Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
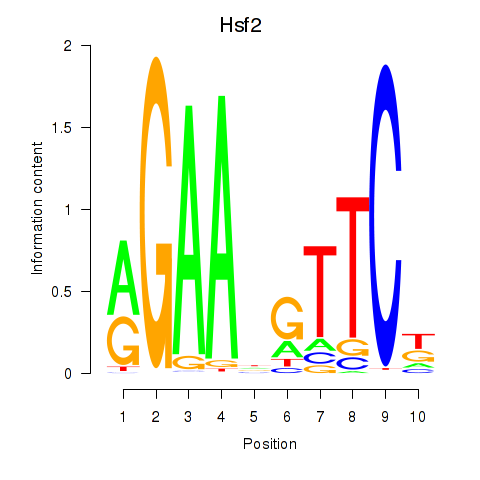
Results for Hsf2
Z-value: 1.04
Transcription factors associated with Hsf2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hsf2
|
ENSRNOG00000000808 | heat shock transcription factor 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hsf2 | rn6_v1_chr20_+_38935820_38935820 | 0.78 | 1.6e-65 | Click! |
Activity profile of Hsf2 motif
Sorted Z-values of Hsf2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_200793571 | 31.98 |
ENSRNOT00000091444
|
Hao2
|
hydroxyacid oxidase 2 |
| chr14_-_6679878 | 25.94 |
ENSRNOT00000075989
ENSRNOT00000067875 |
Spp1
|
secreted phosphoprotein 1 |
| chr1_+_219964429 | 25.93 |
ENSRNOT00000088288
|
Sptbn2
|
spectrin, beta, non-erythrocytic 2 |
| chr10_+_99388130 | 21.62 |
ENSRNOT00000006238
|
Kcnj16
|
potassium voltage-gated channel subfamily J member 16 |
| chr9_-_80166807 | 20.09 |
ENSRNOT00000079493
|
Igfbp5
|
insulin-like growth factor binding protein 5 |
| chr18_+_69841053 | 19.92 |
ENSRNOT00000071545
ENSRNOT00000030613 ENSRNOT00000075543 |
Mro
|
maestro |
| chr18_-_1946840 | 19.07 |
ENSRNOT00000041878
|
Abhd3
|
abhydrolase domain containing 3 |
| chr14_-_64535170 | 17.55 |
ENSRNOT00000082338
|
Gba3
|
glucosidase, beta, acid 3 |
| chr1_+_153861569 | 16.70 |
ENSRNOT00000023329
|
Me3
|
malic enzyme 3 |
| chr10_-_13115294 | 15.78 |
ENSRNOT00000005899
|
NEWGENE_6497122
|
FLYWCH family member 2 |
| chr6_-_23542928 | 15.72 |
ENSRNOT00000083858
|
Spdya
|
speedy/RINGO cell cycle regulator family member A |
| chrX_+_20351486 | 15.65 |
ENSRNOT00000093675
ENSRNOT00000047444 |
Wnk3
|
WNK lysine deficient protein kinase 3 |
| chr8_-_73122740 | 14.78 |
ENSRNOT00000012026
|
Tln2
|
talin 2 |
| chr14_-_78902063 | 14.06 |
ENSRNOT00000088469
|
Ppp2r2c
|
protein phosphatase 2, regulatory subunit B, gamma |
| chr18_-_37096132 | 13.77 |
ENSRNOT00000041188
|
Ppp2r2b
|
protein phosphatase 2, regulatory subunit B, beta |
| chr9_+_8349033 | 13.73 |
ENSRNOT00000073775
|
LOC100360856
|
hypothetical protein LOC100360856 |
| chrX_+_106487870 | 13.65 |
ENSRNOT00000075521
|
Arxes2
|
adipocyte-related X-chromosome expressed sequence 2 |
| chr1_-_173764246 | 13.53 |
ENSRNOT00000019690
ENSRNOT00000086944 |
Lmo1
|
LIM domain only 1 |
| chr6_+_64808238 | 13.40 |
ENSRNOT00000093195
|
Nrcam
|
neuronal cell adhesion molecule |
| chr1_-_188713270 | 13.38 |
ENSRNOT00000082192
ENSRNOT00000065892 |
Gprc5b
|
G protein-coupled receptor, class C, group 5, member B |
| chr1_+_238222521 | 13.34 |
ENSRNOT00000024000
|
Aldh1a1
|
aldehyde dehydrogenase 1 family, member A1 |
| chr8_+_131965134 | 13.14 |
ENSRNOT00000078308
|
Znf660
|
zinc finger protein 660 |
| chrX_-_82743753 | 13.07 |
ENSRNOT00000003512
|
Rps6ka6
|
ribosomal protein S6 kinase A6 |
| chr1_+_32221636 | 12.94 |
ENSRNOT00000022346
ENSRNOT00000089941 |
Slc6a18
|
solute carrier family 6 member 18 |
| chr1_+_201901342 | 12.87 |
ENSRNOT00000036657
|
RGD1559891
|
similar to synaptonemal complex protein 3 |
| chr7_+_129595192 | 12.86 |
ENSRNOT00000071151
|
Zdhhc25
|
zinc finger, DHHC-type containing 25 |
| chr19_+_41482728 | 12.86 |
ENSRNOT00000022943
|
Calb2
|
calbindin 2 |
| chr8_-_63750531 | 12.73 |
ENSRNOT00000009496
|
Neo1
|
neogenin 1 |
| chr6_+_48708368 | 12.67 |
ENSRNOT00000005905
|
Myt1l
|
myelin transcription factor 1-like |
| chr20_-_9855443 | 12.55 |
ENSRNOT00000090275
ENSRNOT00000066266 |
Tff3
|
trefoil factor 3 |
| chr10_-_47825252 | 12.54 |
ENSRNOT00000087102
|
Epn2
|
epsin 2 |
| chr9_+_23503236 | 12.48 |
ENSRNOT00000017996
|
Crisp2
|
cysteine-rich secretory protein 2 |
| chr9_-_27452902 | 12.29 |
ENSRNOT00000018325
|
Gsta1
|
glutathione S-transferase alpha 1 |
| chr3_-_111994337 | 11.93 |
ENSRNOT00000030502
|
Pla2g4e
|
phospholipase A2, group IVE |
| chr4_+_153874852 | 11.74 |
ENSRNOT00000079744
|
Slc6a13
|
solute carrier family 6 member 13 |
| chr11_-_87240833 | 11.66 |
ENSRNOT00000052200
|
Tssk1b
|
testis-specific serine kinase 1B |
| chr1_-_240601744 | 11.58 |
ENSRNOT00000024093
|
Aldh1a7
|
aldehyde dehydrogenase family 1, subfamily A7 |
| chr8_-_73164620 | 11.46 |
ENSRNOT00000031988
|
Tln2
|
talin 2 |
| chr1_+_41323194 | 11.44 |
ENSRNOT00000026350
|
Esr1
|
estrogen receptor 1 |
| chr2_-_181874223 | 11.37 |
ENSRNOT00000035846
|
Rbm46
|
RNA binding motif protein 46 |
| chr10_+_103395511 | 11.28 |
ENSRNOT00000004256
|
Gprc5c
|
G protein-coupled receptor, class C, group 5, member C |
| chr9_-_80167033 | 11.26 |
ENSRNOT00000023530
|
Igfbp5
|
insulin-like growth factor binding protein 5 |
| chr7_-_130827152 | 11.24 |
ENSRNOT00000019406
|
Syt10
|
synaptotagmin 10 |
| chr3_-_51612397 | 11.17 |
ENSRNOT00000081401
|
Scn3a
|
sodium voltage-gated channel alpha subunit 3 |
| chr8_-_82339937 | 11.00 |
ENSRNOT00000080797
|
Mapk6
|
mitogen-activated protein kinase 6 |
| chr11_+_82862695 | 10.98 |
ENSRNOT00000071707
ENSRNOT00000073435 |
Clcn2
|
chloride voltage-gated channel 2 |
| chr1_-_53589844 | 10.97 |
ENSRNOT00000073581
|
Unc93a
|
unc-93 homolog A (C. elegans) |
| chr7_+_23012070 | 10.86 |
ENSRNOT00000042460
|
LOC300308
|
similar to hypothetical protein 4930509O22 |
| chr5_-_62187930 | 10.84 |
ENSRNOT00000011787
|
Coro2a
|
coronin 2A |
| chrX_+_116399611 | 10.80 |
ENSRNOT00000050854
|
Zcchc16
|
zinc finger CCHC-type containing 16 |
| chr2_-_200625434 | 10.73 |
ENSRNOT00000079754
|
Hsd3b5
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 5 |
| chr5_+_71742911 | 10.69 |
ENSRNOT00000047225
|
Zfp462
|
zinc finger protein 462 |
| chr2_+_45182402 | 10.66 |
ENSRNOT00000060673
|
RGD1561161
|
similar to BC067074 protein |
| chrX_-_115175299 | 10.64 |
ENSRNOT00000074322
|
Dcx
|
doublecortin |
| chr7_+_144647587 | 10.62 |
ENSRNOT00000022398
|
Hoxc4
|
homeo box C4 |
| chr8_+_85355766 | 10.53 |
ENSRNOT00000010583
|
Gcm1
|
glial cells missing homolog 1 |
| chr14_+_64686793 | 10.50 |
ENSRNOT00000005894
ENSRNOT00000036646 |
Adgra3
|
adhesion G protein-coupled receptor A3 |
| chr1_+_166739532 | 10.42 |
ENSRNOT00000079846
ENSRNOT00000026665 |
Clpb
|
ClpB homolog, mitochondrial AAA ATPase chaperonin |
| chr1_+_81373340 | 10.34 |
ENSRNOT00000026814
|
Zfp428
|
zinc finger protein 428 |
| chrX_+_33884499 | 10.32 |
ENSRNOT00000090041
|
Reps2
|
RALBP1 associated Eps domain containing protein 2 |
| chr2_-_235177275 | 10.32 |
ENSRNOT00000093153
|
LOC103691699
|
uncharacterized LOC103691699 |
| chr10_-_107539658 | 10.17 |
ENSRNOT00000089346
|
Rbfox3
|
RNA binding protein, fox-1 homolog 3 |
| chr11_+_83884048 | 10.11 |
ENSRNOT00000002376
|
LOC108348101
|
chloride channel protein 2 |
| chr3_-_64554953 | 10.07 |
ENSRNOT00000067452
|
LOC102553814
|
serine/arginine repetitive matrix protein 1-like |
| chrX_+_111122552 | 10.03 |
ENSRNOT00000083566
ENSRNOT00000085078 ENSRNOT00000090928 |
Cldn2
|
claudin 2 |
| chr11_+_67221359 | 9.99 |
ENSRNOT00000086097
|
Casr
|
calcium-sensing receptor |
| chr9_+_4817854 | 9.95 |
ENSRNOT00000040879
|
LOC100910526
|
sulfotransferase 1C2-like |
| chr1_+_217039755 | 9.92 |
ENSRNOT00000091603
|
LOC102552318
|
actin-like |
| chr4_-_14490446 | 9.91 |
ENSRNOT00000009132
|
Sema3c
|
semaphorin 3C |
| chr7_+_140383397 | 9.88 |
ENSRNOT00000090760
|
Ccdc65
|
coiled-coil domain containing 65 |
| chr4_-_100465106 | 9.86 |
ENSRNOT00000086725
ENSRNOT00000057989 |
Elmod3
|
ELMO domain containing 3 |
| chr4_-_128266082 | 9.86 |
ENSRNOT00000039549
|
Nup50
|
nucleoporin 50 |
| chr9_-_92530938 | 9.81 |
ENSRNOT00000064875
|
Slc16a14
|
solute carrier family 16, member 14 |
| chr2_-_195678848 | 9.77 |
ENSRNOT00000028303
ENSRNOT00000075569 |
Oaz3
|
ornithine decarboxylase antizyme 3 |
| chr9_-_63637677 | 9.76 |
ENSRNOT00000049259
|
Satb2
|
SATB homeobox 2 |
| chrX_+_106523278 | 9.73 |
ENSRNOT00000070802
|
MGC109340
|
similar to Microsomal signal peptidase 23 kDa subunit (SPase 22 kDa subunit) (SPC22/23) |
| chr5_+_133896141 | 9.66 |
ENSRNOT00000011434
|
Pdzk1ip1
|
PDZK1 interacting protein 1 |
| chr2_+_237148941 | 9.56 |
ENSRNOT00000015116
|
Dkk2
|
dickkopf WNT signaling pathway inhibitor 2 |
| chr9_-_63291350 | 9.53 |
ENSRNOT00000058831
|
Hsfy2
|
heat shock transcription factor, Y linked 2 |
| chr10_-_40201992 | 9.47 |
ENSRNOT00000075311
|
Lyrm7
|
LYR motif containing 7 |
| chr14_+_12218553 | 9.46 |
ENSRNOT00000003237
|
Prkg2
|
protein kinase, cGMP-dependent, type II |
| chr16_+_66380173 | 9.44 |
ENSRNOT00000048862
|
LOC681180
|
similar to Discs large homolog 5 (Placenta and prostate DLG) (Discs large protein P-dlg) |
| chr1_+_199682688 | 9.42 |
ENSRNOT00000027265
ENSRNOT00000080872 |
Slc5a2
|
solute carrier family 5 member 2 |
| chr6_-_99273033 | 9.36 |
ENSRNOT00000088808
|
Tex21
|
testis expressed 21 |
| chr16_-_15798974 | 9.33 |
ENSRNOT00000046842
ENSRNOT00000065946 |
Nrg3
|
neuregulin 3 |
| chr1_+_61786900 | 9.32 |
ENSRNOT00000090287
|
AABR07001905.1
|
|
| chr2_-_89310946 | 9.30 |
ENSRNOT00000015195
|
Ralyl
|
RALY RNA binding protein-like |
| chr9_+_10172832 | 9.23 |
ENSRNOT00000074555
|
Acsbg2
|
acyl-CoA synthetase bubblegum family member 2 |
| chr1_-_266428239 | 9.13 |
ENSRNOT00000027160
|
Cyp17a1
|
cytochrome P450, family 17, subfamily a, polypeptide 1 |
| chr5_-_78324278 | 9.08 |
ENSRNOT00000082642
ENSRNOT00000048904 |
Wdr31
|
WD repeat domain 31 |
| chr9_+_4107246 | 9.07 |
ENSRNOT00000078212
|
AABR07066160.1
|
|
| chr11_+_45339366 | 8.81 |
ENSRNOT00000002241
|
Tmem30c
|
transmembrane protein 30C |
| chr2_-_231648122 | 8.78 |
ENSRNOT00000014962
|
Ank2
|
ankyrin 2 |
| chr5_-_57372239 | 8.78 |
ENSRNOT00000012975
|
Aqp7
|
aquaporin 7 |
| chr19_-_42180981 | 8.74 |
ENSRNOT00000019577
|
Pmfbp1
|
polyamine modulated factor 1 binding protein 1 |
| chr13_-_111765944 | 8.72 |
ENSRNOT00000073041
|
Syt14
|
synaptotagmin 14 |
| chr6_+_31011687 | 8.68 |
ENSRNOT00000071242
|
AABR07063459.1
|
|
| chr4_+_61850348 | 8.66 |
ENSRNOT00000013423
|
Akr1b7
|
aldo-keto reductase family 1, member B7 |
| chr13_-_84452181 | 8.64 |
ENSRNOT00000005060
|
Mael
|
maelstrom spermatogenic transposon silencer |
| chr14_-_17582823 | 8.62 |
ENSRNOT00000087686
|
LOC100911949
|
uncharacterized LOC100911949 |
| chr1_-_174119815 | 8.55 |
ENSRNOT00000019368
|
Trim66
|
tripartite motif-containing 66 |
| chr7_+_123531682 | 8.48 |
ENSRNOT00000010606
|
Wbp2nl
|
WBP2 N-terminal like |
| chr11_-_82938357 | 8.42 |
ENSRNOT00000035945
|
Map3k13
|
mitogen-activated protein kinase kinase kinase 13 |
| chr1_-_224533219 | 8.35 |
ENSRNOT00000051289
|
Ust5r
|
integral membrane transport protein UST5r |
| chr9_-_65790347 | 8.35 |
ENSRNOT00000028506
|
AABR07067812.1
|
|
| chr10_-_13107771 | 8.33 |
ENSRNOT00000005879
|
Flywch1
|
FLYWCH-type zinc finger 1 |
| chr10_+_93305969 | 8.31 |
ENSRNOT00000008019
|
Efcab3
|
EF-hand calcium binding domain 3 |
| chrX_-_63961527 | 8.27 |
ENSRNOT00000071762
|
Gspt2
|
G1 to S phase transition 2 |
| chr3_+_122928964 | 8.26 |
ENSRNOT00000074724
|
LOC100365450
|
rCG26795-like |
| chr19_-_42180362 | 8.22 |
ENSRNOT00000089515
|
Pmfbp1
|
polyamine modulated factor 1 binding protein 1 |
| chr10_+_91187593 | 8.19 |
ENSRNOT00000004163
|
Acbd4
|
acyl-CoA binding domain containing 4 |
| chr3_+_138974871 | 8.16 |
ENSRNOT00000012524
|
Scp2d1
|
SCP2 sterol-binding domain containing 1 |
| chr13_+_99335020 | 8.11 |
ENSRNOT00000029787
|
AABR07021930.1
|
|
| chr10_-_15166457 | 8.05 |
ENSRNOT00000026676
|
Metrn
|
meteorin, glial cell differentiation regulator |
| chr1_+_217173199 | 8.03 |
ENSRNOT00000075078
|
Shank2
|
SH3 and multiple ankyrin repeat domains 2 |
| chr13_-_112099336 | 8.03 |
ENSRNOT00000009158
ENSRNOT00000044161 |
Camk1g
|
calcium/calmodulin-dependent protein kinase IG |
| chrX_-_105622156 | 7.97 |
ENSRNOT00000029511
|
Armcx2
|
armadillo repeat containing, X-linked 2 |
| chr7_-_83670356 | 7.96 |
ENSRNOT00000005584
|
Sybu
|
syntabulin |
| chr4_-_181348038 | 7.95 |
ENSRNOT00000082879
|
LOC690784
|
hypothetical protein LOC690783 |
| chr6_-_111267734 | 7.90 |
ENSRNOT00000074037
|
Noxred1
|
NADP-dependent oxidoreductase domain containing 1 |
| chr7_+_144623555 | 7.86 |
ENSRNOT00000022217
|
Hoxc6
|
homeo box C6 |
| chr1_+_82174451 | 7.86 |
ENSRNOT00000027783
|
Tmem145
|
transmembrane protein 145 |
| chr13_-_91427575 | 7.80 |
ENSRNOT00000012092
|
Apcs
|
amyloid P component, serum |
| chr1_-_89559960 | 7.70 |
ENSRNOT00000092133
|
Scn1b
|
sodium voltage-gated channel beta subunit 1 |
| chr1_+_72860218 | 7.56 |
ENSRNOT00000024547
|
Syt5
|
synaptotagmin 5 |
| chr2_-_187668677 | 7.54 |
ENSRNOT00000056898
ENSRNOT00000092563 |
Tsacc
|
TSSK6 activating co-chaperone |
| chr8_-_115274165 | 7.37 |
ENSRNOT00000056386
|
LOC102550160
|
IQ domain-containing protein F5-like |
| chr18_-_28444880 | 7.24 |
ENSRNOT00000060696
|
Prob1
|
proline rich basic protein 1 |
| chrX_+_157353577 | 7.19 |
ENSRNOT00000090596
|
Haus7
|
HAUS augmin-like complex, subunit 7 |
| chr1_+_219233750 | 7.18 |
ENSRNOT00000024112
|
Acy3
|
aminoacylase 3 |
| chr15_-_46381967 | 7.12 |
ENSRNOT00000014198
|
Neil2
|
nei-like DNA glycosylase 2 |
| chr10_-_93679974 | 7.11 |
ENSRNOT00000009316
|
March10
|
membrane associated ring-CH-type finger 10 |
| chr4_+_57823411 | 7.10 |
ENSRNOT00000030462
ENSRNOT00000088235 |
Ssmem1
|
serine-rich single-pass membrane protein 1 |
| chr8_-_117211170 | 7.07 |
ENSRNOT00000071189
|
Ccdc36
|
coiled-coil domain containing 36 |
| chr4_+_157057601 | 7.04 |
ENSRNOT00000040592
|
Zfp42l
|
Zinc finger protein 42-like |
| chr10_+_45307007 | 7.03 |
ENSRNOT00000003885
|
Trim17
|
tripartite motif-containing 17 |
| chr4_-_176720012 | 7.00 |
ENSRNOT00000017965
|
Ldhb
|
lactate dehydrogenase B |
| chr8_+_70603249 | 6.98 |
ENSRNOT00000067016
ENSRNOT00000072486 |
Igdcc4
|
immunoglobulin superfamily, DCC subclass, member 4 |
| chr7_+_40316639 | 6.96 |
ENSRNOT00000080150
|
LOC500827
|
similar to hypothetical protein FLJ35821 |
| chr5_+_60528997 | 6.90 |
ENSRNOT00000051445
|
Grhpr
|
glyoxylate and hydroxypyruvate reductase |
| chr10_-_34333305 | 6.87 |
ENSRNOT00000071365
|
Olr1384
|
olfactory receptor gene Olr1384 |
| chr7_+_123482255 | 6.86 |
ENSRNOT00000064487
|
LOC688613
|
hypothetical protein LOC688613 |
| chr20_+_5619012 | 6.85 |
ENSRNOT00000000577
|
Ggnbp1
|
gametogenetin binding protein 1 |
| chr18_-_37776453 | 6.84 |
ENSRNOT00000087876
|
Dpysl3
|
dihydropyrimidinase-like 3 |
| chr15_-_26175645 | 6.84 |
ENSRNOT00000020175
|
Slc35f4
|
solute carrier family 35, member F4 |
| chr8_-_30222036 | 6.83 |
ENSRNOT00000035454
|
Ntm
|
neurotrimin |
| chr1_-_62191818 | 6.81 |
ENSRNOT00000033187
|
AABR07001926.1
|
|
| chr7_-_136853154 | 6.79 |
ENSRNOT00000087376
|
Nell2
|
neural EGFL like 2 |
| chr20_+_1749716 | 6.74 |
ENSRNOT00000048856
|
Olr1735
|
olfactory receptor 1735 |
| chr1_-_193459396 | 6.66 |
ENSRNOT00000020281
|
Zkscan2
|
zinc finger with KRAB and SCAN domains 2 |
| chr9_-_114152716 | 6.66 |
ENSRNOT00000044153
|
AABR07068650.1
|
|
| chr1_+_82169620 | 6.65 |
ENSRNOT00000088955
ENSRNOT00000068251 |
Prr19
|
proline rich 19 |
| chr19_-_34752695 | 6.65 |
ENSRNOT00000052018
|
Nr3c2
|
nuclear receptor subfamily 3, group C, member 2 |
| chr8_-_62332115 | 6.64 |
ENSRNOT00000025783
|
Mpi
|
mannose phosphate isomerase (mapped) |
| chr1_-_18511695 | 6.61 |
ENSRNOT00000075402
|
AABR07000583.1
|
|
| chr20_+_1764794 | 6.59 |
ENSRNOT00000075084
|
Olr1736
|
olfactory receptor 1736 |
| chr10_-_107539465 | 6.53 |
ENSRNOT00000004524
|
Rbfox3
|
RNA binding protein, fox-1 homolog 3 |
| chr3_+_170475831 | 6.53 |
ENSRNOT00000006949
|
Fam209a
|
family with sequence similarity 209, member A |
| chr15_-_57651041 | 6.51 |
ENSRNOT00000072138
|
Spert
|
spermatid associated |
| chr13_-_80862963 | 6.50 |
ENSRNOT00000004864
|
Fmo3
|
flavin containing monooxygenase 3 |
| chr1_-_170397191 | 6.46 |
ENSRNOT00000090181
|
Apbb1
|
amyloid beta precursor protein binding family B member 1 |
| chr1_+_243477493 | 6.40 |
ENSRNOT00000021779
ENSRNOT00000085356 |
Dmrt1
|
doublesex and mab-3 related transcription factor 1 |
| chr17_+_47397558 | 6.39 |
ENSRNOT00000085923
|
Epdr1
|
ependymin related 1 |
| chr20_+_40769586 | 6.37 |
ENSRNOT00000001079
|
Fabp7
|
fatty acid binding protein 7 |
| chr11_-_86920094 | 6.36 |
ENSRNOT00000063878
|
Ccdc188
|
coiled-coil domain containing 188 |
| chr9_+_50526811 | 6.35 |
ENSRNOT00000036990
|
RGD1305645
|
similar to RIKEN cDNA 1500015O10 |
| chr8_-_40883880 | 6.34 |
ENSRNOT00000075593
|
LOC102550797
|
disks large homolog 5-like |
| chr13_+_51218468 | 6.33 |
ENSRNOT00000033636
|
LOC289035
|
similar to UDP-N-acetylglucosamine:a-1,3-D-mannoside beta-1,4-N-acetylgluco |
| chr3_-_3541947 | 6.32 |
ENSRNOT00000024786
|
Nacc2
|
NACC family member 2 |
| chr15_+_28894627 | 6.31 |
ENSRNOT00000017795
|
Olr1643
|
olfactory receptor 1643 |
| chr10_-_98294522 | 6.29 |
ENSRNOT00000005489
|
Abca8
|
ATP binding cassette subfamily A member 8 |
| chr5_-_150949931 | 6.27 |
ENSRNOT00000067905
|
Smpdl3b
|
sphingomyelin phosphodiesterase, acid-like 3B |
| chr4_-_482645 | 6.26 |
ENSRNOT00000062073
ENSRNOT00000071713 |
Cnpy1
|
canopy FGF signaling regulator 1 |
| chr17_-_10766253 | 6.25 |
ENSRNOT00000000117
|
Cplx2
|
complexin 2 |
| chr13_+_49074644 | 6.21 |
ENSRNOT00000000041
|
Klhdc8a
|
kelch domain containing 8A |
| chr10_+_103396155 | 6.21 |
ENSRNOT00000086924
|
Gprc5c
|
G protein-coupled receptor, class C, group 5, member C |
| chr3_+_97723901 | 6.06 |
ENSRNOT00000080416
|
Mpped2
|
metallophosphoesterase domain containing 2 |
| chr20_+_4188766 | 6.02 |
ENSRNOT00000081438
ENSRNOT00000060378 |
Tesb
|
testis specific basic protein |
| chr5_-_32956159 | 6.02 |
ENSRNOT00000078264
|
Cnbd1
|
cyclic nucleotide binding domain containing 1 |
| chr8_+_117836701 | 6.01 |
ENSRNOT00000043345
|
Plxnb1
|
plexin B1 |
| chr10_-_8654892 | 5.99 |
ENSRNOT00000066534
|
Rbfox1
|
RNA binding protein, fox-1 homolog 1 |
| chr2_-_32518643 | 5.98 |
ENSRNOT00000061032
|
Mast4
|
microtubule associated serine/threonine kinase family member 4 |
| chr1_-_81596819 | 5.97 |
ENSRNOT00000074350
|
RGD1564380
|
similar to BC049730 protein |
| chr4_+_163293724 | 5.94 |
ENSRNOT00000077356
|
Gabarapl1
|
GABA type A receptor associated protein like 1 |
| chr4_-_163095614 | 5.94 |
ENSRNOT00000088759
|
RGD1564770
|
similar to CD69 antigen (p60, early T-cell activation antigen) |
| chr16_+_49485256 | 5.83 |
ENSRNOT00000059317
|
RGD1564308
|
similar to LOC495042 protein |
| chr3_+_114176309 | 5.80 |
ENSRNOT00000023350
|
Sord
|
sorbitol dehydrogenase |
| chrX_+_119390013 | 5.79 |
ENSRNOT00000074269
|
Agtr2
|
angiotensin II receptor, type 2 |
| chr18_-_58423196 | 5.79 |
ENSRNOT00000025556
|
Piezo2
|
piezo-type mechanosensitive ion channel component 2 |
| chr20_-_5037022 | 5.78 |
ENSRNOT00000091022
ENSRNOT00000060762 |
Msh5
|
mutS homolog 5 |
| chr18_+_30562178 | 5.74 |
ENSRNOT00000040998
|
LOC108348771
|
protocadherin beta-16-like |
| chr19_+_6046665 | 5.72 |
ENSRNOT00000084126
|
Cdh8
|
cadherin 8 |
| chr2_+_154921999 | 5.69 |
ENSRNOT00000057620
|
LOC691044
|
similar to GTPase activating protein testicular GAP1 |
| chr20_+_4959294 | 5.67 |
ENSRNOT00000074223
|
Hspa1l
|
heat shock protein family A (Hsp70) member 1 like |
| chr7_+_119647375 | 5.66 |
ENSRNOT00000046563
|
Kctd17
|
potassium channel tetramerization domain containing 17 |
| chr16_-_81880502 | 5.63 |
ENSRNOT00000079096
|
Mcf2l
|
MCF.2 cell line derived transforming sequence-like |
| chr4_-_113988246 | 5.61 |
ENSRNOT00000013128
|
LOC103692166
|
WD repeat-containing protein 54 |
| chr7_-_107203897 | 5.61 |
ENSRNOT00000086263
|
Lrrc6
|
leucine rich repeat containing 6 |
| chr8_-_25829569 | 5.58 |
ENSRNOT00000071884
|
Dpy19l2
|
dpy-19 like 2 |
| chr4_-_40385349 | 5.55 |
ENSRNOT00000039005
|
Gpr85
|
G protein-coupled receptor 85 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hsf2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 10.7 | 32.0 | GO:0018924 | mandelate metabolic process(GO:0018924) |
| 8.6 | 25.9 | GO:0006710 | androgen catabolic process(GO:0006710) |
| 7.8 | 31.3 | GO:1904204 | regulation of skeletal muscle hypertrophy(GO:1904204) |
| 6.3 | 25.4 | GO:1903015 | regulation of exo-alpha-sialidase activity(GO:1903015) |
| 5.2 | 15.6 | GO:2000682 | positive regulation of rubidium ion transport(GO:2000682) positive regulation of rubidium ion transmembrane transporter activity(GO:2000688) |
| 3.5 | 10.6 | GO:1990792 | response to glial cell derived neurotrophic factor(GO:1990790) cellular response to glial cell derived neurotrophic factor(GO:1990792) |
| 3.4 | 13.5 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 3.3 | 9.8 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 3.2 | 9.5 | GO:2001226 | negative regulation of chloride transport(GO:2001226) |
| 3.0 | 9.1 | GO:0010034 | response to acetate(GO:0010034) |
| 3.0 | 9.0 | GO:1990091 | sodium-dependent self proteolysis(GO:1990091) |
| 2.9 | 11.4 | GO:1990375 | baculum development(GO:1990375) |
| 2.5 | 7.6 | GO:0018916 | nitrobenzene metabolic process(GO:0018916) |
| 2.5 | 9.9 | GO:0003350 | pulmonary myocardium development(GO:0003350) |
| 2.4 | 12.0 | GO:0060689 | cell differentiation involved in salivary gland development(GO:0060689) |
| 2.4 | 26.2 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 2.3 | 9.3 | GO:0021842 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 2.3 | 11.6 | GO:0035106 | operant conditioning(GO:0035106) |
| 2.3 | 2.3 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 2.3 | 2.3 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 2.2 | 6.7 | GO:1902380 | positive regulation of mRNA cleavage(GO:0031439) positive regulation of endoribonuclease activity(GO:1902380) positive regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904722) |
| 2.2 | 13.3 | GO:0002072 | optic cup morphogenesis involved in camera-type eye development(GO:0002072) |
| 2.2 | 6.6 | GO:0031959 | mineralocorticoid receptor signaling pathway(GO:0031959) |
| 2.2 | 8.8 | GO:0036309 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 2.2 | 25.9 | GO:0016081 | synaptic vesicle docking(GO:0016081) |
| 2.1 | 23.4 | GO:0006465 | signal peptide processing(GO:0006465) |
| 2.1 | 10.5 | GO:0060018 | astrocyte fate commitment(GO:0060018) |
| 2.1 | 8.3 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 2.0 | 8.2 | GO:0010615 | positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) |
| 2.0 | 6.0 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 1.9 | 5.8 | GO:0006059 | hexitol metabolic process(GO:0006059) |
| 1.9 | 5.8 | GO:0035566 | G-protein coupled receptor signaling pathway coupled to cGMP nucleotide second messenger(GO:0007199) regulation of metanephros size(GO:0035566) |
| 1.9 | 18.9 | GO:0046684 | response to pyrethroid(GO:0046684) |
| 1.8 | 8.8 | GO:0015793 | glycerol transport(GO:0015793) |
| 1.7 | 5.2 | GO:0021629 | muscle attachment(GO:0016203) olfactory nerve morphogenesis(GO:0021627) olfactory nerve structural organization(GO:0021629) |
| 1.7 | 6.9 | GO:0006104 | succinyl-CoA metabolic process(GO:0006104) |
| 1.7 | 13.5 | GO:0060631 | regulation of meiosis I(GO:0060631) |
| 1.6 | 27.8 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 1.5 | 4.4 | GO:0051935 | amino acid neurotransmitter reuptake(GO:0051933) glutamate reuptake(GO:0051935) |
| 1.4 | 15.9 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 1.4 | 4.3 | GO:0042726 | flavin-containing compound metabolic process(GO:0042726) |
| 1.4 | 5.6 | GO:0018103 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 1.4 | 4.1 | GO:0098971 | anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) |
| 1.3 | 13.4 | GO:0098870 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 1.3 | 6.6 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 1.3 | 4.0 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 1.3 | 5.2 | GO:0072268 | nephrogenic mesenchyme development(GO:0072076) pattern specification involved in metanephros development(GO:0072268) |
| 1.3 | 3.9 | GO:0015744 | succinate transport(GO:0015744) succinate transmembrane transport(GO:0071422) |
| 1.2 | 14.9 | GO:0006108 | malate metabolic process(GO:0006108) |
| 1.2 | 12.3 | GO:0042178 | xenobiotic catabolic process(GO:0042178) |
| 1.2 | 3.5 | GO:0019255 | glucose 1-phosphate metabolic process(GO:0019255) |
| 1.1 | 3.4 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 1.1 | 10.0 | GO:0060613 | fat pad development(GO:0060613) |
| 1.1 | 15.2 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 1.1 | 4.2 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 1.0 | 4.2 | GO:0060157 | urinary bladder development(GO:0060157) |
| 1.0 | 5.2 | GO:1900620 | acetylcholine biosynthetic process(GO:0008292) acetate ester biosynthetic process(GO:1900620) |
| 1.0 | 5.1 | GO:2000821 | regulation of grooming behavior(GO:2000821) |
| 1.0 | 7.0 | GO:2000653 | regulation of genetic imprinting(GO:2000653) |
| 1.0 | 13.1 | GO:2000381 | negative regulation of mesoderm development(GO:2000381) |
| 1.0 | 4.0 | GO:0090611 | ubiquitin-independent protein catabolic process via the multivesicular body sorting pathway(GO:0090611) |
| 1.0 | 4.9 | GO:0061668 | mitochondrial ribosome assembly(GO:0061668) |
| 1.0 | 3.9 | GO:1903243 | negative regulation of cardiac muscle hypertrophy in response to stress(GO:1903243) |
| 1.0 | 6.8 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.9 | 2.8 | GO:0006532 | aspartate biosynthetic process(GO:0006532) |
| 0.9 | 4.7 | GO:0070120 | ciliary neurotrophic factor-mediated signaling pathway(GO:0070120) |
| 0.9 | 8.4 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.9 | 9.1 | GO:0098703 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.9 | 12.7 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) |
| 0.9 | 2.7 | GO:0002426 | immunoglobulin production in mucosal tissue(GO:0002426) |
| 0.9 | 6.3 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.9 | 13.3 | GO:0042448 | progesterone metabolic process(GO:0042448) |
| 0.9 | 1.8 | GO:1903544 | response to butyrate(GO:1903544) |
| 0.9 | 5.2 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.8 | 8.5 | GO:0007343 | egg activation(GO:0007343) |
| 0.8 | 3.4 | GO:0021586 | pons maturation(GO:0021586) |
| 0.8 | 2.5 | GO:0090247 | cell motility involved in somitogenic axis elongation(GO:0090247) regulation of cell motility involved in somitogenic axis elongation(GO:0090249) |
| 0.8 | 4.1 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.8 | 7.4 | GO:2000344 | positive regulation of acrosome reaction(GO:2000344) |
| 0.8 | 4.9 | GO:0002678 | positive regulation of chronic inflammatory response(GO:0002678) |
| 0.8 | 2.4 | GO:0097037 | heme export(GO:0097037) |
| 0.8 | 3.2 | GO:0061107 | seminal vesicle development(GO:0061107) |
| 0.8 | 7.9 | GO:0006561 | proline biosynthetic process(GO:0006561) |
| 0.8 | 3.2 | GO:0090666 | scaRNA localization to Cajal body(GO:0090666) |
| 0.8 | 7.9 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.8 | 4.7 | GO:0035166 | post-embryonic hemopoiesis(GO:0035166) |
| 0.8 | 3.1 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.8 | 24.5 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.8 | 2.3 | GO:0034499 | late endosome to Golgi transport(GO:0034499) |
| 0.7 | 6.7 | GO:0046543 | development of secondary female sexual characteristics(GO:0046543) |
| 0.7 | 3.6 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.7 | 2.2 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.7 | 2.7 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.7 | 3.9 | GO:0008050 | female courtship behavior(GO:0008050) |
| 0.6 | 2.6 | GO:1900377 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.6 | 3.9 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.6 | 12.3 | GO:0021819 | layer formation in cerebral cortex(GO:0021819) |
| 0.6 | 7.7 | GO:0048642 | negative regulation of skeletal muscle tissue development(GO:0048642) |
| 0.6 | 1.3 | GO:0014873 | response to muscle activity involved in regulation of muscle adaptation(GO:0014873) |
| 0.6 | 8.0 | GO:0098880 | maintenance of postsynaptic specialization structure(GO:0098880) maintenance of postsynaptic density structure(GO:0099562) |
| 0.6 | 2.5 | GO:1903413 | cellular response to bile acid(GO:1903413) |
| 0.6 | 3.1 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.6 | 1.8 | GO:0032053 | ciliary basal body organization(GO:0032053) |
| 0.6 | 6.1 | GO:0014883 | transition between fast and slow fiber(GO:0014883) |
| 0.6 | 5.4 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.6 | 1.8 | GO:0010625 | positive regulation of Schwann cell proliferation(GO:0010625) |
| 0.6 | 5.1 | GO:0055089 | fatty acid homeostasis(GO:0055089) |
| 0.5 | 4.4 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.5 | 4.4 | GO:0070102 | interleukin-6-mediated signaling pathway(GO:0070102) |
| 0.5 | 4.3 | GO:0071318 | cellular response to ATP(GO:0071318) |
| 0.5 | 8.6 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.5 | 4.2 | GO:0032815 | negative regulation of natural killer cell activation(GO:0032815) |
| 0.5 | 6.2 | GO:0031915 | positive regulation of synaptic plasticity(GO:0031915) |
| 0.5 | 2.1 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
| 0.5 | 2.6 | GO:0061709 | reticulophagy(GO:0061709) |
| 0.5 | 27.5 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.5 | 1.5 | GO:0034971 | growth plate cartilage chondrocyte proliferation(GO:0003419) histone H3-R17 methylation(GO:0034971) |
| 0.5 | 3.0 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.5 | 11.4 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
| 0.5 | 6.4 | GO:0060134 | prepulse inhibition(GO:0060134) |
| 0.5 | 4.4 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.5 | 2.0 | GO:0046351 | disaccharide biosynthetic process(GO:0046351) |
| 0.5 | 16.6 | GO:0009395 | phospholipid catabolic process(GO:0009395) |
| 0.5 | 2.4 | GO:0071603 | endothelial cell-cell adhesion(GO:0071603) |
| 0.5 | 5.8 | GO:0099612 | protein localization to axon(GO:0099612) |
| 0.5 | 1.4 | GO:0046947 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.5 | 5.2 | GO:0015747 | urate transport(GO:0015747) |
| 0.5 | 2.8 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.4 | 1.8 | GO:1904640 | response to methionine(GO:1904640) |
| 0.4 | 22.7 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.4 | 4.2 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.4 | 11.7 | GO:0015812 | gamma-aminobutyric acid transport(GO:0015812) |
| 0.4 | 6.2 | GO:0021681 | cerebellar granular layer development(GO:0021681) |
| 0.4 | 0.8 | GO:0009826 | unidimensional cell growth(GO:0009826) |
| 0.4 | 2.8 | GO:0000711 | meiotic DNA repair synthesis(GO:0000711) |
| 0.4 | 12.6 | GO:0046470 | phosphatidylcholine metabolic process(GO:0046470) |
| 0.4 | 1.2 | GO:0021817 | nucleokinesis involved in cell motility in cerebral cortex radial glia guided migration(GO:0021817) nuclear migration along microtubule(GO:0030473) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.4 | 3.0 | GO:0006735 | NADH regeneration(GO:0006735) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.4 | 4.8 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.4 | 1.8 | GO:0033383 | geranyl diphosphate metabolic process(GO:0033383) geranyl diphosphate biosynthetic process(GO:0033384) |
| 0.4 | 3.2 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.3 | 23.5 | GO:0090263 | positive regulation of canonical Wnt signaling pathway(GO:0090263) |
| 0.3 | 1.7 | GO:0036233 | glycine import(GO:0036233) |
| 0.3 | 1.0 | GO:0061643 | chemoattraction of axon(GO:0061642) chemorepulsion of axon(GO:0061643) |
| 0.3 | 2.6 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.3 | 2.6 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.3 | 1.3 | GO:0021779 | oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.3 | 9.4 | GO:1904659 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.3 | 8.4 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.3 | 4.3 | GO:0001967 | suckling behavior(GO:0001967) |
| 0.3 | 4.3 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.3 | 10.1 | GO:1903959 | regulation of anion transmembrane transport(GO:1903959) |
| 0.3 | 2.6 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.3 | 10.5 | GO:0006972 | hyperosmotic response(GO:0006972) |
| 0.3 | 0.8 | GO:0048749 | compound eye development(GO:0048749) |
| 0.3 | 7.0 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.3 | 1.7 | GO:0070459 | prolactin secretion(GO:0070459) |
| 0.3 | 5.5 | GO:0071157 | negative regulation of cell cycle arrest(GO:0071157) |
| 0.3 | 2.4 | GO:0033572 | transferrin transport(GO:0033572) |
| 0.3 | 3.1 | GO:0033141 | regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033139) positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.3 | 0.5 | GO:0014827 | intestine smooth muscle contraction(GO:0014827) |
| 0.3 | 4.6 | GO:0097062 | dendritic spine maintenance(GO:0097062) |
| 0.3 | 0.5 | GO:0031536 | positive regulation of exit from mitosis(GO:0031536) |
| 0.3 | 2.3 | GO:1990001 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) inhibition of cysteine-type endopeptidase activity involved in apoptotic process(GO:1990001) |
| 0.2 | 1.0 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.2 | 2.0 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.2 | 1.0 | GO:0051792 | medium-chain fatty acid biosynthetic process(GO:0051792) |
| 0.2 | 1.7 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.2 | 5.1 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.2 | 4.7 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.2 | 5.7 | GO:0045724 | positive regulation of cilium assembly(GO:0045724) |
| 0.2 | 7.1 | GO:0035036 | sperm-egg recognition(GO:0035036) |
| 0.2 | 1.4 | GO:0007042 | lysosomal lumen acidification(GO:0007042) |
| 0.2 | 3.3 | GO:0051923 | sulfation(GO:0051923) |
| 0.2 | 3.1 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.2 | 0.7 | GO:0098939 | dendritic transport of mitochondrion(GO:0098939) anterograde dendritic transport of mitochondrion(GO:0098972) |
| 0.2 | 11.0 | GO:0060999 | positive regulation of dendritic spine development(GO:0060999) |
| 0.2 | 10.7 | GO:0043392 | negative regulation of DNA binding(GO:0043392) |
| 0.2 | 7.6 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) |
| 0.2 | 1.1 | GO:1900122 | positive regulation of receptor binding(GO:1900122) |
| 0.2 | 2.8 | GO:0055015 | ventricular cardiac muscle cell development(GO:0055015) |
| 0.2 | 2.1 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.2 | 2.3 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 0.2 | 0.6 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) positive regulation of lymphangiogenesis(GO:1901492) |
| 0.2 | 5.6 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.2 | 5.1 | GO:0071526 | semaphorin-plexin signaling pathway(GO:0071526) |
| 0.2 | 5.1 | GO:0072661 | protein targeting to plasma membrane(GO:0072661) |
| 0.2 | 5.0 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.2 | 6.9 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.2 | 0.7 | GO:0021526 | medial motor column neuron differentiation(GO:0021526) |
| 0.2 | 7.2 | GO:0050974 | detection of mechanical stimulus involved in sensory perception(GO:0050974) |
| 0.2 | 3.1 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.2 | 0.7 | GO:1900378 | positive regulation of melanin biosynthetic process(GO:0048023) positive regulation of secondary metabolite biosynthetic process(GO:1900378) |
| 0.2 | 3.1 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.2 | 1.3 | GO:0035989 | tendon development(GO:0035989) integrin biosynthetic process(GO:0045112) |
| 0.2 | 6.8 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.2 | 1.0 | GO:1903054 | negative regulation of extracellular matrix organization(GO:1903054) |
| 0.2 | 1.0 | GO:0038170 | somatostatin receptor signaling pathway(GO:0038169) somatostatin signaling pathway(GO:0038170) |
| 0.2 | 2.2 | GO:0043457 | regulation of cellular respiration(GO:0043457) |
| 0.2 | 1.1 | GO:0015866 | ADP transport(GO:0015866) |
| 0.2 | 1.5 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.2 | 1.5 | GO:1990834 | response to odorant(GO:1990834) |
| 0.2 | 7.2 | GO:0007588 | excretion(GO:0007588) |
| 0.2 | 3.6 | GO:0016242 | negative regulation of macroautophagy(GO:0016242) |
| 0.1 | 3.2 | GO:0010667 | negative regulation of cardiac muscle cell apoptotic process(GO:0010667) |
| 0.1 | 4.6 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.1 | 7.6 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.1 | 3.2 | GO:0060712 | spongiotrophoblast layer development(GO:0060712) |
| 0.1 | 4.2 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.1 | 0.8 | GO:0046836 | glycolipid transport(GO:0046836) |
| 0.1 | 1.6 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.1 | 3.2 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 6.1 | GO:0045739 | positive regulation of DNA repair(GO:0045739) |
| 0.1 | 0.8 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.1 | 5.0 | GO:0048678 | response to axon injury(GO:0048678) |
| 0.1 | 0.6 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.1 | 1.7 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.1 | 0.6 | GO:1904781 | positive regulation of protein localization to centrosome(GO:1904781) |
| 0.1 | 5.3 | GO:0070830 | bicellular tight junction assembly(GO:0070830) |
| 0.1 | 4.8 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.1 | 1.1 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.1 | 2.8 | GO:0036119 | response to platelet-derived growth factor(GO:0036119) cellular response to platelet-derived growth factor stimulus(GO:0036120) |
| 0.1 | 0.5 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.1 | 15.2 | GO:0007286 | spermatid development(GO:0007286) |
| 0.1 | 0.4 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.1 | 0.5 | GO:0042940 | D-amino acid transport(GO:0042940) |
| 0.1 | 0.6 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.1 | 4.3 | GO:0050772 | positive regulation of axonogenesis(GO:0050772) |
| 0.1 | 3.4 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.1 | 0.5 | GO:0003433 | chondrocyte development involved in endochondral bone morphogenesis(GO:0003433) |
| 0.1 | 5.2 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.1 | 3.1 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.1 | 1.0 | GO:0009226 | nucleotide-sugar biosynthetic process(GO:0009226) |
| 0.1 | 0.6 | GO:1901727 | positive regulation of histone deacetylase activity(GO:1901727) |
| 0.1 | 2.9 | GO:0030514 | negative regulation of BMP signaling pathway(GO:0030514) |
| 0.1 | 6.8 | GO:0021549 | cerebellum development(GO:0021549) |
| 0.1 | 2.2 | GO:0045668 | negative regulation of osteoblast differentiation(GO:0045668) |
| 0.1 | 10.4 | GO:0021915 | neural tube development(GO:0021915) |
| 0.1 | 3.5 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.1 | 2.6 | GO:0017144 | drug metabolic process(GO:0017144) |
| 0.1 | 0.8 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.1 | 13.2 | GO:0016358 | dendrite development(GO:0016358) |
| 0.1 | 1.4 | GO:0051481 | negative regulation of cytosolic calcium ion concentration(GO:0051481) |
| 0.1 | 3.2 | GO:0042108 | positive regulation of cytokine biosynthetic process(GO:0042108) |
| 0.1 | 3.0 | GO:0009409 | response to cold(GO:0009409) |
| 0.1 | 2.3 | GO:0035418 | protein localization to synapse(GO:0035418) |
| 0.1 | 2.0 | GO:0002446 | neutrophil mediated immunity(GO:0002446) |
| 0.1 | 0.6 | GO:1901387 | positive regulation of voltage-gated calcium channel activity(GO:1901387) |
| 0.1 | 3.1 | GO:0022904 | respiratory electron transport chain(GO:0022904) |
| 0.1 | 0.6 | GO:0007035 | vacuolar acidification(GO:0007035) |
| 0.1 | 2.1 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.1 | 0.2 | GO:0009597 | detection of virus(GO:0009597) |
| 0.1 | 4.6 | GO:0048706 | embryonic skeletal system development(GO:0048706) |
| 0.1 | 2.1 | GO:0009250 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.1 | 5.1 | GO:0006694 | steroid biosynthetic process(GO:0006694) |
| 0.1 | 2.2 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.1 | 2.5 | GO:0010906 | regulation of glucose metabolic process(GO:0010906) |
| 0.1 | 3.2 | GO:0035383 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.0 | 1.1 | GO:0048635 | negative regulation of muscle organ development(GO:0048635) |
| 0.0 | 0.0 | GO:1904154 | positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 0.0 | 0.4 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 | 1.0 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 47.1 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 3.0 | GO:0007416 | synapse assembly(GO:0007416) |
| 0.0 | 2.3 | GO:1901659 | glycosyl compound biosynthetic process(GO:1901659) |
| 0.0 | 0.9 | GO:0043268 | positive regulation of potassium ion transport(GO:0043268) |
| 0.0 | 3.5 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 1.4 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.0 | 0.7 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 1.0 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.0 | 0.3 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 1.8 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 2.3 | GO:1902600 | hydrogen ion transmembrane transport(GO:1902600) |
| 0.0 | 0.9 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.0 | 0.9 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 0.1 | GO:0086100 | endothelin receptor signaling pathway(GO:0086100) |
| 0.0 | 0.7 | GO:0006900 | membrane budding(GO:0006900) |
| 0.0 | 0.1 | GO:0003105 | negative regulation of glomerular filtration(GO:0003105) |
| 0.0 | 0.3 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.0 | 0.3 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 0.6 | GO:0045103 | intermediate filament-based process(GO:0045103) |
| 0.0 | 0.7 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 0.1 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.0 | 0.2 | GO:0008631 | intrinsic apoptotic signaling pathway in response to oxidative stress(GO:0008631) |
| 0.0 | 1.0 | GO:0090174 | organelle membrane fusion(GO:0090174) |
| 0.0 | 0.6 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.2 | 31.3 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 3.9 | 23.4 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 2.7 | 13.4 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 2.6 | 25.9 | GO:0008091 | spectrin(GO:0008091) |
| 2.1 | 8.3 | GO:0018444 | translation release factor complex(GO:0018444) |
| 2.0 | 6.1 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 1.6 | 26.2 | GO:0005916 | fascia adherens(GO:0005916) |
| 1.6 | 6.5 | GO:1990761 | growth cone lamellipodium(GO:1990761) growth cone filopodium(GO:1990812) |
| 1.6 | 4.7 | GO:0070110 | ciliary neurotrophic factor receptor complex(GO:0070110) |
| 1.4 | 4.3 | GO:0032437 | cuticular plate(GO:0032437) |
| 1.4 | 8.6 | GO:0071547 | piP-body(GO:0071547) |
| 1.3 | 5.2 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 1.2 | 4.9 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 1.2 | 11.8 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 1.1 | 3.4 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 1.1 | 30.3 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 1.1 | 3.4 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 1.0 | 6.2 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) |
| 0.9 | 18.9 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.9 | 17.0 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.9 | 8.0 | GO:0097433 | dense body(GO:0097433) |
| 0.8 | 11.0 | GO:0032156 | septin cytoskeleton(GO:0032156) |
| 0.8 | 2.3 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.7 | 13.4 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.7 | 9.6 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.7 | 12.5 | GO:0030128 | clathrin coat of endocytic vesicle(GO:0030128) |
| 0.7 | 2.2 | GO:0090498 | extrinsic component of Golgi membrane(GO:0090498) |
| 0.7 | 7.2 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.7 | 2.8 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.7 | 2.0 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.6 | 7.9 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.6 | 2.4 | GO:0061474 | phagolysosome membrane(GO:0061474) |
| 0.6 | 2.3 | GO:0043541 | UDP-N-acetylglucosamine transferase complex(GO:0043541) |
| 0.6 | 4.0 | GO:0090543 | Flemming body(GO:0090543) |
| 0.6 | 7.4 | GO:0060091 | kinocilium(GO:0060091) |
| 0.5 | 2.7 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 0.5 | 8.0 | GO:0005883 | neurofilament(GO:0005883) |
| 0.5 | 3.2 | GO:1990393 | Cul7-RING ubiquitin ligase complex(GO:0031467) 3M complex(GO:1990393) |
| 0.5 | 3.0 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.5 | 5.6 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.5 | 11.6 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.5 | 5.9 | GO:0000801 | central element(GO:0000801) |
| 0.5 | 12.3 | GO:0031430 | M band(GO:0031430) |
| 0.5 | 4.1 | GO:1990075 | periciliary membrane compartment(GO:1990075) |
| 0.4 | 3.1 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.4 | 1.8 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 0.4 | 9.8 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.4 | 2.0 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.4 | 5.7 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.4 | 17.4 | GO:0016235 | aggresome(GO:0016235) |
| 0.4 | 10.5 | GO:0005921 | gap junction(GO:0005921) |
| 0.3 | 5.2 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.3 | 44.1 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.3 | 0.6 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.3 | 21.8 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.3 | 1.2 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.3 | 1.2 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.3 | 5.1 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.3 | 2.2 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.3 | 5.2 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.3 | 2.7 | GO:0030062 | mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.3 | 3.2 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.3 | 37.9 | GO:0031514 | motile cilium(GO:0031514) |
| 0.3 | 7.9 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.3 | 14.8 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.2 | 39.1 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.2 | 17.6 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.2 | 6.6 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.2 | 1.4 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.2 | 1.0 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.2 | 9.6 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.2 | 5.8 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.2 | 3.7 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.2 | 4.6 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.2 | 7.6 | GO:0030315 | T-tubule(GO:0030315) |
| 0.2 | 3.1 | GO:1902562 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.1 | 8.2 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.1 | 12.1 | GO:0000794 | condensed nuclear chromosome(GO:0000794) |
| 0.1 | 3.6 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.1 | 7.3 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.1 | 5.6 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.1 | 3.9 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 18.2 | GO:0043204 | perikaryon(GO:0043204) |
| 0.1 | 3.8 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.1 | 4.6 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.1 | 14.1 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.1 | 16.0 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.1 | 39.6 | GO:0045177 | apical part of cell(GO:0045177) |
| 0.1 | 11.8 | GO:0030658 | transport vesicle membrane(GO:0030658) |
| 0.1 | 6.3 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.1 | 0.8 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.1 | 7.4 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.1 | 2.6 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.1 | 1.4 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.1 | 4.3 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.1 | 2.3 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.1 | 11.7 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.1 | 1.1 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.1 | 0.6 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.1 | 3.9 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.1 | 1.5 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.1 | 5.5 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 3.5 | GO:0045202 | synapse(GO:0045202) |
| 0.0 | 1.3 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.6 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.0 | 0.8 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.3 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 1.1 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 3.2 | GO:0070382 | exocytic vesicle(GO:0070382) |
| 0.0 | 1.6 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.8 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 0.4 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 2.5 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 0.7 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 2.2 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 46.1 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.0 | 0.9 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 1.3 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.3 | GO:0030914 | SAGA complex(GO:0000124) STAGA complex(GO:0030914) |
| 0.0 | 1.4 | GO:0031461 | cullin-RING ubiquitin ligase complex(GO:0031461) |
| 0.0 | 4.4 | GO:0005929 | cilium(GO:0005929) |
| 0.0 | 0.6 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 7.4 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 1.9 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 1.7 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 1.8 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 14.2 | GO:0097458 | neuron part(GO:0097458) |
| 0.0 | 0.5 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 10.7 | 32.0 | GO:0052854 | very-long-chain-(S)-2-hydroxy-acid oxidase activity(GO:0052852) long-chain-(S)-2-hydroxy-long-chain-acid oxidase activity(GO:0052853) medium-chain-(S)-2-hydroxy-acid oxidase activity(GO:0052854) |
| 8.3 | 24.9 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 5.9 | 17.6 | GO:0017042 | glycosylceramidase activity(GO:0017042) |
| 3.5 | 31.3 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 3.3 | 9.8 | GO:0015203 | ornithine decarboxylase inhibitor activity(GO:0008073) polyamine transmembrane transporter activity(GO:0015203) putrescine transmembrane transporter activity(GO:0015489) |
| 3.2 | 19.1 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 2.9 | 8.8 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 2.6 | 7.9 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 2.5 | 14.9 | GO:0004473 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 2.4 | 7.2 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 2.3 | 11.4 | GO:0031798 | type 1 metabotropic glutamate receptor binding(GO:0031798) |
| 2.2 | 6.7 | GO:0031249 | denatured protein binding(GO:0031249) |
| 2.0 | 22.3 | GO:0039706 | co-receptor binding(GO:0039706) |
| 2.0 | 11.7 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 1.9 | 17.1 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 1.9 | 9.5 | GO:0004690 | cyclic nucleotide-dependent protein kinase activity(GO:0004690) |
| 1.8 | 10.7 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 1.7 | 7.0 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 1.6 | 4.9 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 1.6 | 12.7 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 1.5 | 7.7 | GO:0086062 | voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 1.5 | 4.4 | GO:0008396 | oxysterol 7-alpha-hydroxylase activity(GO:0008396) |
| 1.4 | 4.3 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 1.3 | 25.5 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 1.3 | 15.6 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 1.3 | 3.9 | GO:0015141 | succinate transmembrane transporter activity(GO:0015141) |
| 1.3 | 5.1 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 1.2 | 3.6 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 1.2 | 5.9 | GO:0004945 | angiotensin type II receptor activity(GO:0004945) |
| 1.2 | 4.7 | GO:0004897 | ciliary neurotrophic factor receptor activity(GO:0004897) |
| 1.2 | 15.0 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 1.1 | 7.8 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 1.1 | 19.9 | GO:0043295 | glutathione binding(GO:0043295) |
| 1.1 | 4.2 | GO:0042806 | fucose binding(GO:0042806) |
| 1.0 | 5.2 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 1.0 | 5.2 | GO:0097016 | L27 domain binding(GO:0097016) |
| 1.0 | 5.1 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 1.0 | 13.3 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 1.0 | 23.4 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 1.0 | 10.0 | GO:0019808 | polyamine binding(GO:0019808) |
| 1.0 | 2.9 | GO:0050429 | calcium-dependent phospholipase C activity(GO:0050429) |
| 0.9 | 2.8 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 0.9 | 9.4 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.9 | 2.7 | GO:0004775 | succinate-CoA ligase activity(GO:0004774) succinate-CoA ligase (ADP-forming) activity(GO:0004775) |
| 0.9 | 6.3 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.9 | 10.6 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.9 | 2.6 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.9 | 6.8 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.8 | 22.7 | GO:0031402 | sodium ion binding(GO:0031402) |
| 0.8 | 7.2 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.8 | 5.5 | GO:0043426 | MRF binding(GO:0043426) |
| 0.8 | 9.2 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.8 | 5.3 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.7 | 9.3 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.7 | 2.8 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.7 | 3.5 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.7 | 2.7 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.7 | 6.6 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.7 | 4.0 | GO:0004144 | 2-acylglycerol O-acyltransferase activity(GO:0003846) diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.7 | 9.2 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.6 | 12.9 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.6 | 7.7 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.6 | 4.4 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.6 | 68.5 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.6 | 8.0 | GO:0098919 | structural constituent of postsynaptic specialization(GO:0098879) structural constituent of synapse(GO:0098918) structural constituent of postsynaptic density(GO:0098919) |
| 0.6 | 3.6 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.6 | 6.9 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.6 | 3.3 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.5 | 2.7 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.5 | 2.1 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.5 | 5.2 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.5 | 3.0 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.5 | 16.1 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.5 | 1.4 | GO:0033823 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) |
| 0.5 | 3.3 | GO:0015526 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.5 | 5.5 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.5 | 5.0 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.5 | 5.4 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.4 | 7.1 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.4 | 13.3 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.4 | 6.2 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.4 | 1.8 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.4 | 3.1 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.4 | 6.9 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.4 | 14.2 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.4 | 6.0 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.4 | 30.2 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.4 | 4.6 | GO:0005549 | odorant binding(GO:0005549) |
| 0.4 | 7.9 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.4 | 6.6 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.4 | 2.4 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.4 | 25.3 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.4 | 3.2 | GO:0003996 | acyl-CoA ligase activity(GO:0003996) |
| 0.4 | 3.9 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.4 | 1.2 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.4 | 2.2 | GO:0051430 | corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.4 | 1.4 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 0.4 | 1.8 | GO:0004161 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.3 | 5.1 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.3 | 8.7 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.3 | 5.7 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.3 | 2.7 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.3 | 1.0 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.3 | 11.1 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.3 | 1.9 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.3 | 8.4 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.3 | 3.1 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.3 | 5.0 | GO:0016671 | oxidoreductase activity, acting on a sulfur group of donors, disulfide as acceptor(GO:0016671) |
| 0.3 | 1.8 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.3 | 4.7 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.3 | 8.1 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.3 | 7.2 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.3 | 8.5 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.3 | 4.7 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.3 | 5.2 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.3 | 0.5 | GO:0004315 | 3-oxoacyl-[acyl-carrier-protein] synthase activity(GO:0004315) |
| 0.3 | 6.1 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.3 | 1.5 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 0.2 | 4.2 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.2 | 5.6 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.2 | 5.6 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.2 | 7.6 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.2 | 11.2 | GO:0098531 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.2 | 1.8 | GO:0008172 | S-methyltransferase activity(GO:0008172) cobalamin binding(GO:0031419) |
| 0.2 | 7.0 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.2 | 0.8 | GO:0017089 | glycolipid transporter activity(GO:0017089) |
| 0.2 | 0.8 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.2 | 5.3 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.2 | 1.7 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.2 | 4.1 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.2 | 4.9 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.2 | 2.4 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.2 | 3.2 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.2 | 1.0 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.2 | 14.4 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.2 | 1.3 | GO:0031432 | titin binding(GO:0031432) |
| 0.2 | 0.6 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.2 | 4.5 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.2 | 8.0 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.1 | 1.3 | GO:0043394 | proteoglycan binding(GO:0043394) |
| 0.1 | 1.9 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.1 | 29.9 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.1 | 7.0 | GO:0051287 | NAD binding(GO:0051287) |
| 0.1 | 2.0 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.1 | 1.1 | GO:0015217 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.1 | 0.7 | GO:0031781 | melanocortin receptor binding(GO:0031779) type 3 melanocortin receptor binding(GO:0031781) type 4 melanocortin receptor binding(GO:0031782) |
| 0.1 | 22.2 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.1 | 0.8 | GO:0016901 | oxidoreductase activity, acting on the CH-OH group of donors, quinone or similar compound as acceptor(GO:0016901) |
| 0.1 | 5.3 | GO:0016706 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, 2-oxoglutarate as one donor, and incorporation of one atom each of oxygen into both donors(GO:0016706) |
| 0.1 | 3.2 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 7.5 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.1 | 1.6 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.1 | 3.1 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 3.4 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.1 | 2.5 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.1 | 7.4 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.1 | 0.3 | GO:0000991 | transcription factor activity, core RNA polymerase II binding(GO:0000991) |
| 0.1 | 0.8 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.1 | 4.7 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.1 | 11.2 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 1.7 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 2.3 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.1 | 3.4 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.1 | 2.0 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.1 | 2.0 | GO:0035250 | UDP-galactosyltransferase activity(GO:0035250) |
| 0.1 | 8.7 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.1 | 2.3 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 1.1 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.1 | 1.1 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.1 | 9.0 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.1 | 16.7 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.1 | 1.2 | GO:0005521 | lamin binding(GO:0005521) |
| 0.1 | 1.0 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.1 | 3.5 | GO:0004812 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.1 | 1.8 | GO:0016645 | oxidoreductase activity, acting on the CH-NH group of donors(GO:0016645) |
| 0.1 | 8.8 | GO:0003774 | motor activity(GO:0003774) |
| 0.1 | 0.6 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.1 | 2.6 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.1 | 0.7 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.1 | 1.5 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 1.5 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.1 | 1.7 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.1 | 1.9 | GO:0030295 | protein kinase activator activity(GO:0030295) |
| 0.0 | 1.0 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 0.6 | GO:0051378 | serotonin binding(GO:0051378) |
| 0.0 | 3.5 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 1.1 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 47.1 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 1.0 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 11.9 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 0.5 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.0 | 4.8 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.3 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.0 | 0.5 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 1.1 | GO:0016874 | ligase activity(GO:0016874) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 27.7 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.5 | 5.6 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.5 | 9.3 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.5 | 11.7 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.3 | 11.8 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.3 | 5.1 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.3 | 11.4 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.3 | 4.4 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.3 | 7.4 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.3 | 13.8 | PID ATR PATHWAY | ATR signaling pathway |
| 0.3 | 3.3 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.3 | 12.5 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.3 | 23.8 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.3 | 7.7 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.2 | 15.6 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.2 | 5.6 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.2 | 2.8 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.1 | 3.1 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.1 | 29.2 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 5.2 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.1 | 1.8 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.1 | 5.1 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.1 | 3.1 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 3.3 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.1 | 1.6 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.1 | 0.8 | PID FOXO PATHWAY | FoxO family signaling |
| 0.1 | 2.0 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.1 | 0.6 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.6 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 0.7 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.6 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 1.7 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 1.2 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.8 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.3 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.8 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 12.7 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 1.5 | 31.3 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 1.3 | 46.9 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.8 | 13.5 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.8 | 9.5 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.7 | 19.9 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.6 | 7.6 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.6 | 20.9 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.6 | 10.1 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.6 | 5.1 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.6 | 12.9 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.5 | 16.2 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.5 | 4.9 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.5 | 6.4 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.5 | 10.0 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.5 | 7.8 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.5 | 13.1 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF RAS | Genes involved in CREB phosphorylation through the activation of Ras |
| 0.5 | 17.6 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.4 | 6.8 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.4 | 5.3 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.4 | 26.5 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.3 | 5.2 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.3 | 7.1 | REACTOME CELL CELL JUNCTION ORGANIZATION | Genes involved in Cell-cell junction organization |
| 0.3 | 5.1 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.3 | 7.0 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.3 | 10.0 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.3 | 8.7 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.3 | 7.0 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.3 | 4.4 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.3 | 5.3 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.3 | 2.8 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.2 | 2.2 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.2 | 7.4 | REACTOME TRIGLYCERIDE BIOSYNTHESIS | Genes involved in Triglyceride Biosynthesis |
| 0.2 | 11.0 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.2 | 4.4 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.2 | 1.8 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.2 | 5.6 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.2 | 11.5 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.2 | 3.9 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.2 | 4.0 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.2 | 7.0 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.2 | 2.7 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.2 | 2.7 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.2 | 3.3 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.2 | 7.6 | REACTOME REGULATION OF INSULIN SECRETION | Genes involved in Regulation of Insulin Secretion |
| 0.1 | 2.0 | REACTOME MEIOSIS | Genes involved in Meiosis |
| 0.1 | 3.6 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 8.7 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.1 | 11.0 | REACTOME SIGNALING BY PDGF | Genes involved in Signaling by PDGF |
| 0.1 | 4.2 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.1 | 2.0 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 1.7 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.1 | 3.0 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.1 | 4.3 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.1 | 2.0 | REACTOME TRNA AMINOACYLATION | Genes involved in tRNA Aminoacylation |
| 0.1 | 1.8 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.1 | 5.1 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.1 | 4.2 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.1 | 1.4 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 1.5 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.6 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 4.7 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 1.8 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 2.6 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 1.0 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.4 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 1.4 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.3 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 0.3 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |