Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
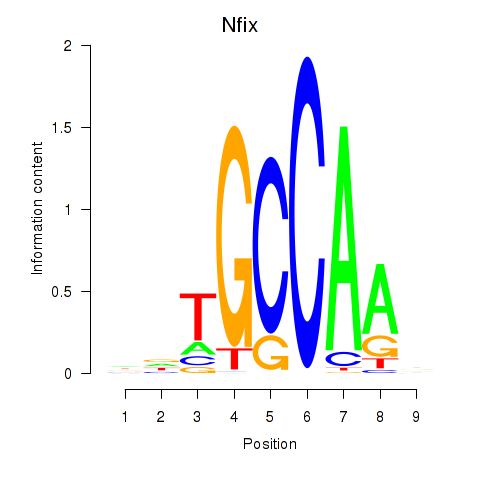
Results for Nfix
Z-value: 0.80
Transcription factors associated with Nfix
| Gene Symbol | Gene ID | Gene Info |
|---|
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nf1x | rn6_v1_chr19_-_25914689_25914696 | 0.35 | 9.0e-11 | Click! |
Activity profile of Nfix motif
Sorted Z-values of Nfix motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_+_53740841 | 53.70 |
ENSRNOT00000004295
|
Myh2
|
myosin heavy chain 2 |
| chr14_-_19191863 | 37.92 |
ENSRNOT00000003921
|
Alb
|
albumin |
| chr8_+_50537009 | 28.10 |
ENSRNOT00000080658
|
Apoa4
|
apolipoprotein A4 |
| chr13_+_83073544 | 27.99 |
ENSRNOT00000066119
ENSRNOT00000079796 ENSRNOT00000077070 |
Dpt
|
dermatopontin |
| chr13_+_83073866 | 27.53 |
ENSRNOT00000075996
|
Dpt
|
dermatopontin |
| chr17_-_389967 | 26.43 |
ENSRNOT00000023865
|
Fbp2
|
fructose-bisphosphatase 2 |
| chr10_+_53778662 | 26.21 |
ENSRNOT00000045718
|
Myh2
|
myosin heavy chain 2 |
| chr2_-_188596222 | 24.27 |
ENSRNOT00000027920
|
Efna1
|
ephrin A1 |
| chr2_-_233743866 | 23.00 |
ENSRNOT00000087062
|
Enpep
|
glutamyl aminopeptidase |
| chr7_+_38819771 | 22.37 |
ENSRNOT00000006109
|
Lum
|
lumican |
| chr15_-_41595345 | 19.14 |
ENSRNOT00000019639
|
Sgcg
|
sarcoglycan, gamma |
| chr13_-_90602365 | 18.52 |
ENSRNOT00000009344
|
Casq1
|
calsequestrin 1 |
| chr14_+_22806132 | 18.36 |
ENSRNOT00000002728
|
Ugt2b10
|
UDP glucuronosyltransferase 2 family, polypeptide B10 |
| chr10_-_88036040 | 18.05 |
ENSRNOT00000018851
|
Krt13
|
keratin 13 |
| chr13_-_82005741 | 17.87 |
ENSRNOT00000076404
|
Mettl11b
|
methyltransferase like 11B |
| chr13_-_82006005 | 15.85 |
ENSRNOT00000039581
|
Mettl11b
|
methyltransferase like 11B |
| chr1_-_162385575 | 15.46 |
ENSRNOT00000016540
|
Thrsp
|
thyroid hormone responsive |
| chr20_+_40769586 | 15.41 |
ENSRNOT00000001079
|
Fabp7
|
fatty acid binding protein 7 |
| chr7_-_143523457 | 14.89 |
ENSRNOT00000012943
ENSRNOT00000082264 |
Krt4
|
keratin 4 |
| chr1_+_282568287 | 13.68 |
ENSRNOT00000015997
|
Ces2i
|
carboxylesterase 2I |
| chr4_+_100218661 | 12.97 |
ENSRNOT00000079415
|
Tmem150a
|
transmembrane protein 150A |
| chr2_-_45518502 | 12.80 |
ENSRNOT00000014627
|
Hspb3
|
heat shock protein family B (small) member 3 |
| chr2_+_188528979 | 12.67 |
ENSRNOT00000087934
|
Thbs3
|
thrombospondin 3 |
| chr14_+_7113544 | 11.66 |
ENSRNOT00000038188
|
Hsd17b13
|
hydroxysteroid (17-beta) dehydrogenase 13 |
| chr1_+_41192824 | 11.49 |
ENSRNOT00000082133
|
Esr1
|
estrogen receptor 1 |
| chrX_+_134940615 | 11.20 |
ENSRNOT00000005604
|
Xpnpep2
|
X-prolyl aminopeptidase 2 |
| chr3_+_58965552 | 11.09 |
ENSRNOT00000002068
|
Map3k20
|
mitogen-activated protein kinase kinase kinase 20 |
| chr8_-_84522588 | 10.98 |
ENSRNOT00000076213
|
Mlip
|
muscular LMNA-interacting protein |
| chr7_-_30105132 | 10.80 |
ENSRNOT00000091227
|
Nr1h4
|
nuclear receptor subfamily 1, group H, member 4 |
| chr16_+_72401887 | 10.71 |
ENSRNOT00000074449
|
LOC100910163
|
uncharacterized LOC100910163 |
| chr17_-_15511972 | 10.33 |
ENSRNOT00000074041
|
Aspnl1
|
asporin-like 1 |
| chr4_+_7076759 | 10.06 |
ENSRNOT00000066598
|
Smarcd3
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3 |
| chr2_-_184951950 | 10.04 |
ENSRNOT00000093486
|
Fam160a1
|
family with sequence similarity 160, member A1 |
| chr16_+_72388880 | 9.91 |
ENSRNOT00000072459
|
LOC684871
|
similar to Protein C8orf4 (Thyroid cancer protein 1) (TC-1) |
| chr16_+_6962722 | 9.65 |
ENSRNOT00000023330
|
Mustn1
|
musculoskeletal, embryonic nuclear protein 1 |
| chr9_-_52238564 | 9.42 |
ENSRNOT00000005073
|
Col5a2
|
collagen type V alpha 2 chain |
| chrX_+_53053609 | 9.26 |
ENSRNOT00000058357
|
Dmd
|
dystrophin |
| chr1_+_144239020 | 8.62 |
ENSRNOT00000032106
|
Adamtsl3
|
ADAMTS-like 3 |
| chr1_+_41323194 | 8.53 |
ENSRNOT00000026350
|
Esr1
|
estrogen receptor 1 |
| chr16_+_2537248 | 8.29 |
ENSRNOT00000017995
|
Asb14
|
ankyrin repeat and SOCS box-containing 14 |
| chr5_-_102786331 | 8.24 |
ENSRNOT00000086635
|
Bnc2
|
basonuclin 2 |
| chrX_+_77278240 | 8.00 |
ENSRNOT00000085653
|
Pgk1
|
phosphoglycerate kinase 1 |
| chr12_-_6740714 | 7.55 |
ENSRNOT00000001205
|
Medag
|
mesenteric estrogen-dependent adipogenesis |
| chr1_+_201620642 | 7.43 |
ENSRNOT00000093674
|
Dmbt1
|
deleted in malignant brain tumors 1 |
| chr2_-_80667481 | 6.99 |
ENSRNOT00000016784
|
Trio
|
trio Rho guanine nucleotide exchange factor |
| chr5_-_93244202 | 6.90 |
ENSRNOT00000075474
|
Ptprd
|
protein tyrosine phosphatase, receptor type, D |
| chr8_-_107952530 | 6.35 |
ENSRNOT00000052043
|
Cldn18
|
claudin 18 |
| chr13_-_50549981 | 6.26 |
ENSRNOT00000003918
ENSRNOT00000080486 |
Golt1a
|
golgi transport 1A |
| chr2_+_222792999 | 6.14 |
ENSRNOT00000036228
|
AABR07013086.1
|
|
| chrX_+_124631881 | 5.87 |
ENSRNOT00000009329
|
Atp1b4
|
ATPase Na+/K+ transporting family member beta 4 |
| chr1_-_101596822 | 5.85 |
ENSRNOT00000028490
|
Fgf21
|
fibroblast growth factor 21 |
| chrX_-_72078551 | 5.82 |
ENSRNOT00000076978
|
Rps4x
|
ribosomal protein S4, X-linked |
| chr2_-_14701903 | 5.46 |
ENSRNOT00000051895
|
Cox7c
|
cytochrome c oxidase subunit 7C |
| chr6_-_25211494 | 5.39 |
ENSRNOT00000009634
|
Xdh
|
xanthine dehydrogenase |
| chr4_-_163214678 | 5.13 |
ENSRNOT00000091602
|
Clec1a
|
C-type lectin domain family 1, member A |
| chr16_+_26859397 | 5.08 |
ENSRNOT00000044171
|
Msmo1
|
methylsterol monooxygenase 1 |
| chr11_-_71284939 | 5.03 |
ENSRNOT00000002421
|
AABR07034428.1
|
|
| chrX_-_75224268 | 4.59 |
ENSRNOT00000089809
|
Abcb7
|
ATP binding cassette subfamily B member 7 |
| chr2_-_193184942 | 4.41 |
ENSRNOT00000032698
|
LOC686143
|
similar to keratinocytes proline-rich protein |
| chr2_+_60920257 | 4.14 |
ENSRNOT00000025170
|
C1qtnf3
|
C1q and tumor necrosis factor related protein 3 |
| chr10_+_104523996 | 4.10 |
ENSRNOT00000065339
ENSRNOT00000086747 |
Itgb4
|
integrin subunit beta 4 |
| chr19_-_12942943 | 4.01 |
ENSRNOT00000064105
ENSRNOT00000090886 |
Large1
|
LARGE xylosyl- and glucuronyltransferase 1 |
| chr1_-_22661377 | 3.79 |
ENSRNOT00000021896
|
Vnn3
|
vanin 3 |
| chr6_-_109205004 | 3.78 |
ENSRNOT00000010512
|
Tmed10
|
transmembrane p24 trafficking protein 10 |
| chr11_+_43465370 | 3.74 |
ENSRNOT00000039939
|
Olr1547
|
olfactory receptor 1547 |
| chr8_-_93390305 | 3.71 |
ENSRNOT00000056930
|
Ibtk
|
inhibitor of Bruton tyrosine kinase |
| chr4_-_71893306 | 3.54 |
ENSRNOT00000077710
|
Olr803
|
olfactory receptor 803 |
| chr2_+_193429176 | 3.28 |
ENSRNOT00000011762
|
Crnn
|
cornulin |
| chr2_-_61692487 | 3.16 |
ENSRNOT00000078544
|
LOC499541
|
LRRGT00045 |
| chr10_-_98018014 | 3.16 |
ENSRNOT00000005367
|
Fam20a
|
FAM20A, golgi associated secretory pathway pseudokinase |
| chr11_-_80826505 | 3.10 |
ENSRNOT00000032383
|
Rtp1
|
receptor (chemosensory) transporter protein 1 |
| chr17_+_47397558 | 3.06 |
ENSRNOT00000085923
|
Epdr1
|
ependymin related 1 |
| chr2_+_233602732 | 3.05 |
ENSRNOT00000044232
|
Pitx2
|
paired-like homeodomain 2 |
| chr1_-_168177139 | 2.87 |
ENSRNOT00000075135
|
Olr72
|
olfactory receptor 72 |
| chr17_-_21705773 | 2.84 |
ENSRNOT00000078010
|
Gcnt2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme |
| chr19_-_38796 | 2.75 |
ENSRNOT00000077592
|
AABR07042565.1
|
|
| chr3_+_75712483 | 2.59 |
ENSRNOT00000049264
|
Olr575
|
olfactory receptor 575 |
| chr11_-_29710849 | 2.56 |
ENSRNOT00000029345
|
Krtap11-1
|
keratin associated protein 11-1 |
| chr5_-_2803855 | 2.54 |
ENSRNOT00000009490
|
LOC297756
|
ribosomal protein S8-like |
| chr6_-_106971250 | 2.54 |
ENSRNOT00000010926
|
Dpf3
|
double PHD fingers 3 |
| chr3_-_103217528 | 2.52 |
ENSRNOT00000074456
|
Olr784
|
olfactory receptor 784 |
| chr5_-_102743417 | 2.51 |
ENSRNOT00000067389
|
Bnc2
|
basonuclin 2 |
| chr11_+_43476319 | 2.46 |
ENSRNOT00000044928
|
Olr1548
|
olfactory receptor 1548 |
| chr1_-_103323476 | 2.29 |
ENSRNOT00000019051
|
Mrgprx3
|
MAS related GPR family member X3 |
| chr17_-_51912496 | 2.28 |
ENSRNOT00000019272
|
Inhba
|
inhibin beta A subunit |
| chr6_-_115769793 | 2.20 |
ENSRNOT00000039910
|
AABR07065217.2
|
|
| chr1_-_82279145 | 2.17 |
ENSRNOT00000057433
|
Cxcl17
|
C-X-C motif chemokine ligand 17 |
| chr10_-_87335823 | 2.13 |
ENSRNOT00000079297
|
Krt12
|
keratin 12 |
| chr6_+_147315328 | 2.13 |
ENSRNOT00000056654
|
Macc1
|
MACC1, MET transcriptional regulator |
| chr1_-_215033460 | 2.10 |
ENSRNOT00000044565
|
Dusp8
|
dual specificity phosphatase 8 |
| chr12_-_35318685 | 2.08 |
ENSRNOT00000074199
|
AABR07036167.1
|
|
| chr1_+_170147300 | 1.99 |
ENSRNOT00000048075
|
Olr200
|
olfactory receptor 200 |
| chr5_-_59198650 | 1.99 |
ENSRNOT00000020958
|
Olr833
|
olfactory receptor 833 |
| chr10_-_94488180 | 1.98 |
ENSRNOT00000015818
|
Gh1
|
growth hormone 1 |
| chr1_+_168244888 | 1.94 |
ENSRNOT00000021102
|
Olr79
|
olfactory receptor 79 |
| chr13_+_91711995 | 1.94 |
ENSRNOT00000040596
|
Olr1582
|
olfactory receptor 1582 |
| chr9_+_81559605 | 1.92 |
ENSRNOT00000089698
|
Gpbar1
|
G protein-coupled bile acid receptor 1 |
| chr4_+_166958466 | 1.92 |
ENSRNOT00000007538
|
Tas2r13
|
taste receptor, type 2, member 13 |
| chr13_-_50497466 | 1.91 |
ENSRNOT00000076072
|
Etnk2
|
ethanolamine kinase 2 |
| chr1_+_263968134 | 1.73 |
ENSRNOT00000018090
|
LOC681458
|
similar to stearoyl-coenzyme A desaturase 3 |
| chr7_-_92996025 | 1.59 |
ENSRNOT00000076327
|
Samd12
|
sterile alpha motif domain containing 12 |
| chr7_+_16394354 | 1.55 |
ENSRNOT00000041157
|
Olr1058
|
olfactory receptor 1058 |
| chr2_-_192381716 | 1.52 |
ENSRNOT00000064950
|
AABR07012291.1
|
|
| chr13_-_62524556 | 1.49 |
ENSRNOT00000033201
|
AABR07021239.1
|
|
| chr1_+_169683311 | 1.48 |
ENSRNOT00000075826
|
Olr162
|
olfactory receptor 162 |
| chr7_+_141642777 | 1.40 |
ENSRNOT00000079811
|
AABR07058884.2
|
|
| chr10_-_78690857 | 1.37 |
ENSRNOT00000089255
|
LOC303448
|
similar to glyceraldehyde-3-phosphate dehydrogenase |
| chr2_+_187322416 | 1.36 |
ENSRNOT00000025183
|
Crabp2
|
cellular retinoic acid binding protein 2 |
| chr10_-_43512287 | 1.34 |
ENSRNOT00000036481
|
Faxdc2
|
fatty acid hydroxylase domain containing 2 |
| chr2_+_192553983 | 1.31 |
ENSRNOT00000085204
|
LOC102550416
|
small proline-rich protein 2I-like |
| chr3_-_74868575 | 1.30 |
ENSRNOT00000051625
|
Olr544
|
olfactory receptor 544 |
| chrX_+_10811478 | 1.28 |
ENSRNOT00000060930
|
LOC307727
|
similar to Probable chromodomain-helicase-DNA-binding protein KIAA1416 |
| chr14_-_45376127 | 1.20 |
ENSRNOT00000059247
|
AABR07015015.2
|
|
| chr14_+_7618022 | 1.09 |
ENSRNOT00000088508
ENSRNOT00000002819 |
Slc10a6
|
solute carrier family 10 member 6 |
| chr7_+_2459141 | 1.01 |
ENSRNOT00000075681
|
Naca
|
nascent polypeptide-associated complex alpha subunit |
| chr7_+_18120715 | 1.01 |
ENSRNOT00000068151
|
Vom1r108
|
vomeronasal 1 receptor 108 |
| chr4_-_144318580 | 1.00 |
ENSRNOT00000007591
|
Ssuh2
|
ssu-2 homolog |
| chr10_+_10875983 | 0.99 |
ENSRNOT00000052265
|
LOC679823
|
similar to 60S ribosomal protein L37a |
| chr14_+_107268128 | 0.96 |
ENSRNOT00000012224
|
Tmem17
|
transmembrane protein 17 |
| chr3_-_134696654 | 0.96 |
ENSRNOT00000006454
|
Flrt3
|
fibronectin leucine rich transmembrane protein 3 |
| chr6_-_21135880 | 0.90 |
ENSRNOT00000051239
|
Rasgrp3
|
RAS guanyl releasing protein 3 |
| chr2_+_192604978 | 0.84 |
ENSRNOT00000084784
|
LOC102550416
|
small proline-rich protein 2I-like |
| chr1_+_264094080 | 0.83 |
ENSRNOT00000047602
ENSRNOT00000017834 |
Scd4
|
stearoyl-coenzyme A desaturase 4 |
| chr19_-_15733412 | 0.80 |
ENSRNOT00000014831
|
Irx6
|
iroquois homeobox 6 |
| chr7_-_122926336 | 0.80 |
ENSRNOT00000000205
|
Chadl
|
chondroadherin-like |
| chr14_+_72889038 | 0.75 |
ENSRNOT00000084261
|
Rab5a
|
RAB5A, member RAS oncogene family |
| chr3_-_77911561 | 0.70 |
ENSRNOT00000084947
|
Olr679
|
olfactory receptor 679 |
| chr3_+_75582317 | 0.70 |
ENSRNOT00000079367
|
Olr569
|
olfactory receptor 569 |
| chr11_+_43340505 | 0.68 |
ENSRNOT00000043299
|
Olr1541
|
olfactory receptor 1541 |
| chrX_+_23416395 | 0.64 |
ENSRNOT00000089821
|
AABR07037545.1
|
|
| chr1_+_168411463 | 0.44 |
ENSRNOT00000072397
|
Olr89
|
olfactory receptor 89 |
| chr1_+_67660337 | 0.39 |
ENSRNOT00000046111
|
LOC102551504
|
vomeronasal type-1 receptor 4-like |
| chr1_-_266428239 | 0.38 |
ENSRNOT00000027160
|
Cyp17a1
|
cytochrome P450, family 17, subfamily a, polypeptide 1 |
| chr14_-_13058172 | 0.36 |
ENSRNOT00000002746
ENSRNOT00000071706 |
Prdm8
|
PR/SET domain 8 |
| chr14_+_22597103 | 0.35 |
ENSRNOT00000048482
|
Ugt2a1
|
UDP glucuronosyltransferase 2 family, polypeptide A1 |
| chr1_-_168082740 | 0.35 |
ENSRNOT00000070789
|
LOC684376
|
similar to olfactory receptor 555 |
| chrX_+_71324365 | 0.31 |
ENSRNOT00000004911
|
Nono
|
non-POU domain containing, octamer-binding |
| chr6_-_77421286 | 0.27 |
ENSRNOT00000011453
|
Nkx2-1
|
NK2 homeobox 1 |
| chr1_+_169074475 | 0.21 |
ENSRNOT00000050248
|
Olr132
|
olfactory receptor 132 |
| chr1_-_168501212 | 0.21 |
ENSRNOT00000014730
|
Olr97
|
olfactory receptor 97 |
| chr1_+_84067938 | 0.20 |
ENSRNOT00000057213
|
Numbl
|
NUMB-like, endocytic adaptor protein |
| chr1_+_80256973 | 0.18 |
ENSRNOT00000024113
|
Ercc1
|
ERCC excision repair 1, endonuclease non-catalytic subunit |
| chr8_+_119214541 | 0.15 |
ENSRNOT00000050117
|
RGD1564138
|
similar to 60S ribosomal protein L29 (P23) |
| chr11_+_85030872 | 0.12 |
ENSRNOT00000074815
|
LOC100911043
|
olfactory receptor 2L3-like |
| chr7_+_18068060 | 0.11 |
ENSRNOT00000065474
|
Vom1r107
|
vomeronasal 1 receptor 107 |
| chr17_+_21600432 | 0.11 |
ENSRNOT00000060340
|
RGD1562963
|
similar to chromosome 6 open reading frame 52 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Nfix
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 11.2 | 33.7 | GO:0006480 | N-terminal protein amino acid methylation(GO:0006480) |
| 9.4 | 28.1 | GO:0034445 | regulation of plasma lipoprotein particle oxidation(GO:0034444) negative regulation of plasma lipoprotein particle oxidation(GO:0034445) multicellular organism lipid catabolic process(GO:0044240) |
| 6.9 | 27.8 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 6.6 | 26.4 | GO:0046351 | disaccharide biosynthetic process(GO:0046351) |
| 6.3 | 37.9 | GO:0044004 | killing by symbiont of host cells(GO:0001907) disruption by symbiont of host cell(GO:0044004) response to platinum ion(GO:0070541) |
| 6.1 | 79.9 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 6.1 | 24.3 | GO:0014028 | notochord formation(GO:0014028) |
| 5.0 | 20.0 | GO:1990375 | baculum development(GO:1990375) |
| 4.5 | 22.5 | GO:0002003 | angiotensin maturation(GO:0002003) |
| 4.5 | 22.4 | GO:0032914 | positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 3.6 | 10.8 | GO:0034971 | histone H3-R17 methylation(GO:0034971) |
| 2.5 | 10.1 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 2.4 | 9.7 | GO:1902730 | positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 2.4 | 9.4 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 2.1 | 6.3 | GO:2001205 | negative regulation of osteoclast development(GO:2001205) |
| 1.6 | 18.1 | GO:0043587 | tongue morphogenesis(GO:0043587) |
| 1.5 | 10.7 | GO:1900019 | regulation of protein kinase C activity(GO:1900019) positive regulation of protein kinase C activity(GO:1900020) |
| 1.5 | 3.1 | GO:0035993 | deltoid tuberosity development(GO:0035993) |
| 1.3 | 12.7 | GO:0060346 | bone trabecula formation(GO:0060346) |
| 1.3 | 3.8 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) |
| 1.2 | 56.3 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 1.2 | 5.8 | GO:1904640 | response to methionine(GO:1904640) |
| 1.1 | 5.4 | GO:0009115 | xanthine catabolic process(GO:0009115) |
| 1.0 | 6.9 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 1.0 | 1.9 | GO:0038183 | bile acid signaling pathway(GO:0038183) |
| 0.8 | 4.1 | GO:0071638 | negative regulation of monocyte chemotactic protein-1 production(GO:0071638) |
| 0.8 | 15.4 | GO:0060134 | prepulse inhibition(GO:0060134) |
| 0.7 | 2.0 | GO:0071469 | cellular response to alkaline pH(GO:0071469) |
| 0.6 | 2.6 | GO:1903964 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.6 | 15.5 | GO:0010866 | regulation of triglyceride biosynthetic process(GO:0010866) |
| 0.6 | 4.1 | GO:0035878 | nail development(GO:0035878) |
| 0.6 | 2.3 | GO:0060279 | positive regulation of ovulation(GO:0060279) |
| 0.6 | 2.8 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.5 | 10.7 | GO:0043586 | tongue development(GO:0043586) |
| 0.5 | 5.5 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.4 | 11.0 | GO:0010614 | negative regulation of cardiac muscle hypertrophy(GO:0010614) |
| 0.4 | 7.4 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.3 | 1.0 | GO:1901227 | negative regulation of transcription from RNA polymerase II promoter involved in heart development(GO:1901227) |
| 0.3 | 11.1 | GO:0007257 | activation of JUN kinase activity(GO:0007257) |
| 0.3 | 13.0 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.3 | 4.0 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.2 | 0.7 | GO:2000286 | receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.2 | 2.1 | GO:0061303 | cornea development in camera-type eye(GO:0061303) |
| 0.2 | 7.0 | GO:0031639 | plasminogen activation(GO:0031639) |
| 0.2 | 10.4 | GO:0046426 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.2 | 3.2 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.2 | 2.2 | GO:0010759 | positive regulation of macrophage chemotaxis(GO:0010759) |
| 0.2 | 12.0 | GO:0046889 | positive regulation of lipid biosynthetic process(GO:0046889) |
| 0.1 | 3.7 | GO:0061099 | negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.1 | 7.5 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.1 | 3.1 | GO:0051205 | protein insertion into membrane(GO:0051205) |
| 0.1 | 1.9 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.1 | 5.1 | GO:0016126 | sterol biosynthetic process(GO:0016126) |
| 0.1 | 3.8 | GO:0006767 | water-soluble vitamin metabolic process(GO:0006767) |
| 0.1 | 1.4 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.1 | 4.6 | GO:0006879 | cellular iron ion homeostasis(GO:0006879) |
| 0.1 | 15.4 | GO:0055001 | muscle cell development(GO:0055001) |
| 0.1 | 2.1 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.1 | 0.8 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.1 | 0.2 | GO:0021849 | neuroblast division in subventricular zone(GO:0021849) |
| 0.1 | 0.3 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.1 | 0.2 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) nucleotide-excision repair, DNA incision, 5'-to lesion(GO:0006296) |
| 0.1 | 2.5 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.1 | 7.8 | GO:0050680 | negative regulation of epithelial cell proliferation(GO:0050680) |
| 0.1 | 0.3 | GO:0060510 | forebrain dorsal/ventral pattern formation(GO:0021798) Clara cell differentiation(GO:0060486) Type II pneumocyte differentiation(GO:0060510) |
| 0.0 | 5.8 | GO:0045727 | positive regulation of translation(GO:0045727) |
| 0.0 | 0.4 | GO:0021540 | corpus callosum morphogenesis(GO:0021540) |
| 0.0 | 0.4 | GO:0019585 | uronic acid metabolic process(GO:0006063) glucuronate metabolic process(GO:0019585) cellular glucuronidation(GO:0052695) |
| 0.0 | 7.0 | GO:0006814 | sodium ion transport(GO:0006814) |
| 0.0 | 1.8 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.0 | 3.1 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.9 | 79.9 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 4.6 | 18.5 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 3.2 | 19.1 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 2.6 | 31.8 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 2.2 | 28.1 | GO:0042627 | chylomicron(GO:0042627) |
| 2.0 | 20.0 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 1.8 | 23.0 | GO:0031983 | vesicle lumen(GO:0031983) |
| 0.8 | 12.6 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.8 | 2.3 | GO:0043511 | inhibin complex(GO:0043511) |
| 0.7 | 9.3 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.7 | 12.0 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.5 | 5.9 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.5 | 35.5 | GO:0045095 | keratin filament(GO:0045095) |
| 0.4 | 2.0 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 0.3 | 10.8 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.3 | 16.4 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.3 | 5.5 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.3 | 38.9 | GO:0005604 | basement membrane(GO:0005604) |
| 0.3 | 10.3 | GO:0046930 | pore complex(GO:0046930) |
| 0.2 | 11.2 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.2 | 26.4 | GO:0030018 | Z disc(GO:0030018) |
| 0.2 | 11.7 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 52.8 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.1 | 4.1 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 5.4 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.1 | 11.0 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 23.8 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.1 | 0.2 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.1 | 1.0 | GO:0036038 | MKS complex(GO:0036038) |
| 0.1 | 0.9 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 2.5 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 6.3 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 2.1 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 16.0 | GO:0005764 | lytic vacuole(GO:0000323) lysosome(GO:0005764) |
| 0.0 | 8.3 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.0 | 8.3 | GO:0005911 | cell-cell junction(GO:0005911) |
| 0.0 | 6.6 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 10.2 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 3.3 | GO:0005667 | transcription factor complex(GO:0005667) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 11.2 | 33.7 | GO:0071885 | N-terminal protein N-methyltransferase activity(GO:0071885) |
| 4.0 | 28.1 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 4.0 | 20.0 | GO:0031798 | type 1 metabotropic glutamate receptor binding(GO:0031798) |
| 3.2 | 12.7 | GO:0038181 | bile acid receptor activity(GO:0038181) |
| 2.7 | 8.0 | GO:0004618 | phosphoglycerate kinase activity(GO:0004618) |
| 2.0 | 37.9 | GO:0015643 | toxic substance binding(GO:0015643) |
| 1.5 | 34.2 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 1.5 | 26.4 | GO:0050308 | sugar-phosphatase activity(GO:0050308) |
| 1.1 | 5.4 | GO:0016726 | xanthine dehydrogenase activity(GO:0004854) oxidoreductase activity, acting on CH or CH2 groups, NAD or NADP as acceptor(GO:0016726) |
| 1.1 | 7.4 | GO:0035375 | zymogen binding(GO:0035375) |
| 0.8 | 4.1 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.7 | 22.7 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.7 | 2.8 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.6 | 2.6 | GO:0032896 | palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 0.6 | 3.1 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.6 | 24.3 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.6 | 2.3 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.5 | 3.7 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.5 | 9.3 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.5 | 10.1 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.4 | 10.3 | GO:0015288 | porin activity(GO:0015288) |
| 0.4 | 10.7 | GO:0005112 | Notch binding(GO:0005112) |
| 0.4 | 11.1 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.4 | 2.0 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.4 | 1.9 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.3 | 22.2 | GO:0005518 | collagen binding(GO:0005518) |
| 0.2 | 12.7 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.2 | 6.8 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.2 | 5.5 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.2 | 73.0 | GO:0003779 | actin binding(GO:0003779) |
| 0.1 | 5.1 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.1 | 9.4 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 2.1 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.1 | 14.0 | GO:0001191 | transcriptional repressor activity, RNA polymerase II transcription factor binding(GO:0001191) |
| 0.1 | 1.4 | GO:0019841 | retinol binding(GO:0019841) |
| 0.1 | 3.2 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.1 | 12.7 | GO:0008201 | heparin binding(GO:0008201) |
| 0.1 | 5.8 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.1 | 0.7 | GO:0051021 | GDP-dissociation inhibitor binding(GO:0051021) |
| 0.1 | 6.9 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 1.5 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.0 | 0.4 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.0 | 1.4 | GO:0016620 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor(GO:0016620) |
| 0.0 | 4.6 | GO:0042626 | ATPase activity, coupled to transmembrane movement of substances(GO:0042626) |
| 0.0 | 4.5 | GO:0008237 | metallopeptidase activity(GO:0008237) |
| 0.0 | 0.4 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.0 | 13.4 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.0 | 1.0 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 3.5 | GO:0030246 | carbohydrate binding(GO:0030246) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 4.1 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 1.7 | 85.7 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.8 | 24.3 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.7 | 38.2 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.5 | 23.2 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.5 | 20.0 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.3 | 71.5 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.3 | 7.0 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.3 | 10.8 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.2 | 8.0 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.2 | 9.4 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.2 | 10.1 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 3.1 | SIG CHEMOTAXIS | Genes related to chemotaxis |
| 0.1 | 2.3 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.1 | 3.7 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 7.7 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.9 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 11.1 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.7 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.3 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.8 | 37.9 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 1.6 | 22.4 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 1.0 | 18.7 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.9 | 34.4 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.5 | 5.4 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.4 | 22.0 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.4 | 10.1 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.3 | 7.0 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.2 | 9.3 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.2 | 2.3 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.2 | 5.1 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.2 | 9.4 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.2 | 6.3 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 1.9 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 5.8 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.1 | 11.7 | REACTOME SIGNALING BY PDGF | Genes involved in Signaling by PDGF |
| 0.1 | 5.5 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 4.1 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.4 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.0 | 1.9 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 0.9 | REACTOME DOWNSTREAM SIGNALING EVENTS OF B CELL RECEPTOR BCR | Genes involved in Downstream Signaling Events Of B Cell Receptor (BCR) |