Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
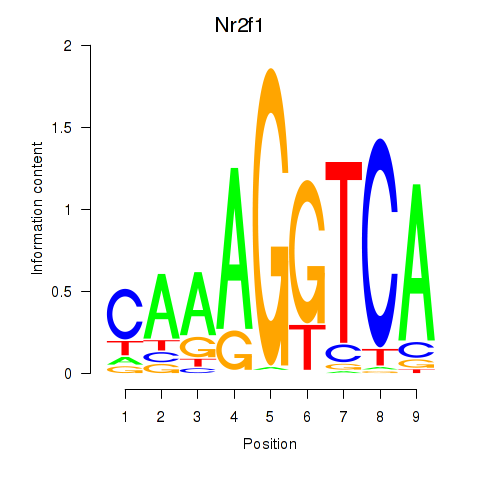
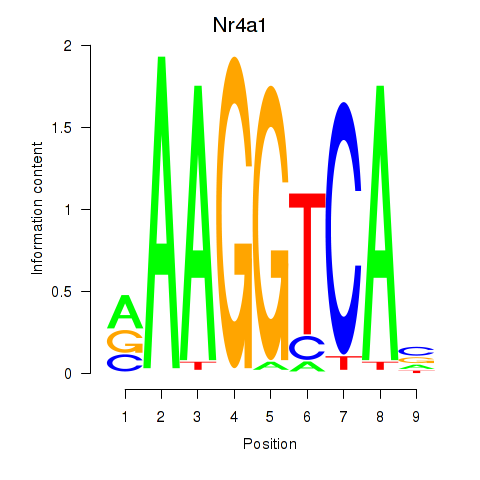
Results for Nr2f1_Nr4a1
Z-value: 1.37
Transcription factors associated with Nr2f1_Nr4a1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nr2f1
|
ENSRNOG00000014795 | nuclear receptor subfamily 2, group F, member 1 |
|
Nr4a1
|
ENSRNOG00000007607 | nuclear receptor subfamily 4, group A, member 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nr4a1 | rn6_v1_chr7_+_142912316_142912316 | -0.27 | 6.4e-07 | Click! |
| Nr2f1 | rn6_v1_chr2_-_5579894_5579894 | -0.03 | 6.3e-01 | Click! |
Activity profile of Nr2f1_Nr4a1 motif
Sorted Z-values of Nr2f1_Nr4a1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_-_50531423 | 83.11 |
ENSRNOT00000090985
ENSRNOT00000074942 |
Apoc3
|
apolipoprotein C3 |
| chr2_-_243407608 | 52.15 |
ENSRNOT00000014631
|
Mttp
|
microsomal triglyceride transfer protein |
| chr5_+_118743632 | 47.87 |
ENSRNOT00000013785
|
Pgm1
|
phosphoglucomutase 1 |
| chr2_-_178389608 | 47.69 |
ENSRNOT00000013262
|
Etfdh
|
electron transfer flavoprotein dehydrogenase |
| chr10_+_63659085 | 46.95 |
ENSRNOT00000005039
|
Rilp
|
Rab interacting lysosomal protein |
| chr17_-_66397653 | 46.23 |
ENSRNOT00000024098
|
Actn2
|
actinin alpha 2 |
| chr2_-_23256158 | 45.75 |
ENSRNOT00000015336
|
Bhmt
|
betaine-homocysteine S-methyltransferase |
| chrX_-_23144324 | 45.34 |
ENSRNOT00000000178
ENSRNOT00000081239 |
Pfkfb1
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 |
| chr19_-_56677084 | 43.77 |
ENSRNOT00000024084
|
Acta1
|
actin, alpha 1, skeletal muscle |
| chr3_-_60813869 | 41.06 |
ENSRNOT00000058234
|
Atp5g3
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C3 (subunit 9) |
| chr12_-_48238887 | 38.88 |
ENSRNOT00000078868
|
Acacb
|
acetyl-CoA carboxylase beta |
| chr11_+_83868655 | 38.71 |
ENSRNOT00000072402
|
NEWGENE_621438
|
thrombopoietin |
| chr17_+_22620721 | 37.81 |
ENSRNOT00000019478
|
Adtrp
|
androgen-dependent TFPI-regulating protein |
| chr2_-_140464607 | 36.03 |
ENSRNOT00000058190
|
Ndufc1
|
NADH:ubiquinone oxidoreductase subunit C1 |
| chr5_+_119097715 | 35.51 |
ENSRNOT00000045987
|
Ror1
|
receptor tyrosine kinase-like orphan receptor 1 |
| chr3_+_171213936 | 34.51 |
ENSRNOT00000031586
|
Pck1
|
phosphoenolpyruvate carboxykinase 1 |
| chr1_+_31545631 | 33.98 |
ENSRNOT00000018336
|
Sdha
|
succinate dehydrogenase complex flavoprotein subunit A |
| chr10_+_61432819 | 33.98 |
ENSRNOT00000003687
ENSRNOT00000092478 |
Cluh
|
clustered mitochondria homolog |
| chr1_-_141579871 | 33.82 |
ENSRNOT00000020002
|
Anpep
|
alanyl aminopeptidase, membrane |
| chr2_+_119139717 | 33.53 |
ENSRNOT00000016051
|
Ndufb5
|
NADH:ubiquinone oxidoreductase subunit B5 |
| chr6_+_8284878 | 32.81 |
ENSRNOT00000009581
|
Slc3a1
|
solute carrier family 3 member 1 |
| chr3_-_8766433 | 32.60 |
ENSRNOT00000021865
|
Kyat1
|
kynurenine aminotransferase 1 |
| chr2_-_53313884 | 32.20 |
ENSRNOT00000046951
|
Ghr
|
growth hormone receptor |
| chr3_-_159775643 | 31.61 |
ENSRNOT00000010939
|
Jph2
|
junctophilin 2 |
| chr5_+_159484370 | 31.58 |
ENSRNOT00000010593
|
Sdhb
|
succinate dehydrogenase complex iron sulfur subunit B |
| chr11_-_77703255 | 31.52 |
ENSRNOT00000083319
|
Cldn16
|
claudin 16 |
| chr5_-_126911520 | 30.96 |
ENSRNOT00000091521
|
Dio1
|
deiodinase, iodothyronine, type I |
| chr1_-_219144610 | 30.79 |
ENSRNOT00000023526
|
Ndufs8
|
NADH:ubiquinone oxidoreductase core subunit S8 |
| chr3_-_80543031 | 30.27 |
ENSRNOT00000022233
|
F2
|
coagulation factor II |
| chr5_-_160179978 | 29.64 |
ENSRNOT00000022820
|
Slc25a34
|
solute carrier family 25, member 34 |
| chr10_+_57278307 | 29.36 |
ENSRNOT00000005612
|
Eno3
|
enolase 3 |
| chr8_-_77398156 | 28.79 |
ENSRNOT00000091858
ENSRNOT00000085349 ENSRNOT00000082763 |
Lipc
|
lipase C, hepatic type |
| chr19_+_568287 | 28.63 |
ENSRNOT00000016419
|
Cdh16
|
cadherin 16 |
| chr10_+_71159869 | 28.20 |
ENSRNOT00000075977
ENSRNOT00000047427 |
Hnf1b
|
HNF1 homeobox B |
| chr13_-_56877611 | 28.05 |
ENSRNOT00000079040
ENSRNOT00000017195 |
Cfhr1
|
complement factor H-related 1 |
| chr19_-_10596851 | 27.73 |
ENSRNOT00000021716
|
Coq9
|
coenzyme Q9 |
| chr16_-_10941414 | 27.41 |
ENSRNOT00000086627
ENSRNOT00000085414 ENSRNOT00000081631 ENSRNOT00000087521 ENSRNOT00000083623 |
Ldb3
|
LIM domain binding 3 |
| chr9_+_95256627 | 27.23 |
ENSRNOT00000025291
|
Ugt1a5
|
UDP glucuronosyltransferase family 1 member A5 |
| chr6_+_23337571 | 26.75 |
ENSRNOT00000011832
|
RGD1304963
|
similar to hypothetical protein MGC38716 |
| chr13_+_56598957 | 26.69 |
ENSRNOT00000016944
ENSRNOT00000080335 ENSRNOT00000089913 |
F13b
|
coagulation factor XIII B chain |
| chr14_-_83741969 | 26.56 |
ENSRNOT00000026293
|
Inpp5j
|
inositol polyphosphate-5-phosphatase J |
| chr15_-_27819376 | 26.13 |
ENSRNOT00000067400
|
A930018M24Rik
|
RIKEN cDNA A930018M24 gene |
| chr10_+_11240138 | 25.97 |
ENSRNOT00000048687
|
Srl
|
sarcalumenin |
| chr5_-_101138427 | 25.85 |
ENSRNOT00000058615
|
Frem1
|
Fras1 related extracellular matrix 1 |
| chr18_+_63016761 | 25.67 |
ENSRNOT00000081432
|
Impa2
|
inositol monophosphatase 2 |
| chr8_-_50526843 | 25.40 |
ENSRNOT00000092188
|
AABR07070085.1
|
|
| chr6_-_7058314 | 25.38 |
ENSRNOT00000045996
|
Haao
|
3-hydroxyanthranilate 3,4-dioxygenase |
| chr1_+_263554453 | 25.24 |
ENSRNOT00000070861
|
Abcc2
|
ATP binding cassette subfamily C member 2 |
| chr1_-_224533219 | 24.99 |
ENSRNOT00000051289
|
Ust5r
|
integral membrane transport protein UST5r |
| chr3_+_55910177 | 24.78 |
ENSRNOT00000009969
|
Klhl41
|
kelch-like family member 41 |
| chr18_+_74156553 | 24.67 |
ENSRNOT00000022892
|
Atp5a1
|
ATP synthase, H+ transporting, mitochondrial F1 complex, alpha subunit 1, cardiac muscle |
| chr4_-_148845267 | 24.61 |
ENSRNOT00000037397
|
Tmem72
|
transmembrane protein 72 |
| chr12_-_48218955 | 24.40 |
ENSRNOT00000067975
ENSRNOT00000080557 ENSRNOT00000000821 |
Acacb
|
acetyl-CoA carboxylase beta |
| chr14_+_22937421 | 24.32 |
ENSRNOT00000065079
|
RGD1559459
|
similar to Expressed sequence AI788959 |
| chr10_-_38782419 | 23.92 |
ENSRNOT00000073964
|
Uqcrq
|
ubiquinol-cytochrome c reductase, complex III subunit VII |
| chr1_-_224389389 | 23.50 |
ENSRNOT00000077408
ENSRNOT00000050010 |
UST4r
|
integral membrane transport protein UST4r |
| chr14_+_22517774 | 23.45 |
ENSRNOT00000047655
|
Ugt2b37
|
UDP-glucuronosyltransferase 2 family, member 37 |
| chr11_-_32088002 | 23.33 |
ENSRNOT00000002732
|
Atp5o
|
ATP synthase, H+ transporting, mitochondrial F1 complex, O subunit |
| chr1_-_183763664 | 23.31 |
ENSRNOT00000044231
|
LOC691427
|
similar to 6.8 kDa mitochondrial proteolipid |
| chr7_-_118108864 | 23.12 |
ENSRNOT00000006184
|
Mb
|
myoglobin |
| chr10_-_104748003 | 22.55 |
ENSRNOT00000042372
ENSRNOT00000046754 |
Acox1
|
acyl-CoA oxidase 1 |
| chr10_+_37724915 | 22.36 |
ENSRNOT00000008477
|
Vdac1
|
voltage-dependent anion channel 1 |
| chr1_-_89488223 | 22.30 |
ENSRNOT00000028624
|
Fxyd1
|
FXYD domain-containing ion transport regulator 1 |
| chr13_-_93677377 | 22.30 |
ENSRNOT00000004917
|
Fh
|
fumarate hydratase |
| chr2_-_119140110 | 22.24 |
ENSRNOT00000058810
|
AABR07009978.1
|
|
| chr5_-_134927235 | 22.17 |
ENSRNOT00000016751
|
Uqcrh
|
ubiquinol-cytochrome c reductase hinge protein |
| chr9_-_9985630 | 22.12 |
ENSRNOT00000071780
|
Crb3
|
crumbs 3, cell polarity complex component |
| chr13_-_56763981 | 22.08 |
ENSRNOT00000087916
|
LOC100361907
|
complement factor H-related protein B |
| chr10_-_38774449 | 21.52 |
ENSRNOT00000049820
|
Leap2
|
liver-expressed antimicrobial peptide 2 |
| chr14_-_91996774 | 21.51 |
ENSRNOT00000005851
|
Ddc
|
dopa decarboxylase |
| chr12_+_9446940 | 21.41 |
ENSRNOT00000074791
|
Urad
|
ureidoimidazoline (2-oxo-4-hydroxy-4-carboxy-5-) decarboxylase |
| chr5_+_159967839 | 21.05 |
ENSRNOT00000051317
|
Hspb7
|
heat shock protein family B (small) member 7 |
| chr12_+_45905371 | 20.91 |
ENSRNOT00000039275
|
Hspb8
|
heat shock protein family B (small) member 8 |
| chr15_+_57290849 | 20.85 |
ENSRNOT00000014909
|
Cpb2
|
carboxypeptidase B2 |
| chr2_+_235738416 | 20.68 |
ENSRNOT00000074209
|
Etnppl
|
ethanolamine-phosphate phospho-lyase |
| chr10_-_15235740 | 20.60 |
ENSRNOT00000027170
|
Mcrip2
|
MAPK regulated co-repressor interacting protein 2 |
| chr8_-_23099042 | 20.49 |
ENSRNOT00000019115
|
Ecsit
|
ECSIT signalling integrator |
| chr15_+_61069581 | 20.49 |
ENSRNOT00000084333
|
Vwa8
|
von Willebrand factor A domain containing 8 |
| chr9_+_47536824 | 20.32 |
ENSRNOT00000049349
|
Tmem182
|
transmembrane protein 182 |
| chr19_+_54245950 | 20.31 |
ENSRNOT00000024033
|
Cox4i1
|
cytochrome c oxidase subunit 4i1 |
| chr5_+_160306727 | 20.22 |
ENSRNOT00000016648
|
Agmat
|
agmatinase |
| chr1_+_224824799 | 19.98 |
ENSRNOT00000024757
|
Slc22a6
|
solute carrier family 22 member 6 |
| chr5_+_90338795 | 19.90 |
ENSRNOT00000077864
ENSRNOT00000058882 |
LOC298139
|
similar to RIKEN cDNA 2310003M01 |
| chr20_-_33323367 | 19.76 |
ENSRNOT00000080444
|
Ros1
|
ROS proto-oncogene 1 , receptor tyrosine kinase |
| chr3_-_172566010 | 19.72 |
ENSRNOT00000071913
|
Atp5e
|
ATP synthase, H+ transporting, mitochondrial F1 complex, epsilon subunit |
| chr2_+_198965685 | 19.54 |
ENSRNOT00000000107
ENSRNOT00000091578 |
Pdzk1
|
PDZ domain containing 1 |
| chr17_-_80807181 | 19.49 |
ENSRNOT00000040052
ENSRNOT00000090064 |
Cubn
|
cubilin |
| chr13_-_82758004 | 19.42 |
ENSRNOT00000003932
|
Atp1b1
|
ATPase Na+/K+ transporting subunit beta 1 |
| chr2_-_14701903 | 19.26 |
ENSRNOT00000051895
|
Cox7c
|
cytochrome c oxidase subunit 7C |
| chr1_+_101884019 | 19.25 |
ENSRNOT00000028650
|
Tmem143
|
transmembrane protein 143 |
| chr14_+_22142364 | 19.20 |
ENSRNOT00000002699
|
Sult1b1
|
sulfotransferase family 1B member 1 |
| chr7_+_11490852 | 19.15 |
ENSRNOT00000044484
|
Creb3l3
|
cAMP responsive element binding protein 3-like 3 |
| chr1_+_107344904 | 18.98 |
ENSRNOT00000082582
|
Gas2
|
growth arrest-specific 2 |
| chr1_+_107262659 | 18.90 |
ENSRNOT00000022499
|
Gas2
|
growth arrest-specific 2 |
| chr1_+_31264755 | 18.85 |
ENSRNOT00000028988
|
LOC679739
|
NADH dehydrogenase (ubiquinone) Fe-S protein 6 |
| chr7_+_2504695 | 18.75 |
ENSRNOT00000003965
|
Atp5b
|
ATP synthase, H+ transporting, mitochondrial F1 complex, beta polypeptide |
| chr9_+_17340341 | 18.68 |
ENSRNOT00000026637
ENSRNOT00000026559 ENSRNOT00000042790 ENSRNOT00000044163 ENSRNOT00000083811 |
Vegfa
|
vascular endothelial growth factor A |
| chrX_+_143097525 | 18.62 |
ENSRNOT00000004559
|
F9
|
coagulation factor IX |
| chr11_+_80736576 | 18.22 |
ENSRNOT00000047678
|
Masp1
|
mannan-binding lectin serine peptidase 1 |
| chr2_+_182006242 | 18.13 |
ENSRNOT00000064091
|
Fga
|
fibrinogen alpha chain |
| chr1_+_213676954 | 17.93 |
ENSRNOT00000050551
|
Nlrp6
|
NLR family, pyrin domain containing 6 |
| chr1_-_263269762 | 17.74 |
ENSRNOT00000022309
|
Got1
|
glutamic-oxaloacetic transaminase 1 |
| chr1_-_222350173 | 17.56 |
ENSRNOT00000030625
|
Flrt1
|
fibronectin leucine rich transmembrane protein 1 |
| chr14_-_84937725 | 17.43 |
ENSRNOT00000083839
|
Uqcr10
|
ubiquinol-cytochrome c reductase, complex III subunit X |
| chr8_-_78233430 | 17.36 |
ENSRNOT00000083220
|
Cgnl1
|
cingulin-like 1 |
| chrX_+_110818716 | 17.24 |
ENSRNOT00000086308
|
Rnf128
|
ring finger protein 128, E3 ubiquitin protein ligase |
| chr9_+_95274707 | 17.21 |
ENSRNOT00000045163
|
Ugt1a5
|
UDP glucuronosyltransferase family 1 member A5 |
| chr5_-_19368431 | 17.18 |
ENSRNOT00000012819
|
Cyp7a1
|
cytochrome P450, family 7, subfamily a, polypeptide 1 |
| chr2_+_187347602 | 17.16 |
ENSRNOT00000025384
|
Nes
|
nestin |
| chr1_-_214263581 | 17.09 |
ENSRNOT00000044238
|
Cdhr5
|
cadherin-related family member 5 |
| chr12_+_10636275 | 17.09 |
ENSRNOT00000001285
|
Cyp3a18
|
cytochrome P450, family 3, subfamily a, polypeptide 18 |
| chr5_-_126080698 | 16.89 |
ENSRNOT00000008739
|
Bsnd
|
barttin CLCNK type accessory beta subunit |
| chr20_-_45126062 | 16.89 |
ENSRNOT00000000720
|
RGD1310495
|
similar to KIAA1919 protein |
| chr7_-_74735650 | 16.88 |
ENSRNOT00000014407
|
Cox6c
|
cytochrome c oxidase subunit 6C |
| chr11_-_71750014 | 16.72 |
ENSRNOT00000072227
|
LOC100911042
|
uncharacterized LOC100911042 |
| chr7_+_38742051 | 16.66 |
ENSRNOT00000006070
|
Dcn
|
decorin |
| chr9_+_61692154 | 16.62 |
ENSRNOT00000082300
|
Hspe1
|
heat shock protein family E member 1 |
| chr9_+_43259709 | 16.56 |
ENSRNOT00000022487
|
Cox5b
|
cytochrome c oxidase subunit 5B |
| chr8_-_50539331 | 16.55 |
ENSRNOT00000088997
|
AABR07073400.1
|
|
| chr2_-_28750237 | 16.54 |
ENSRNOT00000020879
|
Tmem174
|
transmembrane protein 174 |
| chr12_+_47407811 | 16.30 |
ENSRNOT00000001565
|
Hnf1a
|
HNF1 homeobox A |
| chr7_+_117409576 | 16.27 |
ENSRNOT00000017067
|
Cyc1
|
cytochrome c-1 |
| chr5_+_147185474 | 16.27 |
ENSRNOT00000000134
|
Ak2
|
adenylate kinase 2 |
| chr5_+_78222504 | 16.23 |
ENSRNOT00000019544
|
Slc31a1
|
solute carrier family 31 member 1 |
| chr5_+_126334803 | 16.18 |
ENSRNOT00000010288
|
Fam151a
|
family with sequence similarity 151, member A |
| chr8_+_71167305 | 15.93 |
ENSRNOT00000021337
|
Rbpms2
|
RNA binding protein with multiple splicing 2 |
| chr4_-_15505362 | 15.89 |
ENSRNOT00000009763
|
Hgf
|
hepatocyte growth factor |
| chr7_+_12840938 | 15.73 |
ENSRNOT00000039232
|
Polrmt
|
RNA polymerase mitochondrial |
| chr1_-_263762785 | 15.66 |
ENSRNOT00000018221
|
Cpn1
|
carboxypeptidase N subunit 1 |
| chr12_+_39553903 | 15.55 |
ENSRNOT00000001738
|
Atp2a2
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 2 |
| chr3_+_58632476 | 15.40 |
ENSRNOT00000010630
|
Rapgef4
|
Rap guanine nucleotide exchange factor 4 |
| chr17_-_10001901 | 15.40 |
ENSRNOT00000082836
|
Fgfr4
|
fibroblast growth factor receptor 4 |
| chr13_-_56958549 | 15.25 |
ENSRNOT00000017293
ENSRNOT00000083912 |
RGD1564614
|
similar to complement factor H-related protein |
| chr2_+_56426367 | 15.17 |
ENSRNOT00000016036
|
Lifr
|
leukemia inhibitory factor receptor alpha |
| chr1_-_277181345 | 15.15 |
ENSRNOT00000038017
ENSRNOT00000038038 |
Nrap
|
nebulin-related anchoring protein |
| chr3_-_127500709 | 14.97 |
ENSRNOT00000006330
|
Hao1
|
hydroxyacid oxidase 1 |
| chr13_-_89518939 | 14.96 |
ENSRNOT00000004228
|
Sdhc
|
succinate dehydrogenase complex subunit C |
| chr8_+_112594691 | 14.80 |
ENSRNOT00000038383
ENSRNOT00000081281 |
Acad11
|
acyl-CoA dehydrogenase family, member 11 |
| chr5_+_173640780 | 14.76 |
ENSRNOT00000027476
|
Perm1
|
PPARGC1 and ESRR induced regulator, muscle 1 |
| chr7_-_80796670 | 14.65 |
ENSRNOT00000010539
|
Abra
|
actin-binding Rho activating protein |
| chr18_-_28425944 | 14.60 |
ENSRNOT00000084372
|
Slc23a1
|
solute carrier family 23 member 1 |
| chr12_+_47254484 | 14.55 |
ENSRNOT00000001556
|
Acads
|
acyl-CoA dehydrogenase, C-2 to C-3 short chain |
| chr3_+_61613774 | 14.54 |
ENSRNOT00000002148
|
Hoxd10
|
homeo box D10 |
| chr1_+_213686046 | 14.18 |
ENSRNOT00000019808
|
LOC108348167
|
NACHT, LRR and PYD domains-containing protein 6-like |
| chr7_+_53630621 | 14.12 |
ENSRNOT00000067011
ENSRNOT00000080598 |
Csrp2
|
cysteine and glycine-rich protein 2 |
| chr1_+_101884276 | 14.10 |
ENSRNOT00000082917
|
Tmem143
|
transmembrane protein 143 |
| chr8_-_130429132 | 14.05 |
ENSRNOT00000026261
|
Hhatl
|
hedgehog acyltransferase-like |
| chr4_-_38240848 | 13.89 |
ENSRNOT00000007567
|
Ndufa4
|
NADH:ubiquinone oxidoreductase subunit A4 |
| chr10_+_56662561 | 13.83 |
ENSRNOT00000025254
|
Asgr1
|
asialoglycoprotein receptor 1 |
| chr4_-_82215022 | 13.83 |
ENSRNOT00000010256
|
Hoxa11
|
homeobox A11 |
| chr2_+_204512302 | 13.81 |
ENSRNOT00000021846
|
Casq2
|
calsequestrin 2 |
| chr7_+_35125424 | 13.76 |
ENSRNOT00000085978
ENSRNOT00000010117 |
Ndufa12
|
NADH:ubiquinone oxidoreductase subunit A12 |
| chr2_+_263895241 | 13.65 |
ENSRNOT00000014126
ENSRNOT00000014034 |
Ptger3
|
prostaglandin E receptor 3 |
| chr8_-_55037604 | 13.55 |
ENSRNOT00000059169
|
Sdhd
|
succinate dehydrogenase complex subunit D |
| chr11_-_87924816 | 13.54 |
ENSRNOT00000031819
|
Serpind1
|
serpin family D member 1 |
| chr1_-_222178725 | 13.36 |
ENSRNOT00000028697
|
Esrra
|
estrogen related receptor, alpha |
| chr2_+_235264219 | 13.22 |
ENSRNOT00000086245
|
Cfi
|
complement factor I |
| chr1_-_143535583 | 13.15 |
ENSRNOT00000087785
|
Homer2
|
homer scaffolding protein 2 |
| chr10_+_56662242 | 13.07 |
ENSRNOT00000086919
|
Asgr1
|
asialoglycoprotein receptor 1 |
| chr18_-_55891710 | 13.03 |
ENSRNOT00000064686
|
Synpo
|
synaptopodin |
| chr8_-_47339343 | 13.02 |
ENSRNOT00000081007
|
Arhgef12
|
Rho guanine nucleotide exchange factor 12 |
| chr9_+_20213776 | 13.00 |
ENSRNOT00000071439
|
LOC100911515
|
triosephosphate isomerase-like |
| chr4_-_157331905 | 12.64 |
ENSRNOT00000020647
|
Tpi1
|
triosephosphate isomerase 1 |
| chr11_-_38088753 | 12.54 |
ENSRNOT00000002713
|
Tmprss2
|
transmembrane protease, serine 2 |
| chr10_-_18506337 | 12.48 |
ENSRNOT00000043036
|
Gabrp
|
gamma-aminobutyric acid type A receptor pi subunit |
| chr3_+_151126591 | 12.44 |
ENSRNOT00000025859
|
Myh7b
|
myosin heavy chain 7B |
| chr1_+_87009730 | 12.37 |
ENSRNOT00000027537
|
Ech1
|
enoyl-CoA hydratase 1 |
| chr10_-_50574539 | 12.35 |
ENSRNOT00000034261
|
Cox10
|
COX10 heme A:farnesyltransferase cytochrome c oxidase assembly factor |
| chr2_+_93669765 | 12.33 |
ENSRNOT00000045438
|
Slc10a5
|
solute carrier family 10, member 5 |
| chr7_-_44771458 | 12.31 |
ENSRNOT00000006007
|
Alx1
|
ALX homeobox 1 |
| chr7_+_121311024 | 12.27 |
ENSRNOT00000092260
ENSRNOT00000023066 ENSRNOT00000081377 |
Syngr1
|
synaptogyrin 1 |
| chr14_-_106234247 | 12.25 |
ENSRNOT00000081245
|
Ugp2
|
UDP-glucose pyrophosphorylase 2 |
| chrX_+_77263359 | 12.14 |
ENSRNOT00000077604
|
Pgk1
|
phosphoglycerate kinase 1 |
| chrX_-_135342996 | 12.13 |
ENSRNOT00000084848
ENSRNOT00000008503 |
Aifm1
|
apoptosis inducing factor, mitochondria associated 1 |
| chr20_+_28989491 | 12.13 |
ENSRNOT00000074524
|
Pla2g12b
|
phospholipase A2, group XIIB |
| chr1_+_41192824 | 12.09 |
ENSRNOT00000082133
|
Esr1
|
estrogen receptor 1 |
| chr1_-_80617057 | 12.06 |
ENSRNOT00000080453
|
Apoe
|
apolipoprotein E |
| chr6_+_27589657 | 11.91 |
ENSRNOT00000038649
|
Hadha
|
hydroxyacyl-CoA dehydrogenase/3-ketoacyl-CoA thiolase/enoyl-CoA hydratase (trifunctional protein), alpha subunit |
| chr5_+_137357674 | 11.91 |
ENSRNOT00000092813
|
RGD1305347
|
similar to RIKEN cDNA 2610528J11 |
| chr2_+_256609787 | 11.85 |
ENSRNOT00000047564
ENSRNOT00000088437 |
Adgrl4
|
adhesion G protein-coupled receptor L4 |
| chr8_+_117679278 | 11.85 |
ENSRNOT00000042114
|
Uqcrc1
|
ubiquinol-cytochrome c reductase core protein I |
| chr6_+_107460668 | 11.83 |
ENSRNOT00000013515
|
Acot2
|
acyl-CoA thioesterase 2 |
| chr4_-_89281222 | 11.73 |
ENSRNOT00000010898
|
Fam13a
|
family with sequence similarity 13, member A |
| chr1_-_49844547 | 11.65 |
ENSRNOT00000086127
ENSRNOT00000077423 ENSRNOT00000089439 ENSRNOT00000090521 |
AABR07001519.1
|
|
| chr8_+_59164572 | 11.63 |
ENSRNOT00000015102
|
Idh3a
|
isocitrate dehydrogenase 3 (NAD+) alpha |
| chr9_-_69953182 | 11.58 |
ENSRNOT00000015852
|
Ndufs1
|
NADH dehydrogenase (ubiquinone) Fe-S protein 1 |
| chrX_+_106774980 | 11.55 |
ENSRNOT00000046091
|
Tceal7
|
transcription elongation factor A like 7 |
| chr13_+_75175254 | 11.54 |
ENSRNOT00000044008
|
Sec16b
|
SEC16 homolog B, endoplasmic reticulum export factor |
| chr7_-_31824064 | 11.47 |
ENSRNOT00000011494
ENSRNOT00000080824 |
Slc25a3
|
solute carrier family 25 member 3 |
| chr4_+_82214342 | 11.47 |
ENSRNOT00000066360
|
Hoxa11-as
|
homeobox A11, opposite strand |
| chr1_+_242572533 | 11.33 |
ENSRNOT00000035123
|
Tmem252
|
transmembrane protein 252 |
| chr14_-_8432195 | 11.32 |
ENSRNOT00000089800
|
Arhgap24
|
Rho GTPase activating protein 24 |
| chr17_+_32973695 | 11.26 |
ENSRNOT00000065674
|
RGD1565323
|
similar to OTTMUSP00000000621 |
| chr1_+_72882806 | 10.94 |
ENSRNOT00000024640
|
Tnni3
|
troponin I3, cardiac type |
| chr14_+_26662965 | 10.93 |
ENSRNOT00000002621
|
Tecrl
|
trans-2,3-enoyl-CoA reductase-like |
| chr4_+_82300778 | 10.85 |
ENSRNOT00000075254
|
Hoxa11-as
|
homeobox A11, opposite strand |
| chr9_+_64745051 | 10.85 |
ENSRNOT00000021527
|
Spats2l
|
spermatogenesis associated, serine-rich 2-like |
| chr1_-_253186695 | 10.84 |
ENSRNOT00000080928
|
AC096809.1
|
|
| chr4_+_51614676 | 10.84 |
ENSRNOT00000060494
|
Asb15
|
ankyrin repeat and SOCS box containing 15 |
| chr3_+_121490812 | 10.78 |
ENSRNOT00000023913
|
AABR07053707.1
|
|
Network of associatons between targets according to the STRING database.
First level regulatory network of Nr2f1_Nr4a1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 27.7 | 83.1 | GO:0010903 | negative regulation of very-low-density lipoprotein particle remodeling(GO:0010903) regulation of high-density lipoprotein particle clearance(GO:0010982) |
| 15.8 | 47.5 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 15.8 | 63.3 | GO:2001295 | malonyl-CoA biosynthetic process(GO:2001295) |
| 15.6 | 46.9 | GO:0010796 | regulation of multivesicular body size(GO:0010796) |
| 15.4 | 46.2 | GO:2001137 | actin filament uncapping(GO:0051695) phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) positive regulation of endocytic recycling(GO:2001137) |
| 15.2 | 45.7 | GO:0006579 | amino-acid betaine catabolic process(GO:0006579) |
| 15.0 | 15.0 | GO:0009441 | glycolate metabolic process(GO:0009441) |
| 14.0 | 56.2 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 13.3 | 40.0 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 12.9 | 38.7 | GO:0038163 | thrombopoietin-mediated signaling pathway(GO:0038163) |
| 11.5 | 34.5 | GO:0006114 | glycerol biosynthetic process(GO:0006114) |
| 11.1 | 44.5 | GO:0035565 | regulation of pronephros size(GO:0035565) |
| 10.9 | 43.8 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 10.1 | 30.3 | GO:1905225 | response to thyrotropin-releasing hormone(GO:1905225) |
| 8.7 | 26.0 | GO:0014873 | response to muscle activity involved in regulation of muscle adaptation(GO:0014873) |
| 8.5 | 25.4 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 8.4 | 33.6 | GO:0050787 | detoxification of mercury ion(GO:0050787) |
| 8.3 | 41.7 | GO:1903278 | positive regulation of sodium ion export(GO:1903275) positive regulation of sodium ion export from cell(GO:1903278) |
| 8.1 | 32.6 | GO:0097052 | L-kynurenine metabolic process(GO:0097052) |
| 8.0 | 24.0 | GO:0038190 | neuropilin signaling pathway(GO:0038189) VEGF-activated neuropilin signaling pathway(GO:0038190) |
| 8.0 | 23.9 | GO:0021539 | subthalamus development(GO:0021539) |
| 7.8 | 23.3 | GO:1903116 | positive regulation of actin filament-based movement(GO:1903116) |
| 7.7 | 23.1 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 7.7 | 77.0 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 7.7 | 23.0 | GO:0010034 | response to acetate(GO:0010034) |
| 7.5 | 22.5 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 7.0 | 20.9 | GO:0003331 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 6.5 | 52.1 | GO:0034196 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 6.0 | 47.9 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 6.0 | 17.9 | GO:0042412 | taurine biosynthetic process(GO:0042412) |
| 5.7 | 45.3 | GO:0033133 | positive regulation of glucokinase activity(GO:0033133) |
| 5.6 | 5.6 | GO:0044330 | canonical Wnt signaling pathway involved in positive regulation of wound healing(GO:0044330) |
| 5.4 | 21.5 | GO:0052314 | phytoalexin metabolic process(GO:0052314) |
| 5.2 | 15.5 | GO:1903233 | positive regulation of endoplasmic reticulum calcium ion concentration(GO:0032470) regulation of calcium ion-dependent exocytosis of neurotransmitter(GO:1903233) calcium ion import into sarcoplasmic reticulum(GO:1990036) |
| 5.1 | 15.4 | GO:1904457 | positive regulation of neuronal action potential(GO:1904457) |
| 5.0 | 123.9 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 4.9 | 44.4 | GO:0052697 | xenobiotic glucuronidation(GO:0052697) |
| 4.9 | 14.7 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 4.9 | 19.5 | GO:0015879 | carnitine transport(GO:0015879) |
| 4.9 | 14.6 | GO:0015882 | L-ascorbic acid transport(GO:0015882) transepithelial L-ascorbic acid transport(GO:0070904) |
| 4.8 | 33.5 | GO:0060316 | positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) |
| 4.4 | 39.2 | GO:0072338 | creatinine metabolic process(GO:0046449) cellular lactam metabolic process(GO:0072338) |
| 4.3 | 17.2 | GO:0045105 | intermediate filament polymerization or depolymerization(GO:0045105) |
| 4.1 | 12.2 | GO:0006011 | UDP-glucose metabolic process(GO:0006011) glucose 1-phosphate metabolic process(GO:0019255) |
| 4.1 | 44.8 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 4.1 | 16.3 | GO:0006172 | ADP biosynthetic process(GO:0006172) |
| 4.1 | 16.2 | GO:1902861 | copper ion import across plasma membrane(GO:0098705) copper ion import into cell(GO:1902861) |
| 4.0 | 20.2 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 4.0 | 12.1 | GO:1903002 | regulation of lipid transport across blood brain barrier(GO:1903000) positive regulation of lipid transport across blood brain barrier(GO:1903002) |
| 4.0 | 20.0 | GO:0031427 | response to methotrexate(GO:0031427) |
| 4.0 | 8.0 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 4.0 | 15.9 | GO:1902947 | regulation of tau-protein kinase activity(GO:1902947) |
| 3.9 | 31.6 | GO:0006105 | succinate metabolic process(GO:0006105) |
| 3.9 | 19.5 | GO:0015889 | cobalamin transport(GO:0015889) |
| 3.8 | 15.4 | GO:0007037 | vacuolar phosphate transport(GO:0007037) positive regulation of parathyroid hormone secretion(GO:2000830) |
| 3.8 | 15.2 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) leukemia inhibitory factor signaling pathway(GO:0048861) |
| 3.6 | 18.1 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 3.5 | 14.1 | GO:1903059 | regulation of protein lipidation(GO:1903059) |
| 3.4 | 17.1 | GO:1904970 | brush border assembly(GO:1904970) |
| 3.4 | 34.0 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 3.3 | 26.8 | GO:1903546 | protein localization to photoreceptor outer segment(GO:1903546) |
| 3.3 | 9.9 | GO:0019747 | regulation of isoprenoid metabolic process(GO:0019747) |
| 3.3 | 19.8 | GO:0010966 | regulation of phosphate transport(GO:0010966) |
| 3.3 | 13.0 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 3.2 | 25.7 | GO:0006021 | inositol biosynthetic process(GO:0006021) |
| 3.2 | 9.5 | GO:0045041 | protein import into mitochondrial intermembrane space(GO:0045041) |
| 3.1 | 15.7 | GO:0030070 | insulin processing(GO:0030070) |
| 3.0 | 12.1 | GO:1990375 | baculum development(GO:1990375) |
| 2.9 | 8.6 | GO:2000546 | positive regulation of cell chemotaxis to fibroblast growth factor(GO:1904849) positive regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000546) |
| 2.8 | 13.8 | GO:0071313 | cellular response to caffeine(GO:0071313) |
| 2.7 | 19.2 | GO:0009812 | flavonoid metabolic process(GO:0009812) |
| 2.7 | 8.1 | GO:0042822 | pyridoxal phosphate metabolic process(GO:0042822) |
| 2.7 | 21.3 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 2.7 | 8.0 | GO:1902445 | regulation of mitochondrial membrane permeability involved in programmed necrotic cell death(GO:1902445) |
| 2.6 | 10.4 | GO:1902031 | regulation of NADP metabolic process(GO:1902031) |
| 2.6 | 7.8 | GO:0000105 | histidine biosynthetic process(GO:0000105) |
| 2.5 | 12.6 | GO:0019563 | glycerol catabolic process(GO:0019563) |
| 2.5 | 7.4 | GO:0021627 | muscle attachment(GO:0016203) olfactory nerve morphogenesis(GO:0021627) olfactory nerve structural organization(GO:0021629) |
| 2.4 | 12.1 | GO:1904045 | cellular response to aldosterone(GO:1904045) |
| 2.4 | 7.2 | GO:1902037 | negative regulation of hematopoietic stem cell differentiation(GO:1902037) |
| 2.4 | 16.7 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 2.4 | 7.1 | GO:0009249 | protein lipoylation(GO:0009249) |
| 2.3 | 23.5 | GO:0015747 | urate transport(GO:0015747) |
| 2.3 | 41.3 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 2.3 | 6.8 | GO:0042222 | interleukin-1 biosynthetic process(GO:0042222) |
| 2.3 | 31.5 | GO:0070633 | transepithelial transport(GO:0070633) |
| 2.0 | 6.1 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 2.0 | 26.3 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 1.9 | 11.6 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 1.9 | 1.9 | GO:0010841 | positive regulation of circadian sleep/wake cycle, wakefulness(GO:0010841) |
| 1.9 | 7.7 | GO:0019072 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 1.9 | 24.8 | GO:2000291 | regulation of myoblast proliferation(GO:2000291) |
| 1.9 | 24.2 | GO:0006851 | mitochondrial calcium ion transport(GO:0006851) |
| 1.8 | 7.3 | GO:0046351 | disaccharide biosynthetic process(GO:0046351) |
| 1.8 | 21.8 | GO:0035814 | negative regulation of renal sodium excretion(GO:0035814) |
| 1.8 | 3.6 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 1.8 | 21.2 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 1.7 | 3.5 | GO:0060066 | oviduct development(GO:0060066) |
| 1.7 | 17.2 | GO:0070857 | regulation of bile acid biosynthetic process(GO:0070857) |
| 1.7 | 27.5 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 1.7 | 11.8 | GO:0042760 | very long-chain fatty acid catabolic process(GO:0042760) |
| 1.7 | 32.0 | GO:0010257 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 1.7 | 5.0 | GO:2001205 | negative regulation of osteoclast development(GO:2001205) |
| 1.7 | 29.9 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 1.6 | 31.3 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 1.6 | 4.9 | GO:0009644 | response to high light intensity(GO:0009644) |
| 1.6 | 22.8 | GO:0060272 | embryonic skeletal joint morphogenesis(GO:0060272) |
| 1.6 | 6.5 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 1.6 | 12.5 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 1.5 | 18.2 | GO:0045916 | negative regulation of complement activation(GO:0045916) negative regulation of protein activation cascade(GO:2000258) |
| 1.4 | 2.9 | GO:0098923 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) trans-synaptic signaling by soluble gas(GO:0099543) |
| 1.4 | 9.8 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 1.4 | 7.0 | GO:0061727 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 1.4 | 4.1 | GO:0051012 | microtubule sliding(GO:0051012) |
| 1.4 | 6.8 | GO:0086048 | membrane depolarization during Purkinje myocyte cell action potential(GO:0086047) membrane depolarization during bundle of His cell action potential(GO:0086048) |
| 1.4 | 5.4 | GO:1905233 | carbohydrate export(GO:0033231) daunorubicin transport(GO:0043215) response to borneol(GO:1905230) cellular response to borneol(GO:1905231) response to codeine(GO:1905233) response to quercetin(GO:1905235) drug transport across blood-brain barrier(GO:1990962) establishment of blood-retinal barrier(GO:1990963) |
| 1.4 | 4.1 | GO:0071579 | regulation of zinc ion transport(GO:0071579) |
| 1.3 | 9.4 | GO:0070508 | sterol import(GO:0035376) cholesterol import(GO:0070508) |
| 1.3 | 1.3 | GO:1904054 | regulation of cholangiocyte proliferation(GO:1904054) positive regulation of cholangiocyte proliferation(GO:1904056) |
| 1.3 | 10.5 | GO:0060613 | fat pad development(GO:0060613) |
| 1.3 | 6.5 | GO:0000960 | mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 1.3 | 16.8 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 1.3 | 19.2 | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 1.3 | 3.8 | GO:0017062 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 1.2 | 3.7 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 1.2 | 14.5 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 1.2 | 2.4 | GO:0003130 | BMP signaling pathway involved in heart induction(GO:0003130) endodermal-mesodermal cell signaling(GO:0003133) endodermal-mesodermal cell signaling involved in heart induction(GO:0003134) |
| 1.2 | 26.0 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 1.2 | 2.3 | GO:0033552 | response to vitamin B3(GO:0033552) |
| 1.2 | 9.3 | GO:0055089 | fatty acid homeostasis(GO:0055089) |
| 1.1 | 17.9 | GO:0070255 | regulation of mucus secretion(GO:0070255) |
| 1.1 | 3.3 | GO:0015680 | intracellular copper ion transport(GO:0015680) |
| 1.1 | 5.5 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 1.1 | 30.4 | GO:0014002 | astrocyte development(GO:0014002) |
| 1.1 | 32.6 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 1.0 | 12.5 | GO:0046598 | positive regulation of viral entry into host cell(GO:0046598) |
| 1.0 | 16.6 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 1.0 | 20.9 | GO:0019682 | pentose-phosphate shunt(GO:0006098) glyceraldehyde-3-phosphate metabolic process(GO:0019682) |
| 1.0 | 10.9 | GO:0010882 | regulation of cardiac muscle contraction by calcium ion signaling(GO:0010882) |
| 1.0 | 2.9 | GO:0046078 | dUMP metabolic process(GO:0046078) |
| 1.0 | 18.3 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.9 | 4.7 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.9 | 3.8 | GO:0016103 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) |
| 0.9 | 14.8 | GO:0014850 | response to muscle activity(GO:0014850) |
| 0.9 | 6.4 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.9 | 3.7 | GO:0002084 | protein depalmitoylation(GO:0002084) |
| 0.9 | 2.7 | GO:0072076 | nephrogenic mesenchyme development(GO:0072076) pattern specification involved in metanephros development(GO:0072268) |
| 0.9 | 12.4 | GO:0008535 | respiratory chain complex IV assembly(GO:0008535) |
| 0.9 | 29.4 | GO:0043403 | skeletal muscle tissue regeneration(GO:0043403) |
| 0.9 | 5.2 | GO:0030046 | parallel actin filament bundle assembly(GO:0030046) |
| 0.8 | 4.2 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.8 | 12.5 | GO:0010666 | positive regulation of striated muscle cell apoptotic process(GO:0010663) positive regulation of cardiac muscle cell apoptotic process(GO:0010666) |
| 0.8 | 2.5 | GO:0032053 | ciliary basal body organization(GO:0032053) positive regulation of protein localization to cilium(GO:1903566) |
| 0.8 | 7.4 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.8 | 2.4 | GO:0034241 | positive regulation of macrophage fusion(GO:0034241) |
| 0.8 | 4.8 | GO:0032914 | positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 0.8 | 12.1 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.8 | 8.0 | GO:0051560 | mitochondrial calcium ion homeostasis(GO:0051560) |
| 0.8 | 5.6 | GO:0044351 | macropinocytosis(GO:0044351) |
| 0.8 | 10.3 | GO:0016188 | synaptic vesicle maturation(GO:0016188) |
| 0.8 | 11.4 | GO:0006688 | glycosphingolipid biosynthetic process(GO:0006688) |
| 0.8 | 3.8 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.7 | 3.7 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.7 | 13.1 | GO:0035584 | calcium-mediated signaling using intracellular calcium source(GO:0035584) |
| 0.7 | 9.5 | GO:0006086 | acetyl-CoA biosynthetic process from pyruvate(GO:0006086) |
| 0.7 | 12.3 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.7 | 4.2 | GO:0070885 | negative regulation of calcineurin-NFAT signaling cascade(GO:0070885) |
| 0.7 | 9.2 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.7 | 33.9 | GO:0042446 | hormone biosynthetic process(GO:0042446) |
| 0.7 | 9.9 | GO:0090140 | regulation of mitochondrial fission(GO:0090140) |
| 0.7 | 13.2 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.6 | 6.4 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.6 | 7.0 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.6 | 2.5 | GO:0018214 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.6 | 8.6 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.6 | 11.5 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.6 | 23.7 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.6 | 1.7 | GO:0046947 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.5 | 2.1 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.5 | 13.4 | GO:1900078 | positive regulation of cellular response to insulin stimulus(GO:1900078) |
| 0.5 | 12.1 | GO:0070328 | acylglycerol homeostasis(GO:0055090) triglyceride homeostasis(GO:0070328) |
| 0.5 | 4.5 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.5 | 2.9 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.5 | 2.4 | GO:1902162 | regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) |
| 0.5 | 6.2 | GO:0071340 | skeletal muscle acetylcholine-gated channel clustering(GO:0071340) |
| 0.5 | 3.8 | GO:0071318 | cellular response to ATP(GO:0071318) |
| 0.5 | 2.4 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.5 | 15.0 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.5 | 12.1 | GO:0031639 | plasminogen activation(GO:0031639) |
| 0.5 | 3.2 | GO:0035879 | plasma membrane lactate transport(GO:0035879) |
| 0.5 | 0.9 | GO:0032252 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) secretory granule localization(GO:0032252) |
| 0.4 | 1.3 | GO:2000721 | positive regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000721) |
| 0.4 | 2.6 | GO:2000973 | regulation of pro-B cell differentiation(GO:2000973) |
| 0.4 | 3.8 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.4 | 2.9 | GO:0046133 | pyrimidine ribonucleoside catabolic process(GO:0046133) |
| 0.4 | 2.0 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.4 | 18.9 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.4 | 1.2 | GO:2000054 | negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 0.4 | 12.3 | GO:0010718 | positive regulation of epithelial to mesenchymal transition(GO:0010718) |
| 0.4 | 5.9 | GO:0090201 | negative regulation of release of cytochrome c from mitochondria(GO:0090201) |
| 0.4 | 17.6 | GO:0046426 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.4 | 5.0 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.4 | 5.7 | GO:0043950 | positive regulation of cAMP-mediated signaling(GO:0043950) |
| 0.4 | 10.5 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.4 | 1.1 | GO:0061589 | calcium activated phosphatidylserine scrambling(GO:0061589) |
| 0.4 | 4.1 | GO:0044331 | cell-cell adhesion mediated by cadherin(GO:0044331) |
| 0.4 | 1.1 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.4 | 5.1 | GO:0001967 | suckling behavior(GO:0001967) |
| 0.4 | 1.1 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.4 | 0.7 | GO:0097018 | positive regulation of glomerular filtration(GO:0003104) renal albumin absorption(GO:0097018) regulation of renal albumin absorption(GO:2000532) |
| 0.3 | 5.9 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.3 | 52.8 | GO:0007596 | blood coagulation(GO:0007596) |
| 0.3 | 5.0 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.3 | 1.6 | GO:0070309 | lens fiber cell morphogenesis(GO:0070309) |
| 0.3 | 3.5 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.3 | 2.5 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.3 | 2.2 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.3 | 1.2 | GO:1904431 | positive regulation of t-circle formation(GO:1904431) |
| 0.3 | 0.9 | GO:0044376 | RNA polymerase II complex import to nucleus(GO:0044376) RNA polymerase III complex localization to nucleus(GO:1990022) |
| 0.3 | 11.3 | GO:0071806 | protein transmembrane transport(GO:0071806) |
| 0.3 | 1.2 | GO:0007182 | common-partner SMAD protein phosphorylation(GO:0007182) |
| 0.3 | 2.9 | GO:0061051 | positive regulation of cell growth involved in cardiac muscle cell development(GO:0061051) |
| 0.3 | 3.7 | GO:0031424 | keratinization(GO:0031424) |
| 0.3 | 6.2 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.3 | 1.7 | GO:2000074 | regulation of type B pancreatic cell development(GO:2000074) negative regulation of glucocorticoid receptor signaling pathway(GO:2000323) |
| 0.3 | 3.8 | GO:0007097 | nuclear migration(GO:0007097) |
| 0.3 | 7.1 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.3 | 11.1 | GO:0006805 | xenobiotic metabolic process(GO:0006805) |
| 0.3 | 3.0 | GO:2000505 | regulation of energy homeostasis(GO:2000505) |
| 0.3 | 4.3 | GO:0042761 | very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.2 | 8.2 | GO:0042246 | tissue regeneration(GO:0042246) |
| 0.2 | 6.4 | GO:0042036 | negative regulation of cytokine biosynthetic process(GO:0042036) |
| 0.2 | 1.1 | GO:0090274 | positive regulation of pancreatic juice secretion(GO:0090187) positive regulation of somatostatin secretion(GO:0090274) |
| 0.2 | 0.9 | GO:0031547 | brain-derived neurotrophic factor receptor signaling pathway(GO:0031547) |
| 0.2 | 4.7 | GO:0061436 | establishment of skin barrier(GO:0061436) |
| 0.2 | 4.7 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.2 | 2.5 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.2 | 1.4 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.2 | 7.0 | GO:0045214 | sarcomere organization(GO:0045214) |
| 0.2 | 7.1 | GO:0005978 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.2 | 1.1 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.2 | 12.8 | GO:0045445 | myoblast differentiation(GO:0045445) |
| 0.2 | 1.6 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.2 | 1.9 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.2 | 8.1 | GO:0001707 | mesoderm formation(GO:0001707) |
| 0.2 | 1.2 | GO:0070601 | centromeric sister chromatid cohesion(GO:0070601) |
| 0.2 | 5.4 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.2 | 0.9 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.1 | 1.5 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.1 | 0.4 | GO:0098972 | dendritic transport of mitochondrion(GO:0098939) anterograde dendritic transport of mitochondrion(GO:0098972) |
| 0.1 | 0.5 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.1 | 8.8 | GO:0098869 | cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.1 | 2.9 | GO:0045736 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) |
| 0.1 | 5.7 | GO:0006360 | transcription from RNA polymerase I promoter(GO:0006360) |
| 0.1 | 1.2 | GO:2000773 | negative regulation of cellular senescence(GO:2000773) |
| 0.1 | 4.2 | GO:0009268 | response to pH(GO:0009268) |
| 0.1 | 1.6 | GO:0050892 | intestinal absorption(GO:0050892) |
| 0.1 | 2.4 | GO:0060259 | regulation of feeding behavior(GO:0060259) |
| 0.1 | 2.5 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.1 | 2.5 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.1 | 0.5 | GO:0030222 | response to nematode(GO:0009624) eosinophil differentiation(GO:0030222) |
| 0.1 | 4.2 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.1 | 2.0 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.1 | 0.2 | GO:0032298 | positive regulation of DNA-dependent DNA replication initiation(GO:0032298) |
| 0.1 | 1.4 | GO:0007635 | chemosensory behavior(GO:0007635) |
| 0.1 | 0.6 | GO:1902969 | mitotic DNA replication(GO:1902969) |
| 0.1 | 1.4 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.1 | 6.1 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.1 | 9.4 | GO:0006865 | amino acid transport(GO:0006865) |
| 0.1 | 1.7 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.1 | 69.4 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.1 | 0.6 | GO:0050907 | detection of chemical stimulus involved in sensory perception(GO:0050907) |
| 0.1 | 8.7 | GO:0007606 | sensory perception of chemical stimulus(GO:0007606) |
| 0.0 | 0.3 | GO:0032460 | negative regulation of protein oligomerization(GO:0032460) |
| 0.0 | 4.2 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 2.1 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.0 | 1.3 | GO:0048488 | synaptic vesicle endocytosis(GO:0048488) |
| 0.0 | 3.3 | GO:0043433 | negative regulation of sequence-specific DNA binding transcription factor activity(GO:0043433) |
| 0.0 | 2.1 | GO:0045582 | positive regulation of T cell differentiation(GO:0045582) |
| 0.0 | 0.8 | GO:0030857 | negative regulation of epithelial cell differentiation(GO:0030857) |
| 0.0 | 2.1 | GO:0043488 | regulation of mRNA stability(GO:0043488) |
| 0.0 | 3.2 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.0 | 0.8 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.0 | 1.7 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.3 | GO:2000738 | positive regulation of stem cell differentiation(GO:2000738) |
| 0.0 | 0.4 | GO:0007032 | endosome organization(GO:0007032) |
| 0.0 | 1.5 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 26.4 | 79.1 | GO:0045257 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) fumarate reductase complex(GO:0045283) |
| 15.9 | 95.2 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 15.0 | 15.0 | GO:0045281 | respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) |
| 10.4 | 82.9 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 6.7 | 74.2 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) |
| 6.4 | 32.2 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 5.8 | 92.2 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 5.7 | 17.1 | GO:0044214 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 5.2 | 15.5 | GO:0090534 | longitudinal sarcoplasmic reticulum(GO:0014801) calcium ion-transporting ATPase complex(GO:0090534) |
| 5.0 | 64.4 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 4.9 | 29.4 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 4.5 | 40.2 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 4.3 | 13.0 | GO:0097444 | spine apparatus(GO:0097444) |
| 3.9 | 31.3 | GO:0005753 | mitochondrial proton-transporting ATP synthase complex(GO:0005753) |
| 3.9 | 11.6 | GO:0005962 | mitochondrial isocitrate dehydrogenase complex (NAD+)(GO:0005962) isocitrate dehydrogenase complex (NAD+)(GO:0045242) |
| 3.3 | 167.5 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 3.1 | 9.4 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 3.1 | 15.4 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 3.0 | 15.1 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 3.0 | 12.0 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 2.9 | 71.7 | GO:0031528 | microvillus membrane(GO:0031528) |
| 2.7 | 10.9 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 2.6 | 33.8 | GO:0031983 | vesicle lumen(GO:0031983) |
| 2.5 | 12.4 | GO:0070069 | cytochrome complex(GO:0070069) |
| 2.4 | 30.7 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 2.3 | 18.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 2.3 | 24.8 | GO:0031143 | pseudopodium(GO:0031143) |
| 2.2 | 24.3 | GO:0070469 | respiratory chain(GO:0070469) |
| 2.1 | 43.8 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 2.1 | 12.4 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 1.8 | 5.4 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 1.8 | 19.5 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 1.7 | 8.6 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 1.7 | 49.6 | GO:0031304 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 1.6 | 19.4 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 1.4 | 18.3 | GO:0032426 | stereocilium tip(GO:0032426) |
| 1.4 | 17.9 | GO:0061702 | inflammasome complex(GO:0061702) |
| 1.2 | 29.8 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 1.2 | 14.3 | GO:0045239 | tricarboxylic acid cycle enzyme complex(GO:0045239) |
| 1.1 | 12.5 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 1.1 | 1.1 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 1.1 | 54.8 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 1.1 | 9.5 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 1.0 | 4.0 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.9 | 9.4 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.9 | 0.9 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.9 | 4.4 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.9 | 18.3 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.8 | 125.4 | GO:0030018 | Z disc(GO:0030018) |
| 0.8 | 191.2 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.8 | 2.4 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.8 | 8.7 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.8 | 18.8 | GO:0045120 | pronucleus(GO:0045120) |
| 0.8 | 21.1 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.8 | 3.8 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.7 | 78.4 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.7 | 157.2 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.7 | 28.2 | GO:0016235 | aggresome(GO:0016235) |
| 0.7 | 5.3 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.6 | 28.9 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.5 | 11.5 | GO:0098644 | complex of collagen trimers(GO:0098644) |
| 0.5 | 7.1 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.5 | 5.4 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.5 | 19.3 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.5 | 31.6 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.5 | 49.4 | GO:0005604 | basement membrane(GO:0005604) |
| 0.5 | 2.0 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.5 | 101.3 | GO:0005770 | late endosome(GO:0005770) |
| 0.5 | 17.4 | GO:0016459 | myosin complex(GO:0016459) |
| 0.5 | 6.2 | GO:0099634 | postsynaptic specialization membrane(GO:0099634) |
| 0.5 | 4.7 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.5 | 4.2 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.5 | 5.7 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.5 | 25.5 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.4 | 8.6 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.4 | 3.4 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.4 | 1.2 | GO:0033557 | Slx1-Slx4 complex(GO:0033557) |
| 0.4 | 6.4 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.4 | 18.3 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.4 | 1.5 | GO:0043541 | UDP-N-acetylglucosamine transferase complex(GO:0043541) |
| 0.4 | 1.1 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
| 0.3 | 3.8 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.3 | 39.1 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.3 | 9.7 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.3 | 2.5 | GO:0031211 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.3 | 12.9 | GO:0030017 | sarcomere(GO:0030017) |
| 0.3 | 1.7 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.3 | 1.4 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.3 | 1.3 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.3 | 14.1 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.3 | 13.3 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.2 | 1.6 | GO:0045179 | apical cortex(GO:0045179) |
| 0.2 | 14.5 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.2 | 2.2 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.2 | 38.2 | GO:0031966 | mitochondrial membrane(GO:0031966) |
| 0.2 | 7.8 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.2 | 3.3 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.2 | 1.2 | GO:0031415 | NatA complex(GO:0031415) |
| 0.2 | 8.1 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.2 | 7.1 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.2 | 4.7 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.2 | 64.3 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.2 | 29.8 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.2 | 0.3 | GO:0097451 | glial limiting end-foot(GO:0097451) |
| 0.1 | 4.6 | GO:0005903 | brush border(GO:0005903) |
| 0.1 | 3.4 | GO:0005902 | microvillus(GO:0005902) |
| 0.1 | 2.9 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.1 | 1.8 | GO:0044292 | dendrite terminus(GO:0044292) |
| 0.1 | 1.7 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.1 | 4.2 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.1 | 2.0 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 1.4 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.1 | 3.6 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.1 | 22.2 | GO:0005911 | cell-cell junction(GO:0005911) |
| 0.1 | 1.7 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.1 | 16.0 | GO:0045177 | apical part of cell(GO:0045177) |
| 0.1 | 1.3 | GO:0031985 | Golgi cisterna(GO:0031985) |
| 0.1 | 1.7 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.1 | 39.4 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.1 | 59.5 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.1 | 9.8 | GO:0061695 | transferase complex, transferring phosphorus-containing groups(GO:0061695) |
| 0.0 | 0.3 | GO:0032590 | dendrite membrane(GO:0032590) |
| 0.0 | 79.5 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.0 | 1.1 | GO:0045095 | keratin filament(GO:0045095) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 27.7 | 83.1 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 26.4 | 79.1 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) |
| 15.8 | 63.3 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 15.0 | 15.0 | GO:0000104 | succinate dehydrogenase activity(GO:0000104) |
| 12.6 | 50.3 | GO:0070546 | L-phenylalanine aminotransferase activity(GO:0070546) |
| 11.9 | 47.7 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 11.1 | 33.3 | GO:0004807 | triose-phosphate isomerase activity(GO:0004807) |
| 9.6 | 47.9 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 9.2 | 46.2 | GO:0051373 | FATZ binding(GO:0051373) |
| 8.9 | 26.6 | GO:0052743 | phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase activity(GO:0034485) inositol tetrakisphosphate phosphatase activity(GO:0052743) |
| 8.7 | 43.4 | GO:0043532 | angiostatin binding(GO:0043532) |
| 8.1 | 32.2 | GO:0004903 | growth hormone receptor activity(GO:0004903) |
| 7.2 | 21.5 | GO:0036468 | aromatic-L-amino-acid decarboxylase activity(GO:0004058) L-dopa decarboxylase activity(GO:0036468) |
| 6.5 | 45.3 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 5.8 | 103.8 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 5.8 | 46.1 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 5.7 | 45.7 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 5.6 | 22.5 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 5.1 | 15.2 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 5.0 | 49.9 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 4.9 | 14.8 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 4.9 | 29.4 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 4.9 | 14.7 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 4.9 | 14.6 | GO:0008520 | L-ascorbate:sodium symporter activity(GO:0008520) L-ascorbic acid transporter activity(GO:0015229) sodium-dependent L-ascorbate transmembrane transporter activity(GO:0070890) |
| 4.8 | 28.8 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 4.3 | 43.5 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 4.2 | 12.5 | GO:0047025 | 3-oxoacyl-[acyl-carrier-protein] reductase (NADH) activity(GO:0047025) |
| 4.1 | 20.7 | GO:0008453 | alanine-glyoxylate transaminase activity(GO:0008453) |
| 4.0 | 12.1 | GO:0004618 | phosphoglycerate kinase activity(GO:0004618) |
| 4.0 | 12.1 | GO:0046911 | metal chelating activity(GO:0046911) |
| 4.0 | 11.9 | GO:0016509 | long-chain-enoyl-CoA hydratase activity(GO:0016508) long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 3.9 | 31.0 | GO:0008430 | selenium binding(GO:0008430) |
| 3.8 | 30.3 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 3.7 | 15.0 | GO:0003973 | (S)-2-hydroxy-acid oxidase activity(GO:0003973) |
| 3.7 | 25.7 | GO:0052832 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) |
| 3.5 | 10.4 | GO:0008948 | oxaloacetate decarboxylase activity(GO:0008948) |
| 3.5 | 104.0 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 3.3 | 16.3 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 3.2 | 19.5 | GO:0031419 | cobalamin binding(GO:0031419) |
| 3.1 | 9.2 | GO:0008489 | UDP-galactose:glucosylceramide beta-1,4-galactosyltransferase activity(GO:0008489) |
| 3.0 | 92.2 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 2.9 | 11.6 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 2.8 | 75.2 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 2.8 | 8.3 | GO:0036478 | tyrosine 3-monooxygenase activator activity(GO:0036470) L-dopa decarboxylase activator activity(GO:0036478) |
| 2.7 | 19.2 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 2.7 | 10.9 | GO:0030172 | troponin C binding(GO:0030172) |
| 2.7 | 42.6 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 2.6 | 7.7 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 2.5 | 49.7 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 2.5 | 9.9 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 2.4 | 12.1 | GO:0031798 | type 1 metabotropic glutamate receptor binding(GO:0031798) |
| 2.4 | 7.1 | GO:0031177 | phosphopantetheine binding(GO:0031177) |
| 2.2 | 2.2 | GO:1990763 | arrestin family protein binding(GO:1990763) |
| 2.1 | 18.7 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 2.0 | 6.1 | GO:0004359 | glutaminase activity(GO:0004359) |
| 2.0 | 17.9 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 2.0 | 8.0 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 2.0 | 7.8 | GO:0004487 | methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 1.9 | 13.6 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 1.9 | 15.4 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 1.9 | 13.1 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 1.9 | 9.4 | GO:0032557 | pyrimidine ribonucleotide binding(GO:0032557) |
| 1.9 | 5.6 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 1.8 | 48.5 | GO:0019825 | oxygen binding(GO:0019825) |
| 1.8 | 5.4 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 1.8 | 25.0 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 1.8 | 19.6 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 1.7 | 34.5 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 1.7 | 6.8 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 1.7 | 5.0 | GO:0004140 | dephospho-CoA kinase activity(GO:0004140) |
| 1.7 | 5.0 | GO:0004307 | ethanolaminephosphotransferase activity(GO:0004307) |
| 1.6 | 14.6 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 1.6 | 9.3 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 1.5 | 19.2 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 1.5 | 33.8 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 1.4 | 17.2 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 1.4 | 9.9 | GO:0004064 | arylesterase activity(GO:0004064) |
| 1.4 | 7.0 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 1.4 | 6.8 | GO:0086062 | voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 1.4 | 35.5 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 1.4 | 36.5 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 1.3 | 12.1 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 1.3 | 18.6 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 1.3 | 19.2 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 1.3 | 3.8 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 1.2 | 6.1 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 1.2 | 4.9 | GO:0045174 | oxidoreductase activity, acting on a sulfur group of donors, quinone or similar compound as acceptor(GO:0016672) oxidoreductase activity, acting on phosphorus or arsenic in donors(GO:0030613) oxidoreductase activity, acting on phosphorus or arsenic in donors, disulfide as acceptor(GO:0030614) glutathione dehydrogenase (ascorbate) activity(GO:0045174) methylarsonate reductase activity(GO:0050610) |
| 1.2 | 9.5 | GO:0034604 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 1.1 | 8.0 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 1.1 | 5.7 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 1.1 | 30.9 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 1.1 | 5.4 | GO:0090554 | phosphatidylcholine-translocating ATPase activity(GO:0090554) phosphatidylethanolamine-translocating ATPase activity(GO:0090555) |
| 1.1 | 8.6 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 1.1 | 10.8 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 1.0 | 10.5 | GO:0019808 | polyamine binding(GO:0019808) |
| 1.0 | 18.8 | GO:0031005 | filamin binding(GO:0031005) |
| 1.0 | 9.4 | GO:0032052 | bile acid binding(GO:0032052) |
| 1.0 | 6.2 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 1.0 | 7.1 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.9 | 3.7 | GO:0070568 | guanylyltransferase activity(GO:0070568) |
| 0.9 | 22.4 | GO:0008308 | voltage-gated anion channel activity(GO:0008308) |
| 0.9 | 6.4 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.9 | 10.9 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.9 | 16.9 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.9 | 16.8 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.9 | 26.2 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.9 | 7.8 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.9 | 15.5 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.8 | 4.2 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.8 | 2.5 | GO:0047057 | oxidoreductase activity, acting on the CH-OH group of donors, disulfide as acceptor(GO:0016900) vitamin-K-epoxide reductase (warfarin-sensitive) activity(GO:0047057) |
| 0.8 | 52.7 | GO:0038024 | cargo receptor activity(GO:0038024) |
| 0.8 | 11.0 | GO:0010181 | FMN binding(GO:0010181) |
| 0.8 | 6.2 | GO:0043495 | protein anchor(GO:0043495) |
| 0.7 | 5.7 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.7 | 25.7 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.7 | 2.0 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.7 | 16.3 | GO:0009055 | electron carrier activity(GO:0009055) |
| 0.7 | 22.3 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.7 | 5.3 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) vascular endothelial growth factor binding(GO:0038085) |
| 0.7 | 2.6 | GO:0055100 | adiponectin binding(GO:0055100) |
| 0.6 | 4.5 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.6 | 3.7 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.6 | 4.8 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.6 | 2.4 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.6 | 23.2 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.6 | 8.1 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.6 | 3.4 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.6 | 1.7 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) |
| 0.5 | 2.2 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) |
| 0.5 | 10.6 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.5 | 11.5 | GO:0015114 | phosphate ion transmembrane transporter activity(GO:0015114) |
| 0.5 | 4.1 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.5 | 5.6 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.5 | 2.5 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.5 | 2.5 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.5 | 7.3 | GO:0050308 | sugar-phosphatase activity(GO:0050308) |
| 0.5 | 20.5 | GO:0016651 | oxidoreductase activity, acting on NAD(P)H(GO:0016651) |
| 0.5 | 3.4 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.5 | 3.8 | GO:0046790 | virion binding(GO:0046790) |
| 0.5 | 3.7 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.5 | 6.5 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.5 | 26.1 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.5 | 6.4 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.4 | 4.9 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.4 | 1.3 | GO:0042903 | tubulin deacetylase activity(GO:0042903) |
| 0.4 | 12.1 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.4 | 19.8 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.4 | 2.9 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.4 | 3.2 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.4 | 1.6 | GO:0004082 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.4 | 17.9 | GO:0030170 | pyridoxal phosphate binding(GO:0030170) |
| 0.4 | 15.4 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.4 | 3.7 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.4 | 24.4 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.4 | 1.1 | GO:0004040 | amidase activity(GO:0004040) glutaminyl-tRNA synthase (glutamine-hydrolyzing) activity(GO:0050567) |
| 0.4 | 1.1 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.4 | 8.2 | GO:0046965 | retinoid X receptor binding(GO:0046965) |
| 0.3 | 2.4 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.3 | 3.0 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.3 | 10.9 | GO:0016627 | oxidoreductase activity, acting on the CH-CH group of donors(GO:0016627) |
| 0.3 | 2.0 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.3 | 2.9 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.3 | 16.3 | GO:0001221 | transcription cofactor binding(GO:0001221) |
| 0.3 | 1.6 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.3 | 1.4 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.3 | 7.2 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.3 | 4.8 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.3 | 0.8 | GO:0031783 | corticotropin hormone receptor binding(GO:0031780) type 5 melanocortin receptor binding(GO:0031783) |
| 0.2 | 3.6 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.2 | 4.3 | GO:0033549 | MAP kinase phosphatase activity(GO:0033549) |
| 0.2 | 1.6 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.2 | 2.2 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.2 | 7.1 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.2 | 2.3 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.2 | 3.8 | GO:0005521 | lamin binding(GO:0005521) |
| 0.2 | 4.9 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.2 | 0.9 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.2 | 6.4 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.2 | 15.9 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.2 | 25.5 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.2 | 2.9 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.2 | 2.7 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.2 | 6.5 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.2 | 2.2 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.2 | 6.6 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.1 | 12.5 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.1 | 1.0 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.1 | 4.5 | GO:0034062 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.1 | 7.8 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.1 | 3.6 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.1 | 4.7 | GO:0043531 | ADP binding(GO:0043531) |
| 0.1 | 13.2 | GO:0008201 | heparin binding(GO:0008201) |
| 0.1 | 0.7 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.1 | 2.6 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.1 | 1.2 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.1 | 8.0 | GO:0005496 | steroid binding(GO:0005496) |
| 0.1 | 0.9 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.1 | 1.8 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.1 | 3.5 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.1 | 2.0 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 2.2 | GO:0008198 | ferrous iron binding(GO:0008198) |
| 0.1 | 9.5 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.1 | 37.6 | GO:0005549 | odorant binding(GO:0005549) |
| 0.1 | 6.1 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.1 | 3.6 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.1 | 11.9 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.1 | 1.6 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 6.8 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.1 | 2.9 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.1 | 5.3 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.1 | 0.9 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.0 | 0.2 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.6 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.0 | 31.8 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.3 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.0 | 0.4 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 1.6 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 2.5 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 3.4 | GO:0016853 | isomerase activity(GO:0016853) |
| 0.0 | 2.5 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 4.4 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.0 | 2.8 | GO:0008514 | organic anion transmembrane transporter activity(GO:0008514) |
| 0.0 | 0.1 | GO:0016933 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.0 | 0.4 | GO:0001190 | transcriptional activator activity, RNA polymerase II transcription factor binding(GO:0001190) transcriptional repressor activity, RNA polymerase II activating transcription factor binding(GO:0098811) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 24.0 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 2.0 | 107.7 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.9 | 56.5 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.8 | 2.4 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.8 | 54.8 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.7 | 9.9 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.6 | 18.1 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.5 | 26.2 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.5 | 40.7 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.5 | 21.0 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.4 | 14.6 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.4 | 17.2 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.4 | 19.5 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.4 | 8.6 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.4 | 12.1 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.4 | 10.5 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.4 | 12.9 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.4 | 14.6 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.3 | 2.9 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.3 | 71.4 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.2 | 3.4 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.2 | 5.7 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.2 | 5.6 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.2 | 10.3 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.2 | 5.2 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.2 | 3.8 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.2 | 2.4 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.2 | 8.3 | PID FGF PATHWAY | FGF signaling pathway |
| 0.2 | 2.0 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.1 | 4.2 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.1 | 0.9 | PID S1P S1P3 PATHWAY | S1P3 pathway |
| 0.1 | 6.5 | PID E2F PATHWAY | E2F transcription factor network |
| 0.1 | 5.9 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.1 | 2.6 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.1 | 1.7 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.1 | 1.5 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.1 | 5.0 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.1 | 1.3 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 1.3 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.1 | 2.7 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.1 | 0.9 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.9 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.9 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 2.1 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.5 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 1.2 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 1.9 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 10.4 | 176.1 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 7.3 | 471.4 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 6.6 | 106.2 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 4.9 | 48.9 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 4.2 | 168.3 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 3.7 | 44.8 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 3.2 | 51.1 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 3.1 | 69.1 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 3.0 | 56.7 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 2.8 | 25.4 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 2.7 | 21.6 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 2.6 | 33.9 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 2.6 | 46.2 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 2.4 | 66.1 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 2.2 | 44.9 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 2.0 | 24.0 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 1.9 | 56.2 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 1.9 | 26.5 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 1.8 | 20.2 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 1.8 | 60.3 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 1.8 | 20.0 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 1.7 | 13.6 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 1.7 | 18.2 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 1.6 | 19.5 | REACTOME LIPOPROTEIN METABOLISM | Genes involved in Lipoprotein metabolism |
| 1.6 | 33.6 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 1.5 | 19.6 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 1.5 | 57.9 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 1.5 | 19.2 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 1.3 | 17.2 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 1.1 | 15.4 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 1.1 | 22.7 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 1.1 | 13.9 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 1.1 | 41.2 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 1.0 | 12.2 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.9 | 8.1 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.8 | 32.8 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.8 | 11.7 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.8 | 27.8 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.8 | 16.3 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.7 | 13.4 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.7 | 11.5 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.7 | 6.9 | REACTOME N GLYCAN TRIMMING IN THE ER AND CALNEXIN CALRETICULIN CYCLE | Genes involved in N-glycan trimming in the ER and Calnexin/Calreticulin cycle |
| 0.7 | 9.5 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.7 | 0.7 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.6 | 20.6 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.6 | 15.5 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.5 | 18.4 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.5 | 9.3 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.4 | 8.1 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.4 | 5.0 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.4 | 6.1 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.4 | 3.8 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.4 | 5.7 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.4 | 13.0 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.4 | 17.3 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.3 | 7.1 | REACTOME GLUCOSE METABOLISM | Genes involved in Glucose metabolism |
| 0.3 | 5.3 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.3 | 2.9 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.3 | 3.3 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.3 | 4.0 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.3 | 3.7 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.3 | 8.3 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.2 | 3.7 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.2 | 23.8 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.2 | 12.5 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.2 | 2.9 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.2 | 22.9 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.2 | 2.5 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.2 | 10.2 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.2 | 7.9 | REACTOME ASPARAGINE N LINKED GLYCOSYLATION | Genes involved in Asparagine N-linked glycosylation |
| 0.1 | 9.3 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.1 | 1.0 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.1 | 2.9 | REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.1 | 7.4 | REACTOME RNA POL I RNA POL III AND MITOCHONDRIAL TRANSCRIPTION | Genes involved in RNA Polymerase I, RNA Polymerase III, and Mitochondrial Transcription |
| 0.1 | 2.6 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 1.4 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.9 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.1 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.0 | 0.5 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.9 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |