 6 member 7 |
| chr2_-_178389608 | 14.59 |
ENSRNOT00000013262
|
Etfdh
|
electron transfer flavoprotein dehydrogenase |
| chr13_+_52667969 | 14.53 |
ENSRNOT00000084986
ENSRNOT00000050284 |
Tnnt2
|
troponin T2, cardiac type |
| chr1_-_56168351 | 14.48 |
ENSRNOT00000035125
|
Dact2
|
dishevelled-binding antagonist of beta-catenin 2 |
| chr4_-_79989572 | 14.46 |
ENSRNOT00000013264
ENSRNOT00000086453 |
Dfna5
|
deafness, autosomal dominant 5 (human) |
| chr13_-_107831014 | 14.44 |
ENSRNOT00000003684
|
Kcnk2
|
potassium two pore domain channel subfamily K member 2 |
| chr3_+_161425988 | 14.13 |
ENSRNOT00000065184
|
Slc12a5
|
solute carrier family 12 member 5 |
| chr4_+_99753500 | 14.07 |
ENSRNOT00000086291
ENSRNOT00000043199 |
Immt
|
inner membrane mitochondrial protein |
| chr5_-_166726794 | 14.03 |
ENSRNOT00000022799
|
Slc25a33
|
solute carrier family 25 member 33 |
| chr1_-_112947399 | 14.03 |
ENSRNOT00000093306
ENSRNOT00000093259 |
Gabra5
|
gamma-aminobutyric acid type A receptor alpha 5 subunit |
| chr16_-_55002322 | 13.99 |
ENSRNOT00000036092
|
Zdhhc2
|
zinc finger, DHHC-type containing 2 |
| chr2_-_179704629 | 13.97 |
ENSRNOT00000083361
ENSRNOT00000077941 |
Gria2
|
glutamate ionotropic receptor AMPA type subunit 2 |
| chr6_-_132747828 | 13.89 |
ENSRNOT00000005753
|
Slc25a29
|
solute carrier family 25 member 29 |
| chr20_-_14573519 | 13.76 |
ENSRNOT00000001772
|
Rab36
|
RAB36, member RAS oncogene family |
| chr11_-_38103290 | 13.61 |
ENSRNOT00000066413
|
Tmprss2
|
transmembrane protease, serine 2 |
| chr7_-_117788550 | 13.46 |
ENSRNOT00000021775
|
MGC94207
|
similar to RIKEN cDNA C030006K11 |
| chr5_-_40867023 | 13.26 |
ENSRNOT00000011576
|
Manea
|
mannosidase, endo-alpha |
| chr7_-_2786856 | 13.21 |
ENSRNOT00000047530
ENSRNOT00000086939 |
Coq10a
|
coenzyme Q10A |
| chrX_-_139464798 | 13.19 |
ENSRNOT00000003282
|
Gpc4
|
glypican 4 |
| chr16_+_20962227 | 13.14 |
ENSRNOT00000027615
|
Slc25a42
|
solute carrier family 25, member 42 |
| chr3_+_56862691 | 13.06 |
ENSRNOT00000087712
|
Gad1
|
glutamate decarboxylase 1 |
| chr1_-_112947162 | 13.06 |
ENSRNOT00000014573
ENSRNOT00000083894 |
Gabra5
|
gamma-aminobutyric acid type A receptor alpha 5 subunit |
| chr11_+_83884048 | 13.00 |
ENSRNOT00000002376
|
LOC108348101
|
chloride channel protein 2 |
| chr11_+_83883879 | 12.98 |
ENSRNOT00000050927
|
LOC108348101
|
chloride channel protein 2 |
| chr1_+_221801524 | 12.92 |
ENSRNOT00000031227
|
Nrxn2
|
neurexin 2 |
| chr2_+_98252925 | 12.91 |
ENSRNOT00000011579
|
Pex2
|
peroxisomal biogenesis factor 2 |
| chr14_-_9456990 | 12.90 |
ENSRNOT00000002918
|
Cds1
|
CDP-diacylglycerol synthase 1 |
| chr6_+_36107147 | 12.88 |
ENSRNOT00000006020
|
Rdh14
|
retinol dehydrogenase 14 (all-trans/9-cis/11-cis) |
| chr11_-_64952687 | 12.87 |
ENSRNOT00000087892
|
Popdc2
|
popeye domain containing 2 |
| chr11_+_86512797 | 12.85 |
ENSRNOT00000051680
|
Gp1bb
|
glycoprotein Ib platelet beta subunit |
| chr4_+_147692028 | 12.68 |
ENSRNOT00000014207
|
Cand2
|
cullin-associated and neddylation-dissociated 2 (putative) |
| chr9_-_71900044 | 12.67 |
ENSRNOT00000020322
|
Idh1
|
isocitrate dehydrogenase (NADP(+)) 1, cytosolic |
| chr2_+_112868707 | 12.56 |
ENSRNOT00000017805
|
Nceh1
|
neutral cholesterol ester hydrolase 1 |
| chr5_-_136965191 | 12.51 |
ENSRNOT00000056842
|
St3gal3
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 3 |
| chr16_-_64716224 | 12.49 |
ENSRNOT00000071166
|
Fut10
|
fucosyltransferase 10 |
| chr1_+_102414625 | 12.44 |
ENSRNOT00000089488
|
Kcnc1
|
potassium voltage-gated channel subfamily C member 1 |
| chr10_+_75495817 | 12.31 |
ENSRNOT00000013743
|
Cuedc1
|
CUE domain containing 1 |
| chr1_-_72509431 | 12.21 |
ENSRNOT00000071971
|
LOC684270
|
similar to isochorismatase domain containing 2 |
| chr5_+_137189473 | 12.19 |
ENSRNOT00000056815
|
Hyi
|
hydroxypyruvate isomerase |
| chr15_-_55424024 | 12.09 |
ENSRNOT00000077733
|
Nudt15
|
nudix hydrolase 15 |
| chr10_-_57653359 | 12.08 |
ENSRNOT00000089638
|
Derl2
|
derlin 2 |
| chr7_+_77763512 | 12.02 |
ENSRNOT00000006411
|
Baalc
|
brain and acute leukemia, cytoplasmic |
| chr3_+_8928534 | 11.98 |
ENSRNOT00000088509
ENSRNOT00000065300 |
Miga2
|
mitoguardin 2 |
| chr10_-_56558487 | 11.92 |
ENSRNOT00000023256
|
Slc2a4
|
solute carrier family 2 member 4 |
| chr7_-_12519154 | 11.70 |
ENSRNOT00000093376
ENSRNOT00000077681 |
Gpx4
|
glutathione peroxidase 4 |
| chrX_-_14910727 | 11.59 |
ENSRNOT00000085079
ENSRNOT00000030146 |
Ssx1
|
SSX family member 1 |
| chr16_+_81587446 | 11.36 |
ENSRNOT00000092680
|
NEWGENE_1582994
|
DCN1, defective in cullin neddylation 1, domain containing 2 |
| chr3_-_114307250 | 11.26 |
ENSRNOT00000064592
|
Shf
|
Src homology 2 domain containing F |
| chr13_-_105684374 | 11.09 |
ENSRNOT00000073142
|
Spata17
|
spermatogenesis associated 17 |
| chr5_-_88629491 | 11.09 |
ENSRNOT00000058906
|
Tle1
|
transducin like enhancer of split 1 |
| chr11_+_82862695 | 11.06 |
ENSRNOT00000071707
ENSRNOT00000073435 |
Clcn2
|
chloride voltage-gated channel 2 |
| chr6_+_50528823 | 10.96 |
ENSRNOT00000008321
|
Lamb1
|
laminin subunit beta 1 |
| chrX_-_131343853 | 10.93 |
ENSRNOT00000038653
|
Dcaf12l1
|
DDB1 and CUL4 associated factor 12-like 1 |
| chr16_+_74531564 | 10.79 |
ENSRNOT00000078971
|
Slc25a15
|
solute carrier family 25 member 15 |
| chr5_+_169213115 | 10.76 |
ENSRNOT00000013535
|
Nol9
|
nucleolar protein 9 |
| chr1_+_101541266 | 10.74 |
ENSRNOT00000028436
|
Hsd17b14
|
hydroxysteroid (17-beta) dehydrogenase 14 |
| chr18_+_53727209 | 10.73 |
ENSRNOT00000026706
|
Isoc1
|
isochorismatase domain containing 1 |
| chr7_-_136853154 | 10.70 |
ENSRNOT00000087376
|
Nell2
|
neural EGFL like 2 |
| chr2_-_52282548 | 10.65 |
ENSRNOT00000033627
|
Nnt
|
nicotinamide nucleotide transhydrogenase |
| chr1_-_24056373 | 10.65 |
ENSRNOT00000015566
|
Slc2a12
|
solute carrier family 2 member 12 |
| chr3_-_2459385 | 10.63 |
ENSRNOT00000014080
|
Cysrt1
|
cysteine rich tail 1 |
| chr2_-_30127269 | 10.63 |
ENSRNOT00000023869
|
Cartpt
|
CART prepropeptide |
| chr11_-_70499200 | 10.61 |
ENSRNOT00000002439
|
Slc12a8
|
solute carrier family 12, member 8 |
| chr1_-_2846200 | 10.58 |
ENSRNOT00000017688
|
Sash1
|
SAM and SH3 domain containing 1 |
| chr1_+_28301922 | 10.57 |
ENSRNOT00000017831
|
Rnf217
|
ring finger protein 217 |
| chr4_+_84423653 | 10.55 |
ENSRNOT00000012655
|
Chn2
|
chimerin 2 |
| chr5_-_57267002 | 10.51 |
ENSRNOT00000011455
|
Bag1
|
Bcl2 associated athanogene 1 |
| chr1_-_204605552 | 10.46 |
ENSRNOT00000022904
|
Nkx1-2
|
NK1 homeobox 2 |
| chr1_+_31264755 | 10.41 |
ENSRNOT00000028988
|
LOC679739
|
NADH dehydrogenase (ubiquinone) Fe-S protein 6 |
| chr1_+_97632473 | 10.36 |
ENSRNOT00000023671
|
Vstm2b
|
V-set and transmembrane domain containing 2B |
| chrX_-_63961527 | 10.16 |
ENSRNOT00000071762
|
Gspt2
|
G1 to S phase transition 2 |
| chr15_+_33822730 | 10.09 |
ENSRNOT00000067607
|
RGD1564324
|
similar to dehydrogenase/reductase member 2 |
| chr5_+_24146171 | 10.04 |
ENSRNOT00000037547
|
MGC94199
|
similar to RIKEN cDNA 2610301B20; EST AI428449 |
| chr7_+_125058897 | 9.98 |
ENSRNOT00000016520
|
Samm50
|
SAMM50 sorting and assembly machinery component |
| chr1_-_99870024 | 9.90 |
ENSRNOT00000085556
|
AABR07003241.2
|
|
| chr2_+_21931493 | 9.82 |
ENSRNOT00000018259
|
Dhfr
|
dihydrofolate reductase |
| chr9_+_92435896 | 9.80 |
ENSRNOT00000022901
|
Fbxo36
|
F-box protein 36 |
| chr3_+_151310598 | 9.76 |
ENSRNOT00000092194
|
Mmp24
|
matrix metallopeptidase 24 |
| chr1_+_280633938 | 9.73 |
ENSRNOT00000012564
|
Emx2
|
empty spiracles homeobox 2 |
| chr5_+_169659188 | 9.68 |
ENSRNOT00000066623
|
Nphp4
|
nephrocystin 4 |
| chr11_-_64583994 | 9.39 |
ENSRNOT00000004289
|
B4galt4
|
beta-1,4-galactosyltransferase 4 |
| chr12_-_24710019 | 9.39 |
ENSRNOT00000049601
|
Stx1a
|
syntaxin 1A |
| chr2_+_93669765 | 9.35 |
ENSRNOT00000045438
|
Slc10a5
|
solute carrier family 10, member 5 |
| chr3_+_55369214 | 9.28 |
ENSRNOT00000067161
|
Nostrin
|
nitric oxide synthase trafficking |
| chr12_+_22082057 | 9.21 |
ENSRNOT00000077711
ENSRNOT00000064572 |
Agfg2
|
ArfGAP with FG repeats 2 |
| chr3_-_12029877 | 9.18 |
ENSRNOT00000022327
|
Slc2a8
|
solute carrier family 2 member 8 |
| chr2_-_187133993 | 9.15 |
ENSRNOT00000019502
|
Pear1
|
platelet endothelial aggregation receptor 1 |
| chr1_+_95397991 | 9.01 |
ENSRNOT00000039649
|
Zfp939
|
zinc finger protein 939 |
| chr9_-_16647458 | 9.00 |
ENSRNOT00000024380
|
Mrpl2
|
mitochondrial ribosomal protein L2 |
| chr3_+_81814261 | 8.99 |
ENSRNOT00000011459
|
Syt13
|
synaptotagmin 13 |
| chr10_-_85084850 | 8.98 |
ENSRNOT00000012462
|
Tbkbp1
|
TBK1 binding protein 1 |
| chr3_-_138462063 | 8.88 |
ENSRNOT00000065553
|
Ovol2
|
ovo-like zinc finger 2 |
| chr1_+_86948918 | 8.83 |
ENSRNOT00000084839
|
Sirt2
|
sirtuin 2 |
| chr8_-_58542844 | 8.82 |
ENSRNOT00000012041
|
Elmod1
|
ELMO domain containing 1 |
| chr9_-_101388833 | 8.82 |
ENSRNOT00000071817
|
Chchd2
|
coiled-coil-helix-coiled-coil-helix domain containing 2 |
| chr6_-_136145837 | 8.82 |
ENSRNOT00000015122
|
Ckb
|
creatine kinase B |
Network of associatons between targets according to the STRING database.
First level regulatory network of Nr2f2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 19.5 | 58.6 | GO:0033512 | L-lysine catabolic process to acetyl-CoA via saccharopine(GO:0033512) |
| 10.0 | 39.8 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 8.8 | 26.4 | GO:0042732 | D-xylose metabolic process(GO:0042732) |
| 8.3 | 25.0 | GO:0046167 | glycerol-3-phosphate biosynthetic process(GO:0046167) |
| 8.0 | 31.9 | GO:0046351 | disaccharide biosynthetic process(GO:0046351) |
| 7.7 | 23.1 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 7.6 | 22.8 | GO:0097089 | methyl-branched fatty acid metabolic process(GO:0097089) |
| 7.2 | 57.4 | GO:0090080 | positive regulation of MAPKKK cascade by fibroblast growth factor receptor signaling pathway(GO:0090080) |
| 6.7 | 20.2 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 6.6 | 86.2 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 6.4 | 25.7 | GO:0010609 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) |
| 6.2 | 18.7 | GO:0034769 | basement membrane disassembly(GO:0034769) |
| 6.2 | 30.9 | GO:0072061 | inner medullary collecting duct development(GO:0072061) |
| 6.2 | 18.5 | GO:0071469 | cellular response to alkaline pH(GO:0071469) |
| 5.8 | 40.7 | GO:0045196 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 5.5 | 16.6 | GO:0019417 | sulfur oxidation(GO:0019417) |
| 5.5 | 16.4 | GO:0021550 | medulla oblongata development(GO:0021550) musculoskeletal movement, spinal reflex action(GO:0050883) |
| 5.2 | 15.7 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 5.1 | 15.2 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 5.0 | 20.1 | GO:0019074 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 5.0 | 19.8 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 4.9 | 38.9 | GO:0090258 | negative regulation of mitochondrial fission(GO:0090258) |
| 4.8 | 14.5 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 4.8 | 14.4 | GO:1900039 | positive regulation of cellular response to hypoxia(GO:1900039) |
| 4.8 | 38.0 | GO:0071267 | amino acid salvage(GO:0043102) L-methionine salvage(GO:0071267) |
| 4.5 | 22.5 | GO:0001582 | detection of chemical stimulus involved in sensory perception of sweet taste(GO:0001582) |
| 4.5 | 17.9 | GO:0061107 | seminal vesicle development(GO:0061107) |
| 4.3 | 12.9 | GO:0060931 | sinoatrial node cell development(GO:0060931) |
| 4.3 | 21.3 | GO:0044691 | tooth eruption(GO:0044691) |
| 4.3 | 17.0 | GO:0045105 | intermediate filament polymerization or depolymerization(GO:0045105) |
| 4.2 | 12.7 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 4.2 | 25.3 | GO:0045113 | regulation of integrin biosynthetic process(GO:0045113) |
| 3.9 | 23.2 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 3.8 | 15.2 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 3.8 | 7.6 | GO:2000813 | negative regulation of barbed-end actin filament capping(GO:2000813) |
| 3.8 | 11.3 | GO:0060160 | negative regulation of dopamine receptor signaling pathway(GO:0060160) |
| 3.7 | 11.0 | GO:0021812 | neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 3.6 | 10.8 | GO:0000448 | cleavage in ITS2 between 5.8S rRNA and LSU-rRNA of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000448) |
| 3.2 | 9.7 | GO:1903348 | positive regulation of bicellular tight junction assembly(GO:1903348) |
| 3.2 | 19.0 | GO:1902746 | regulation of lens fiber cell differentiation(GO:1902746) |
| 3.1 | 18.7 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 3.1 | 15.5 | GO:0006545 | glycine biosynthetic process(GO:0006545) |
| 3.1 | 30.5 | GO:2001023 | regulation of response to drug(GO:2001023) |
| 3.0 | 36.5 | GO:0006108 | malate metabolic process(GO:0006108) |
| 2.9 | 8.8 | GO:2000777 | positive regulation of oocyte maturation(GO:1900195) positive regulation of proteasomal ubiquitin-dependent protein catabolic process involved in cellular response to hypoxia(GO:2000777) |
| 2.9 | 14.7 | GO:0018243 | protein O-linked glycosylation via threonine(GO:0018243) |
| 2.9 | 29.3 | GO:0036376 | sodium ion export from cell(GO:0036376) |
| 2.9 | 22.9 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 2.9 | 5.7 | GO:0061642 | chemoattraction of axon(GO:0061642) |
| 2.7 | 21.6 | GO:1901727 | positive regulation of histone deacetylase activity(GO:1901727) |
| 2.7 | 10.6 | GO:0070093 | negative regulation of glucagon secretion(GO:0070093) |
| 2.6 | 10.6 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 2.6 | 13.1 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 2.5 | 10.2 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 2.5 | 29.4 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 2.4 | 14.7 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 2.4 | 12.1 | GO:0042262 | dGTP catabolic process(GO:0006203) DNA protection(GO:0042262) |
| 2.4 | 7.1 | GO:0032417 | positive regulation of sodium:proton antiporter activity(GO:0032417) |
| 2.3 | 7.0 | GO:0034241 | positive regulation of macrophage fusion(GO:0034241) PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 2.3 | 7.0 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 2.3 | 20.8 | GO:0044351 | macropinocytosis(GO:0044351) phagosome-lysosome fusion(GO:0090385) |
| 2.3 | 25.2 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 2.2 | 8.9 | GO:0060214 | endocardium formation(GO:0060214) |
| 2.2 | 11.1 | GO:0060689 | cell differentiation involved in salivary gland development(GO:0060689) |
| 2.2 | 13.1 | GO:0015860 | purine nucleoside transmembrane transport(GO:0015860) |
| 2.2 | 10.8 | GO:0000066 | mitochondrial ornithine transport(GO:0000066) |
| 2.2 | 12.9 | GO:0097116 | gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 2.1 | 6.4 | GO:0036102 | leukotriene catabolic process(GO:0036100) leukotriene B4 catabolic process(GO:0036101) leukotriene B4 metabolic process(GO:0036102) vitamin E metabolic process(GO:0042360) icosanoid catabolic process(GO:1901523) fatty acid derivative catabolic process(GO:1901569) |
| 2.1 | 12.7 | GO:0006102 | isocitrate metabolic process(GO:0006102) glyoxylate metabolic process(GO:0046487) |
| 2.1 | 10.5 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 2.1 | 6.3 | GO:0009644 | response to high light intensity(GO:0009644) |
| 2.1 | 12.4 | GO:0021759 | globus pallidus development(GO:0021759) |
| 2.1 | 8.2 | GO:1902774 | late endosome to lysosome transport(GO:1902774) |
| 1.9 | 7.7 | GO:1902202 | proteoglycan catabolic process(GO:0030167) regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) |
| 1.9 | 15.4 | GO:0017062 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) |
| 1.7 | 24.4 | GO:1901386 | negative regulation of voltage-gated calcium channel activity(GO:1901386) |
| 1.7 | 8.6 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) |
| 1.7 | 5.0 | GO:0080120 | prenylated protein catabolic process(GO:0030327) CAAX-box protein processing(GO:0071586) CAAX-box protein maturation(GO:0080120) |
| 1.7 | 15.0 | GO:0060174 | limb bud formation(GO:0060174) |
| 1.7 | 10.0 | GO:0045040 | protein import into mitochondrial outer membrane(GO:0045040) |
| 1.7 | 5.0 | GO:0032510 | endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) |
| 1.6 | 4.9 | GO:0042245 | RNA repair(GO:0042245) |
| 1.6 | 4.9 | GO:0071934 | thiamine transmembrane transport(GO:0071934) |
| 1.6 | 14.6 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 1.6 | 9.4 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) |
| 1.5 | 19.5 | GO:0097091 | synaptic vesicle clustering(GO:0097091) |
| 1.5 | 14.6 | GO:0035524 | proline transmembrane transport(GO:0035524) |
| 1.5 | 1.5 | GO:0097033 | mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 1.4 | 12.9 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 1.3 | 4.0 | GO:0000105 | histidine biosynthetic process(GO:0000105) |
| 1.3 | 9.2 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 1.3 | 6.5 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 1.3 | 20.6 | GO:0045056 | transcytosis(GO:0045056) |
| 1.3 | 23.8 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 1.3 | 15.0 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 1.2 | 17.3 | GO:0015693 | magnesium ion transport(GO:0015693) magnesium ion transmembrane transport(GO:1903830) |
| 1.2 | 13.6 | GO:0046598 | positive regulation of viral entry into host cell(GO:0046598) |
| 1.2 | 4.6 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 1.1 | 8.0 | GO:0048311 | mitochondrion distribution(GO:0048311) |
| 1.1 | 2.3 | GO:0090210 | regulation of establishment of blood-brain barrier(GO:0090210) |
| 1.1 | 12.1 | GO:0070862 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 1.1 | 2.2 | GO:0038189 | neuropilin signaling pathway(GO:0038189) VEGF-activated neuropilin signaling pathway(GO:0038190) |
| 1.1 | 6.4 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 1.0 | 6.3 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 1.0 | 3.1 | GO:0032900 | negative regulation of neurotrophin production(GO:0032900) nerve growth factor production(GO:0032902) negative regulation of receptor catabolic process(GO:2000645) |
| 1.0 | 17.4 | GO:0048642 | negative regulation of skeletal muscle tissue development(GO:0048642) |
| 1.0 | 31.7 | GO:0035428 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 1.0 | 38.7 | GO:0002675 | positive regulation of acute inflammatory response(GO:0002675) |
| 1.0 | 14.7 | GO:0014052 | regulation of gamma-aminobutyric acid secretion(GO:0014052) |
| 1.0 | 18.6 | GO:0051597 | response to methylmercury(GO:0051597) |
| 1.0 | 3.9 | GO:0071442 | RNA 5'-end processing(GO:0000966) regulation of histone H3-K14 acetylation(GO:0071440) positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 1.0 | 9.8 | GO:0044331 | cell-cell adhesion mediated by cadherin(GO:0044331) |
| 1.0 | 10.7 | GO:0046543 | development of secondary female sexual characteristics(GO:0046543) |
| 0.9 | 11.4 | GO:0042407 | cristae formation(GO:0042407) |
| 0.9 | 7.6 | GO:0032308 | positive regulation of prostaglandin secretion(GO:0032308) |
| 0.9 | 17.0 | GO:0017004 | cytochrome complex assembly(GO:0017004) |
| 0.9 | 28.3 | GO:0060384 | innervation(GO:0060384) |
| 0.9 | 15.9 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.9 | 11.1 | GO:2000811 | negative regulation of anoikis(GO:2000811) |
| 0.9 | 5.5 | GO:0031580 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.9 | 12.5 | GO:0036065 | fucosylation(GO:0036065) |
| 0.9 | 5.3 | GO:0048597 | post-embryonic camera-type eye morphogenesis(GO:0048597) |
| 0.9 | 7.9 | GO:0034651 | cortisol metabolic process(GO:0034650) cortisol biosynthetic process(GO:0034651) |
| 0.8 | 24.4 | GO:0014002 | astrocyte development(GO:0014002) |
| 0.8 | 7.5 | GO:1902414 | protein localization to cell junction(GO:1902414) |
| 0.8 | 6.7 | GO:0090261 | positive regulation of inclusion body assembly(GO:0090261) |
| 0.8 | 13.2 | GO:1901663 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.8 | 36.7 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.8 | 5.5 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.7 | 21.3 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.7 | 3.5 | GO:0072733 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.7 | 26.0 | GO:0065002 | intracellular protein transmembrane transport(GO:0065002) |
| 0.7 | 32.6 | GO:0090505 | wound healing, spreading of cells(GO:0044319) epiboly involved in wound healing(GO:0090505) |
| 0.7 | 2.7 | GO:0060856 | establishment of blood-brain barrier(GO:0060856) |
| 0.7 | 26.0 | GO:1903959 | regulation of anion transmembrane transport(GO:1903959) |
| 0.7 | 5.3 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.6 | 8.8 | GO:1900037 | regulation of cellular response to hypoxia(GO:1900037) |
| 0.6 | 10.5 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.6 | 2.5 | GO:0043418 | homocysteine catabolic process(GO:0043418) |
| 0.6 | 3.6 | GO:1903862 | positive regulation of oxidative phosphorylation(GO:1903862) |
| 0.6 | 3.6 | GO:0009912 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.6 | 3.6 | GO:0002943 | tRNA dihydrouridine synthesis(GO:0002943) |
| 0.6 | 6.5 | GO:0060013 | righting reflex(GO:0060013) |
| 0.6 | 2.9 | GO:0050747 | positive regulation of lipoprotein metabolic process(GO:0050747) |
| 0.6 | 2.9 | GO:0043570 | maintenance of DNA repeat elements(GO:0043570) |
| 0.6 | 6.8 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.6 | 12.5 | GO:0097503 | sialylation(GO:0097503) |
| 0.6 | 7.8 | GO:2000060 | positive regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000060) |
| 0.6 | 10.0 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.5 | 9.7 | GO:0021542 | dentate gyrus development(GO:0021542) |
| 0.5 | 5.6 | GO:0043584 | nose development(GO:0043584) |
| 0.5 | 4.5 | GO:0070072 | vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.5 | 1.5 | GO:1901662 | phylloquinone metabolic process(GO:0042374) phylloquinone catabolic process(GO:0042376) quinone catabolic process(GO:1901662) |
| 0.5 | 5.2 | GO:0033603 | positive regulation of dopamine secretion(GO:0033603) |
| 0.5 | 14.0 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.4 | 31.0 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.4 | 10.7 | GO:0006706 | steroid catabolic process(GO:0006706) |
| 0.4 | 4.0 | GO:0009650 | UV protection(GO:0009650) |
| 0.4 | 6.2 | GO:0090201 | negative regulation of release of cytochrome c from mitochondria(GO:0090201) |
| 0.4 | 20.1 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.4 | 1.2 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.4 | 1.2 | GO:0090283 | regulation of protein glycosylation in Golgi(GO:0090283) |
| 0.4 | 12.9 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.4 | 9.4 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.4 | 24.3 | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.4 | 2.6 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.4 | 2.9 | GO:1901642 | nucleoside transmembrane transport(GO:1901642) |
| 0.4 | 16.0 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.4 | 27.7 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.3 | 10.8 | GO:0060292 | long term synaptic depression(GO:0060292) |
| 0.3 | 4.3 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.3 | 13.1 | GO:0006739 | NADP metabolic process(GO:0006739) |
| 0.3 | 1.5 | GO:0060024 | rhythmic synaptic transmission(GO:0060024) |
| 0.3 | 3.0 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.3 | 2.5 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.3 | 3.3 | GO:1901897 | regulation of relaxation of cardiac muscle(GO:1901897) |
| 0.3 | 1.9 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.3 | 12.6 | GO:0006805 | xenobiotic metabolic process(GO:0006805) |
| 0.3 | 3.2 | GO:0033141 | regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033139) positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.3 | 5.4 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.2 | 2.9 | GO:0061051 | positive regulation of cell growth involved in cardiac muscle cell development(GO:0061051) |
| 0.2 | 16.3 | GO:1901379 | regulation of potassium ion transmembrane transport(GO:1901379) |
| 0.2 | 7.7 | GO:0009268 | response to pH(GO:0009268) |
| 0.2 | 2.7 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.2 | 3.6 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.2 | 5.2 | GO:0044342 | type B pancreatic cell proliferation(GO:0044342) |
| 0.2 | 1.9 | GO:0003417 | growth plate cartilage development(GO:0003417) |
| 0.2 | 3.1 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.2 | 1.7 | GO:0006777 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) |
| 0.2 | 1.8 | GO:0006390 | DNA-templated transcription, termination(GO:0006353) transcription from mitochondrial promoter(GO:0006390) |
| 0.2 | 8.1 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.1 | 1.8 | GO:0043923 | positive regulation by host of viral transcription(GO:0043923) |
| 0.1 | 4.4 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.1 | 19.4 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.1 | 6.1 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.1 | 1.5 | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.1 | 3.4 | GO:0040020 | regulation of meiotic nuclear division(GO:0040020) |
| 0.1 | 6.2 | GO:0016126 | sterol biosynthetic process(GO:0016126) |
| 0.1 | 12.7 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.1 | 1.4 | GO:0045217 | cell-cell junction maintenance(GO:0045217) |
| 0.1 | 5.2 | GO:0042775 | mitochondrial ATP synthesis coupled electron transport(GO:0042775) |
| 0.1 | 1.7 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.1 | 1.5 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.1 | 2.8 | GO:0045880 | positive regulation of smoothened signaling pathway(GO:0045880) |
| 0.1 | 0.2 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.1 | 3.2 | GO:0030195 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.1 | 4.2 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.1 | 0.4 | GO:0060449 | bud elongation involved in lung branching(GO:0060449) |
| 0.1 | 5.6 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.1 | 6.5 | GO:0070301 | cellular response to hydrogen peroxide(GO:0070301) |
| 0.1 | 8.5 | GO:0050796 | regulation of insulin secretion(GO:0050796) |
| 0.1 | 0.8 | GO:0033599 | regulation of mammary gland epithelial cell proliferation(GO:0033599) |
| 0.0 | 1.5 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.0 | 3.2 | GO:0033574 | response to testosterone(GO:0033574) |
| 0.0 | 5.7 | GO:0050773 | regulation of dendrite development(GO:0050773) |
| 0.0 | 0.5 | GO:0042026 | protein refolding(GO:0042026) |
| 0.0 | 4.0 | GO:0071805 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.0 | 2.3 | GO:0042552 | myelination(GO:0042552) |
| 0.0 | 2.8 | GO:0008585 | female gonad development(GO:0008585) |
| 0.0 | 3.6 | GO:0006814 | sodium ion transport(GO:0006814) |
| 0.0 | 0.6 | GO:0036465 | synaptic vesicle recycling(GO:0036465) |
| 0.0 | 0.3 | GO:0045124 | regulation of bone resorption(GO:0045124) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 10.7 | 85.8 | GO:0005587 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) |
| 9.7 | 38.9 | GO:0045298 | tubulin complex(GO:0045298) |
| 5.8 | 23.2 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 4.9 | 29.3 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 4.5 | 22.5 | GO:1903768 | sweet taste receptor complex(GO:1903767) taste receptor complex(GO:1903768) |
| 4.0 | 12.0 | GO:0045203 | intrinsic component of cell outer membrane(GO:0031230) integral component of cell outer membrane(GO:0045203) |
| 3.6 | 14.5 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 3.6 | 25.4 | GO:0045293 | mRNA editing complex(GO:0045293) |
| 2.7 | 11.0 | GO:0005607 | laminin-1 complex(GO:0005606) laminin-2 complex(GO:0005607) laminin-10 complex(GO:0043259) |
| 2.5 | 10.2 | GO:0018444 | translation release factor complex(GO:0018444) |
| 2.5 | 57.6 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 2.5 | 17.3 | GO:0061617 | MICOS complex(GO:0061617) |
| 2.3 | 4.6 | GO:1990246 | uniplex complex(GO:1990246) |
| 2.3 | 16.0 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 2.1 | 6.4 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 2.0 | 38.7 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 2.0 | 12.1 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 2.0 | 10.0 | GO:0098799 | outer mitochondrial membrane protein complex(GO:0098799) |
| 1.9 | 15.3 | GO:0005638 | lamin filament(GO:0005638) |
| 1.9 | 7.7 | GO:1990745 | EARP complex(GO:1990745) |
| 1.9 | 9.4 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 1.8 | 14.4 | GO:0044305 | calyx of Held(GO:0044305) |
| 1.7 | 6.8 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 1.5 | 7.7 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 1.4 | 55.9 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 1.3 | 25.2 | GO:0005916 | fascia adherens(GO:0005916) |
| 1.3 | 26.5 | GO:0043220 | Schmidt-Lanterman incisure(GO:0043220) |
| 1.3 | 3.9 | GO:1990923 | PET complex(GO:1990923) |
| 1.2 | 29.3 | GO:0031304 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 1.2 | 42.2 | GO:0032590 | dendrite membrane(GO:0032590) |
| 1.2 | 14.0 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 1.2 | 23.1 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 1.1 | 29.3 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 1.1 | 8.7 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 1.1 | 7.6 | GO:0071203 | WASH complex(GO:0071203) |
| 1.0 | 24.1 | GO:0034706 | sodium channel complex(GO:0034706) |
| 0.9 | 5.6 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.9 | 2.8 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.9 | 25.7 | GO:0098827 | endoplasmic reticulum subcompartment(GO:0098827) |
| 0.9 | 11.9 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.9 | 5.5 | GO:1990696 | stereocilia ankle link complex(GO:0002142) USH2 complex(GO:1990696) |
| 0.9 | 2.7 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.8 | 3.2 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.8 | 40.2 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.8 | 51.7 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.8 | 69.1 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.8 | 6.1 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.7 | 9.7 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.7 | 15.3 | GO:0005753 | mitochondrial proton-transporting ATP synthase complex(GO:0005753) |
| 0.7 | 5.1 | GO:0036454 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
| 0.7 | 70.5 | GO:0005604 | basement membrane(GO:0005604) |
| 0.7 | 6.7 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.7 | 33.6 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.6 | 5.1 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.6 | 5.6 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.6 | 25.5 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.6 | 5.0 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.6 | 53.2 | GO:0031985 | Golgi cisterna(GO:0031985) |
| 0.6 | 5.8 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.6 | 1.7 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.5 | 54.0 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.5 | 13.1 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.5 | 5.0 | GO:0005798 | Golgi-associated vesicle(GO:0005798) |
| 0.5 | 9.8 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.5 | 4.2 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.4 | 4.0 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.4 | 26.4 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.4 | 24.0 | GO:0005844 | polysome(GO:0005844) |
| 0.4 | 40.9 | GO:0034705 | potassium channel complex(GO:0034705) |
| 0.4 | 6.3 | GO:0034709 | methylosome(GO:0034709) |
| 0.4 | 10.7 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.4 | 3.2 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.4 | 16.6 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.4 | 5.4 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.4 | 109.0 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.4 | 12.6 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.4 | 7.0 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.4 | 2.6 | GO:0048500 | signal recognition particle(GO:0048500) |
| 0.4 | 10.0 | GO:0097546 | ciliary base(GO:0097546) |
| 0.4 | 16.5 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.3 | 9.2 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.3 | 2.2 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.3 | 3.0 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.3 | 23.5 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.3 | 5.2 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.3 | 25.7 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.3 | 2.9 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.3 | 10.5 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.3 | 2.8 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.3 | 100.5 | GO:0045177 | apical part of cell(GO:0045177) |
| 0.3 | 8.8 | GO:0031970 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.2 | 21.8 | GO:0043204 | perikaryon(GO:0043204) |
| 0.2 | 19.7 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.2 | 1.5 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.2 | 258.0 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.2 | 10.7 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.2 | 13.2 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.2 | 14.1 | GO:0030496 | midbody(GO:0030496) |
| 0.2 | 62.7 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.2 | 2.8 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.1 | 0.4 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) TEAD-2-YAP complex(GO:0071149) |
| 0.1 | 2.5 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 14.9 | GO:0001726 | ruffle(GO:0001726) |
| 0.1 | 12.2 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.1 | 2.9 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.1 | 2.0 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.1 | 0.4 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.1 | 3.7 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 6.9 | GO:0005814 | centriole(GO:0005814) |
| 0.1 | 13.5 | GO:0098793 | presynapse(GO:0098793) |
| 0.1 | 1.8 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 1.5 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 3.5 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 1.2 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 2.2 | GO:0070160 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 6.9 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 0.6 | GO:0048475 | membrane coat(GO:0030117) coated membrane(GO:0048475) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 19.1 | 57.4 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 14.6 | 58.6 | GO:0016212 | kynurenine-oxoglutarate transaminase activity(GO:0016212) kynurenine aminotransferase activity(GO:0036137) |
| 9.7 | 38.9 | GO:0099609 | microtubule lateral binding(GO:0099609) |
| 9.1 | 36.5 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 8.3 | 25.0 | GO:0004370 | glycerol kinase activity(GO:0004370) |
| 7.4 | 22.2 | GO:1902379 | chemoattractant activity involved in axon guidance(GO:1902379) |
| 6.7 | 20.1 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 6.2 | 18.5 | GO:0097603 | temperature-gated ion channel activity(GO:0097603) |
| 5.8 | 46.6 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 5.8 | 23.2 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 5.8 | 23.1 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 5.6 | 22.5 | GO:0003842 | 1-pyrroline-5-carboxylate dehydrogenase activity(GO:0003842) |
| 5.3 | 37.1 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 5.2 | 20.7 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 4.7 | 18.7 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 4.6 | 27.8 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 4.6 | 36.7 | GO:0008430 | selenium binding(GO:0008430) |
| 4.3 | 12.9 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 4.2 | 12.7 | GO:0051990 | (R)-2-hydroxyglutarate dehydrogenase activity(GO:0051990) |
| 4.0 | 20.2 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 4.0 | 12.1 | GO:0035539 | 8-oxo-7,8-dihydroguanosine triphosphate pyrophosphatase activity(GO:0008413) 8-oxo-7,8-dihydrodeoxyguanosine triphosphate pyrophosphatase activity(GO:0035539) |
| 3.9 | 15.7 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 3.7 | 40.7 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 3.7 | 22.2 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 3.6 | 14.5 | GO:0030172 | troponin C binding(GO:0030172) |
| 3.6 | 10.7 | GO:0047045 | testosterone 17-beta-dehydrogenase (NADP+) activity(GO:0047045) |
| 3.5 | 10.5 | GO:0001565 | phorbol ester receptor activity(GO:0001565) |
| 3.4 | 20.6 | GO:0042954 | lipoprotein transporter activity(GO:0042954) |
| 3.1 | 9.4 | GO:0003945 | N-acetyllactosamine synthase activity(GO:0003945) |
| 3.1 | 12.5 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 3.0 | 15.2 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 2.9 | 8.8 | GO:0042903 | tubulin deacetylase activity(GO:0042903) |
| 2.9 | 14.4 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 2.9 | 31.4 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 2.8 | 14.0 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 2.7 | 10.8 | GO:0051731 | polynucleotide 5'-hydroxyl-kinase activity(GO:0051731) |
| 2.7 | 24.1 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 2.7 | 18.6 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 2.6 | 13.1 | GO:0051185 | adenosine-diphosphatase activity(GO:0043262) coenzyme transporter activity(GO:0051185) |
| 2.6 | 13.1 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 2.5 | 20.1 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 2.5 | 9.8 | GO:0051870 | methotrexate binding(GO:0051870) |
| 2.3 | 31.9 | GO:0016208 | AMP binding(GO:0016208) |
| 2.3 | 22.8 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 2.2 | 6.7 | GO:0031531 | thyrotropin-releasing hormone receptor binding(GO:0031531) |
| 2.2 | 15.6 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 2.2 | 57.6 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 2.2 | 12.9 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 2.1 | 6.4 | GO:0052871 | leukotriene-B4 20-monooxygenase activity(GO:0050051) tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) |
| 2.1 | 29.3 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) D1 dopamine receptor binding(GO:0031748) |
| 2.1 | 12.5 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 2.1 | 24.7 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 1.9 | 17.3 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 1.9 | 5.7 | GO:0008732 | threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 1.9 | 15.2 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 1.9 | 35.6 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 1.8 | 14.7 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 1.8 | 62.9 | GO:0016655 | oxidoreductase activity, acting on NAD(P)H, quinone or similar compound as acceptor(GO:0016655) |
| 1.7 | 17.0 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 1.6 | 4.9 | GO:0015234 | thiamine transmembrane transporter activity(GO:0015234) |
| 1.6 | 8.0 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 1.6 | 9.4 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 1.6 | 18.7 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 1.5 | 98.6 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 1.5 | 7.6 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 1.5 | 44.4 | GO:0005501 | retinoid binding(GO:0005501) |
| 1.5 | 14.6 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 1.4 | 27.4 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 1.4 | 17.0 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 1.4 | 11.3 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 1.4 | 19.2 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 1.4 | 15.0 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 1.4 | 19.1 | GO:0005351 | sugar:proton symporter activity(GO:0005351) |
| 1.3 | 10.8 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 1.3 | 14.7 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 1.3 | 11.9 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 1.3 | 17.2 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 1.2 | 20.8 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 1.2 | 12.2 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 1.2 | 4.9 | GO:0035514 | DNA demethylase activity(GO:0035514) oxidative RNA demethylase activity(GO:0035515) |
| 1.2 | 6.1 | GO:0043532 | angiostatin binding(GO:0043532) |
| 1.2 | 12.9 | GO:0008106 | alcohol dehydrogenase (NADP+) activity(GO:0008106) |
| 1.2 | 17.3 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 1.0 | 12.6 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 1.0 | 4.2 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 1.0 | 40.7 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 1.0 | 21.4 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 1.0 | 4.0 | GO:0004487 | methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 1.0 | 3.9 | GO:0034584 | piRNA binding(GO:0034584) |
| 1.0 | 16.4 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 1.0 | 7.6 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 1.0 | 2.9 | GO:0032139 | dinucleotide insertion or deletion binding(GO:0032139) single guanine insertion binding(GO:0032142) |
| 0.9 | 9.4 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.9 | 15.0 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.8 | 15.3 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.8 | 10.2 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.8 | 6.5 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.8 | 5.3 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.8 | 16.6 | GO:0071949 | FAD binding(GO:0071949) |
| 0.7 | 16.1 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.7 | 7.8 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.7 | 2.8 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 0.7 | 13.2 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.7 | 13.3 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.6 | 8.4 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.6 | 2.6 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.6 | 51.7 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.6 | 3.6 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.6 | 8.2 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.6 | 7.9 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.5 | 9.8 | GO:0015215 | nucleotide transmembrane transporter activity(GO:0015215) |
| 0.5 | 37.0 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.5 | 6.5 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.5 | 12.4 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.5 | 2.5 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.5 | 16.5 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.5 | 12.0 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.5 | 5.5 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.4 | 4.3 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.4 | 3.8 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.4 | 2.9 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.4 | 44.1 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.4 | 3.2 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.4 | 5.0 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.4 | 10.6 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.3 | 1.0 | GO:0031714 | C5a anaphylatoxin chemotactic receptor binding(GO:0031714) |
| 0.3 | 31.0 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.3 | 4.4 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.3 | 2.9 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.3 | 13.6 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.3 | 6.3 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.3 | 5.2 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.3 | 6.3 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.2 | 1.4 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.2 | 45.1 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.2 | 7.1 | GO:0019212 | phosphatase inhibitor activity(GO:0019212) |
| 0.2 | 10.6 | GO:0050661 | NADP binding(GO:0050661) |
| 0.2 | 15.1 | GO:0035254 | glutamate receptor binding(GO:0035254) |
| 0.2 | 14.1 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.2 | 1.2 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.2 | 2.1 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.2 | 3.6 | GO:0005112 | Notch binding(GO:0005112) |
| 0.2 | 9.6 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.2 | 8.0 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.2 | 2.0 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.2 | 7.4 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.2 | 24.0 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.1 | 4.4 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.1 | 12.5 | GO:0005178 | integrin binding(GO:0005178) |
| 0.1 | 2.5 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.1 | 35.9 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.1 | 5.5 | GO:0017022 | myosin binding(GO:0017022) |
| 0.1 | 9.8 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 3.3 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.1 | 2.9 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.1 | 1.5 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 2.5 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 5.0 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.1 | 0.3 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.1 | 8.5 | GO:0031072 | heat shock protein binding(GO:0031072) |
| 0.1 | 2.5 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.1 | 0.3 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.1 | 1.5 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.1 | 2.2 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.1 | 4.4 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.1 | 1.3 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.1 | 1.9 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.1 | 3.6 | GO:0015293 | symporter activity(GO:0015293) |
| 0.0 | 0.9 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 1.3 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 1.3 | GO:0051213 | dioxygenase activity(GO:0051213) |
| 0.0 | 1.0 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 5.3 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.2 | GO:0016866 | intramolecular transferase activity(GO:0016866) |
| 0.0 | 0.8 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.0 | 5.6 | GO:0005509 | calcium ion binding(GO:0005509) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.6 | 46.5 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 2.7 | 133.1 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 1.4 | 40.0 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 1.3 | 5.3 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.9 | 20.6 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.8 | 22.2 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.6 | 18.8 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.6 | 18.3 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.6 | 24.8 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.6 | 4.6 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.6 | 11.3 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.5 | 17.3 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.3 | 11.1 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.3 | 8.8 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.3 | 17.1 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.3 | 3.3 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.2 | 13.6 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.2 | 27.1 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.2 | 5.2 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.2 | 10.5 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.1 | 7.0 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.1 | 28.1 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 19.4 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 1.3 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.1 | 2.8 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.1 | 1.0 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 3.1 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.1 | 2.5 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.1 | 1.1 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 1.7 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 1.2 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.0 | 5.6 | NABA MATRISOME ASSOCIATED | Ensemble of genes encoding ECM-associated proteins including ECM-affilaited proteins, ECM regulators and secreted factors |
| 0.0 | 0.4 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.3 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.5 | 58.6 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 3.4 | 57.6 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 2.6 | 26.4 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 2.5 | 33.0 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 2.4 | 55.9 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 2.3 | 50.6 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 2.2 | 96.1 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 2.2 | 88.5 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 1.9 | 31.7 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 1.8 | 47.8 | REACTOME FGFR LIGAND BINDING AND ACTIVATION | Genes involved in FGFR ligand binding and activation |
| 1.7 | 51.7 | REACTOME KINESINS | Genes involved in Kinesins |
| 1.4 | 20.1 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 1.4 | 32.9 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 1.3 | 36.7 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 1.2 | 12.8 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 1.0 | 22.5 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 1.0 | 16.5 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 1.0 | 17.2 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.9 | 19.1 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.9 | 22.4 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.8 | 54.8 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.8 | 12.5 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.8 | 10.6 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.8 | 14.6 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.7 | 14.0 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.7 | 8.0 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.6 | 9.4 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.6 | 20.6 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.5 | 18.8 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.5 | 16.4 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.5 | 13.3 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.5 | 12.9 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.5 | 22.5 | REACTOME TRIGLYCERIDE BIOSYNTHESIS | Genes involved in Triglyceride Biosynthesis |
| 0.4 | 5.2 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.4 | 14.7 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.4 | 6.4 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.4 | 7.7 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 0.4 | 6.3 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.4 | 6.1 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.4 | 7.6 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.3 | 5.1 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.3 | 15.9 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.3 | 3.6 | REACTOME SIGNALING BY NOTCH4 | Genes involved in Signaling by NOTCH4 |
| 0.3 | 11.3 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.3 | 21.3 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.3 | 29.7 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.3 | 14.2 | REACTOME REGULATION OF INSULIN SECRETION | Genes involved in Regulation of Insulin Secretion |
| 0.3 | 3.6 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.2 | 18.4 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.2 | 1.7 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.2 | 2.4 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.2 | 5.0 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.2 | 3.2 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.2 | 39.3 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.2 | 1.3 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.2 | 6.2 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.2 | 3.7 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 2.3 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.1 | 2.5 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.1 | 11.3 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.1 | 2.0 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.1 | 2.5 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 4.5 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.1 | 1.0 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 1.2 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.1 | 4.4 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.0 | 2.1 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 1.8 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 1.6 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 1.2 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.3 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.3 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |