Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
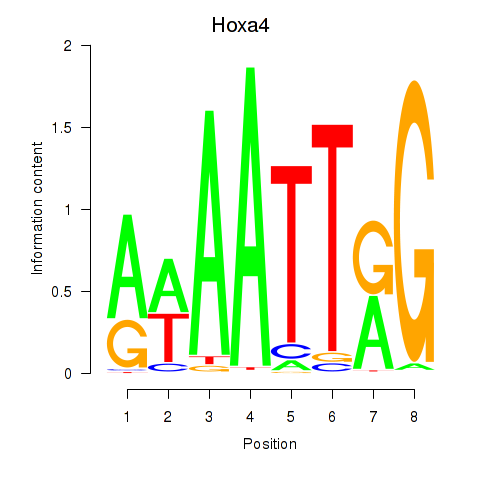
Results for Hoxa4
Z-value: 1.50
Transcription factors associated with Hoxa4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hoxa4
|
ENSRNOG00000027365 | homeo box A4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hoxa4 | rn6_v1_chr4_-_82160240_82160240 | 0.06 | 2.9e-01 | Click! |
Activity profile of Hoxa4 motif
Sorted Z-values of Hoxa4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_63836017 | 127.06 |
ENSRNOT00000030978
|
AABR07052585.1
|
|
| chr12_+_49761120 | 78.12 |
ENSRNOT00000070961
|
Myo18b
|
myosin XVIIIb |
| chr11_+_66713888 | 76.43 |
ENSRNOT00000003340
|
Fbxo40
|
F-box protein 40 |
| chr10_+_53740841 | 65.70 |
ENSRNOT00000004295
|
Myh2
|
myosin heavy chain 2 |
| chr15_-_27819376 | 63.01 |
ENSRNOT00000067400
|
A930018M24Rik
|
RIKEN cDNA A930018M24 gene |
| chr8_-_130429132 | 62.93 |
ENSRNOT00000026261
|
Hhatl
|
hedgehog acyltransferase-like |
| chr13_+_52889737 | 56.35 |
ENSRNOT00000074366
|
Cacna1s
|
calcium voltage-gated channel subunit alpha1 S |
| chr10_+_53818818 | 54.67 |
ENSRNOT00000057260
|
Myh8
|
myosin heavy chain 8 |
| chr10_+_53778662 | 52.75 |
ENSRNOT00000045718
|
Myh2
|
myosin heavy chain 2 |
| chr4_-_15859132 | 51.63 |
ENSRNOT00000082161
|
Cacna2d1
|
calcium voltage-gated channel auxiliary subunit alpha2delta 1 |
| chr15_+_4064706 | 43.97 |
ENSRNOT00000011956
|
Synpo2l
|
synaptopodin 2-like |
| chr10_-_8654892 | 43.00 |
ENSRNOT00000066534
|
Rbfox1
|
RNA binding protein, fox-1 homolog 1 |
| chr9_-_53315915 | 42.37 |
ENSRNOT00000038093
|
Mstn
|
myostatin |
| chr2_+_219598162 | 40.82 |
ENSRNOT00000020297
|
Lrrc39
|
leucine rich repeat containing 39 |
| chr3_+_168345152 | 38.05 |
ENSRNOT00000017654
|
Dok5
|
docking protein 5 |
| chr12_-_5685448 | 35.68 |
ENSRNOT00000077167
|
Fry
|
FRY microtubule binding protein |
| chrX_-_23144324 | 34.05 |
ENSRNOT00000000178
ENSRNOT00000081239 |
Pfkfb1
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 |
| chr4_+_172942020 | 33.02 |
ENSRNOT00000072450
|
Lmo3
|
LIM domain only 3 |
| chr13_-_98478327 | 32.55 |
ENSRNOT00000030135
|
Coq8a
|
coenzyme Q8A |
| chr8_+_110982777 | 31.27 |
ENSRNOT00000010992
|
Ky
|
kyphoscoliosis peptidase |
| chr15_+_2734068 | 29.58 |
ENSRNOT00000017663
|
Dusp13
|
dual specificity phosphatase 13 |
| chr15_+_11298478 | 29.20 |
ENSRNOT00000007672
|
Lrrc3b
|
leucine rich repeat containing 3B |
| chr2_-_98610368 | 28.57 |
ENSRNOT00000011641
|
Zfhx4
|
zinc finger homeobox 4 |
| chr10_-_34439470 | 27.98 |
ENSRNOT00000072081
|
Btnl9
|
butyrophilin-like 9 |
| chr7_+_70807867 | 26.15 |
ENSRNOT00000010639
|
Stac3
|
SH3 and cysteine rich domain 3 |
| chr6_-_8344574 | 24.78 |
ENSRNOT00000009660
|
Prepl
|
prolyl endopeptidase-like |
| chr1_-_99632845 | 24.66 |
ENSRNOT00000049123
|
LOC103691107
|
NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 4 pseudogene |
| chr3_-_48536235 | 24.56 |
ENSRNOT00000083536
|
Fap
|
fibroblast activation protein, alpha |
| chr1_+_90729274 | 23.42 |
ENSRNOT00000047207
|
RGD1560088
|
similar to NADH:ubiquinone oxidoreductase B15 subunit |
| chr1_-_192057612 | 21.11 |
ENSRNOT00000024471
|
Ndufab1
|
NADH:ubiquinone oxidoreductase subunit AB1 |
| chrX_-_42329232 | 20.61 |
ENSRNOT00000045705
|
LOC100361934
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex 4 |
| chr4_+_100166863 | 20.41 |
ENSRNOT00000014505
|
Sftpb
|
surfactant protein B |
| chrX_+_157759624 | 20.37 |
ENSRNOT00000003192
|
LOC686087
|
similar to motile sperm domain containing 1 |
| chr2_-_104461863 | 20.08 |
ENSRNOT00000016953
|
Crh
|
corticotropin releasing hormone |
| chr2_+_42829413 | 19.17 |
ENSRNOT00000065528
|
Actbl2
|
actin, beta-like 2 |
| chr2_+_252771017 | 18.74 |
ENSRNOT00000075452
ENSRNOT00000055318 |
Ttll7
|
tubulin tyrosine ligase like 7 |
| chr1_-_275882444 | 18.67 |
ENSRNOT00000083215
|
Gpam
|
glycerol-3-phosphate acyltransferase, mitochondrial |
| chr16_+_54332660 | 18.55 |
ENSRNOT00000037685
|
Mtus1
|
microtubule associated tumor suppressor 1 |
| chrM_+_5323 | 17.96 |
ENSRNOT00000050156
|
Mt-co1
|
mitochondrially encoded cytochrome c oxidase 1 |
| chrM_+_3904 | 17.86 |
ENSRNOT00000040993
|
Mt-nd2
|
mitochondrially encoded NADH dehydrogenase 2 |
| chr18_+_30527705 | 16.76 |
ENSRNOT00000027168
|
Pcdhb14
|
protocadherin beta 14 |
| chr14_-_13058172 | 16.47 |
ENSRNOT00000002746
ENSRNOT00000071706 |
Prdm8
|
PR/SET domain 8 |
| chr16_-_27472687 | 16.15 |
ENSRNOT00000046984
|
AABR07025074.1
|
|
| chr20_+_25990656 | 15.95 |
ENSRNOT00000081254
|
Lrrtm3
|
leucine rich repeat transmembrane neuronal 3 |
| chr14_+_7949239 | 15.86 |
ENSRNOT00000044617
|
Ndufb4l1
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 4-like 1 |
| chr7_-_69982592 | 15.22 |
ENSRNOT00000040010
|
RGD1564306
|
similar to developmental pluripotency associated 5 |
| chr10_-_98469799 | 15.19 |
ENSRNOT00000087502
ENSRNOT00000088646 |
Abca9
|
ATP binding cassette subfamily A member 9 |
| chr13_-_36290531 | 14.47 |
ENSRNOT00000071388
|
Steap3
|
STEAP3 metalloreductase |
| chr16_-_74700815 | 14.17 |
ENSRNOT00000081297
|
Thsd1
|
thrombospondin type 1 domain containing 1 |
| chr2_+_196334626 | 13.69 |
ENSRNOT00000050914
ENSRNOT00000028645 ENSRNOT00000090729 |
Sema6c
|
semaphorin 6C |
| chr4_+_119225040 | 13.45 |
ENSRNOT00000012365
|
Bmp10
|
bone morphogenetic protein 10 |
| chr4_+_129574264 | 13.32 |
ENSRNOT00000010185
|
Arl6ip5
|
ADP-ribosylation factor like GTPase 6 interacting protein 5 |
| chrX_+_107496072 | 13.22 |
ENSRNOT00000003283
|
Plp1
|
proteolipid protein 1 |
| chr2_-_46544457 | 13.20 |
ENSRNOT00000015680
|
Fst
|
follistatin |
| chr11_-_782954 | 13.19 |
ENSRNOT00000040065
|
Epha3
|
Eph receptor A3 |
| chr13_+_90943255 | 13.12 |
ENSRNOT00000011539
|
Vsig8
|
V-set and immunoglobulin domain containing 8 |
| chr1_-_255557055 | 12.63 |
ENSRNOT00000033780
|
Fgfbp3
|
fibroblast growth factor binding protein 3 |
| chr11_-_4397361 | 12.52 |
ENSRNOT00000046370
|
Cadm2
|
cell adhesion molecule 2 |
| chr10_+_13836128 | 12.22 |
ENSRNOT00000012720
|
Pgp
|
phosphoglycolate phosphatase |
| chr4_-_166869399 | 12.16 |
ENSRNOT00000007521
|
Tas2r121
|
taste receptor, type 2, member 121 |
| chr10_+_89251370 | 12.07 |
ENSRNOT00000076820
|
Aoc3
|
amine oxidase, copper containing 3 |
| chr7_-_143408276 | 11.92 |
ENSRNOT00000013122
ENSRNOT00000091540 |
Krt73
|
keratin 73 |
| chr15_-_62200837 | 10.80 |
ENSRNOT00000017599
|
Pcdh8
|
protocadherin 8 |
| chr1_-_174620064 | 10.35 |
ENSRNOT00000016224
|
Tmem41b
|
transmembrane protein 41B |
| chr13_-_97838228 | 10.25 |
ENSRNOT00000003618
|
Tfb2m
|
transcription factor B2, mitochondrial |
| chr7_+_12619774 | 10.05 |
ENSRNOT00000015257
|
Med16
|
mediator complex subunit 16 |
| chr4_+_56744561 | 9.98 |
ENSRNOT00000009737
|
Atp6v1f
|
ATPase H+ transporting V1 subunit F |
| chr20_+_25990304 | 9.95 |
ENSRNOT00000033980
|
Lrrtm3
|
leucine rich repeat transmembrane neuronal 3 |
| chr9_+_81566074 | 9.88 |
ENSRNOT00000074131
ENSRNOT00000046229 ENSRNOT00000090383 |
Pnkd
|
paroxysmal nonkinesigenic dyskinesia |
| chr20_-_11638483 | 9.77 |
ENSRNOT00000067800
|
Krtap12-2
|
keratin associated protein 12-2 |
| chr14_-_10395047 | 9.43 |
ENSRNOT00000002936
|
Gpat3
|
glycerol-3-phosphate acyltransferase 3 |
| chr1_+_228395558 | 9.00 |
ENSRNOT00000065411
|
Osbp
|
oxysterol binding protein |
| chr3_-_71798531 | 8.70 |
ENSRNOT00000088170
|
Calcrl
|
calcitonin receptor like receptor |
| chr11_-_71136673 | 8.64 |
ENSRNOT00000042240
|
Fyttd1
|
forty-two-three domain containing 1 |
| chr5_-_105582375 | 8.59 |
ENSRNOT00000083373
|
Slc24a2
|
solute carrier family 24 member 2 |
| chr14_-_2032593 | 8.41 |
ENSRNOT00000000037
|
Fgfrl1
|
fibroblast growth factor receptor-like 1 |
| chr18_+_30820321 | 8.41 |
ENSRNOT00000060472
|
Pcdhga3
|
protocadherin gamma subfamily A, 3 |
| chr5_-_711033 | 8.40 |
ENSRNOT00000024199
|
Crispld1
|
cysteine-rich secretory protein LCCL domain containing 1 |
| chr20_-_34679779 | 8.29 |
ENSRNOT00000078420
|
Cep85l
|
centrosomal protein 85-like |
| chr10_+_39850818 | 8.24 |
ENSRNOT00000012671
|
Fnip1
|
folliculin interacting protein 1 |
| chr8_+_48718329 | 7.96 |
ENSRNOT00000089763
|
Slc37a4
|
solute carrier family 37 member 4 |
| chr18_+_30509393 | 7.87 |
ENSRNOT00000043846
|
Pcdhb12
|
protocadherin beta 12 |
| chr18_-_17716880 | 7.73 |
ENSRNOT00000061151
|
Celf4
|
CUGBP, Elav-like family member 4 |
| chr11_-_62451149 | 7.65 |
ENSRNOT00000093686
ENSRNOT00000081443 |
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr10_-_87954055 | 7.59 |
ENSRNOT00000018048
|
Krt34
|
keratin 34 |
| chr10_-_87248572 | 7.43 |
ENSRNOT00000066637
ENSRNOT00000085677 |
Krt26
|
keratin 26 |
| chr12_+_24367199 | 7.16 |
ENSRNOT00000001971
|
Fkbp6
|
FK506 binding protein 6 |
| chr6_-_51019407 | 7.07 |
ENSRNOT00000011659
|
Gpr22
|
G protein-coupled receptor 22 |
| chr1_-_173469699 | 6.84 |
ENSRNOT00000040345
|
Olr281
|
olfactory receptor 281 |
| chr6_+_73358112 | 6.76 |
ENSRNOT00000041373
|
Arhgap5
|
Rho GTPase activating protein 5 |
| chrX_+_76083549 | 6.73 |
ENSRNOT00000003573
|
Magee1
|
MAGE family member E1 |
| chr3_+_95715193 | 6.65 |
ENSRNOT00000089525
|
Pax6
|
paired box 6 |
| chrX_-_13279082 | 6.51 |
ENSRNOT00000051898
ENSRNOT00000060857 |
Tspan7
|
tetraspanin 7 |
| chr3_+_56862691 | 6.37 |
ENSRNOT00000087712
|
Gad1
|
glutamate decarboxylase 1 |
| chr18_+_30387937 | 6.23 |
ENSRNOT00000027210
|
Pcdhb4
|
protocadherin beta 4 |
| chr3_-_161115313 | 6.20 |
ENSRNOT00000043716
|
Wfdc9
|
WAP four-disulfide core domain 9 |
| chr6_+_8219385 | 6.16 |
ENSRNOT00000040509
|
Ppm1b
|
protein phosphatase, Mg2+/Mn2+ dependent, 1B |
| chr16_+_8823872 | 6.02 |
ENSRNOT00000027186
|
Drgx
|
dorsal root ganglia homeobox |
| chr1_-_149603272 | 5.74 |
ENSRNOT00000045582
|
Olr13
|
olfactory receptor 13 |
| chr2_+_207930796 | 5.39 |
ENSRNOT00000047827
|
Kcnd3
|
potassium voltage-gated channel subfamily D member 3 |
| chr17_+_60287203 | 5.27 |
ENSRNOT00000025585
|
Armc4
|
armadillo repeat containing 4 |
| chr4_+_86135050 | 5.01 |
ENSRNOT00000081561
|
Ccdc129
|
coiled-coil domain containing 129 |
| chr8_-_122311431 | 5.00 |
ENSRNOT00000033126
|
Fbxl2
|
F-box and leucine-rich repeat protein 2 |
| chr17_+_38457783 | 4.87 |
ENSRNOT00000059757
|
Prl5a2
|
prolactin family 5, subfamily a, member 2 |
| chr6_-_51018050 | 4.76 |
ENSRNOT00000082691
|
Gpr22
|
G protein-coupled receptor 22 |
| chr6_+_97168453 | 4.72 |
ENSRNOT00000085790
|
Syt16
|
synaptotagmin 16 |
| chr20_-_11620945 | 4.56 |
ENSRNOT00000079725
|
Krtap12-2
|
keratin associated protein 12-2 |
| chr3_-_66417741 | 4.50 |
ENSRNOT00000007662
|
Neurod1
|
neuronal differentiation 1 |
| chr1_+_167857056 | 4.39 |
ENSRNOT00000074214
|
LOC684208
|
similar to olfactory receptor 557 |
| chr1_+_173012247 | 4.28 |
ENSRNOT00000073864
|
LOC100912217
|
olfactory receptor 10A3-like |
| chr9_+_3896337 | 4.08 |
ENSRNOT00000079166
|
LOC103693189
|
protein tyrosine phosphatase type IVA 1 |
| chr2_+_3750094 | 4.04 |
ENSRNOT00000064743
|
Mctp1
|
multiple C2 and transmembrane domain containing 1 |
| chr13_+_109909053 | 3.83 |
ENSRNOT00000090541
|
AC125873.1
|
|
| chr18_-_786674 | 3.82 |
ENSRNOT00000021955
|
Cetn1
|
centrin 1 |
| chr10_-_87286387 | 3.76 |
ENSRNOT00000044206
|
Krt28
|
keratin 28 |
| chr8_+_94686938 | 3.72 |
ENSRNOT00000013285
|
Ripply2
|
ripply transcriptional repressor 2 |
| chr12_-_47482961 | 3.70 |
ENSRNOT00000001600
|
Oasl2
|
2'-5' oligoadenylate synthetase-like 2 |
| chr1_-_22404002 | 3.63 |
ENSRNOT00000044098
|
Taar8a
|
trace amine-associated receptor 8a |
| chr3_-_74991168 | 3.51 |
ENSRNOT00000049273
|
Olr550
|
olfactory receptor 550 |
| chr7_-_4216833 | 3.40 |
ENSRNOT00000003174
|
Olr982
|
olfactory receptor 982 |
| chr4_+_87167514 | 3.01 |
ENSRNOT00000007821
|
Fkbp9
|
FK506 binding protein 9 |
| chrX_+_9436707 | 2.97 |
ENSRNOT00000004187
|
Cask
|
calcium/calmodulin dependent serine protein kinase |
| chr4_+_134398354 | 2.94 |
ENSRNOT00000077970
|
AABR07061577.1
|
|
| chr2_-_1561464 | 2.88 |
ENSRNOT00000062055
ENSRNOT00000062054 ENSRNOT00000062052 |
Cast
|
calpastatin |
| chr6_-_67084234 | 2.80 |
ENSRNOT00000050372
|
Nova1
|
NOVA alternative splicing regulator 1 |
| chr7_+_144647587 | 2.79 |
ENSRNOT00000022398
|
Hoxc4
|
homeo box C4 |
| chr7_-_7753978 | 2.76 |
ENSRNOT00000048223
|
Olr1029
|
olfactory receptor 1029 |
| chr1_-_163611299 | 2.72 |
ENSRNOT00000043788
|
Emsy
|
EMSY BRCA2-interacting transcriptional repressor |
| chr8_+_115511974 | 2.61 |
ENSRNOT00000067683
|
Timm8a1
|
translocase of inner mitochondrial membrane 8 homolog A1 (yeast) |
| chr10_+_78111050 | 2.60 |
ENSRNOT00000079364
|
Cox11
|
COX11 cytochrome c oxidase copper chaperone |
| chr7_+_15621989 | 2.55 |
ENSRNOT00000051928
|
Olr1095
|
olfactory receptor 1095 |
| chr7_-_6754349 | 2.50 |
ENSRNOT00000085061
|
Olr956
|
olfactory receptor 956 |
| chr3_+_31802999 | 2.43 |
ENSRNOT00000041305
|
AABR07051996.1
|
|
| chr4_+_108301129 | 2.37 |
ENSRNOT00000007993
|
LRRTM1
|
leucine rich repeat transmembrane neuronal 1 |
| chr4_-_167145096 | 2.24 |
ENSRNOT00000028970
|
Tas2r116
|
taste receptor, type 2, member 116 |
| chr1_-_173456488 | 2.11 |
ENSRNOT00000044753
|
Olr282
|
olfactory receptor 282 |
| chr3_+_76468294 | 2.10 |
ENSRNOT00000037779
|
Olr619
|
olfactory receptor 619 |
| chr16_+_60925093 | 1.82 |
ENSRNOT00000015813
|
Tnks
|
tankyrase |
| chr2_+_193627243 | 1.82 |
ENSRNOT00000082934
|
AABR07012331.1
|
|
| chr15_-_20783063 | 1.80 |
ENSRNOT00000083268
|
Bmp4
|
bone morphogenetic protein 4 |
| chr6_+_48452369 | 1.67 |
ENSRNOT00000044310
|
Myt1l
|
myelin transcription factor 1-like |
| chr18_+_12056113 | 1.64 |
ENSRNOT00000038450
|
Dsg4
|
desmoglein 4 |
| chr2_-_177924970 | 1.63 |
ENSRNOT00000029340
|
Rapgef2
|
Rap guanine nucleotide exchange factor 2 |
| chr9_-_4945352 | 1.50 |
ENSRNOT00000082530
|
Sult1c3
|
sulfotransferase family 1C member 3 |
| chr2_+_62198562 | 1.49 |
ENSRNOT00000087636
|
Zfr
|
zinc finger RNA binding protein |
| chr13_-_76049363 | 1.35 |
ENSRNOT00000075865
ENSRNOT00000007455 |
Brinp2
|
BMP/retinoic acid inducible neural specific 2 |
| chrX_+_156552528 | 1.20 |
ENSRNOT00000086429
|
Tex28
|
testis expressed 28 |
| chr20_-_32628953 | 1.16 |
ENSRNOT00000000451
|
Gprc6a
|
G protein-coupled receptor, class C, group 6, member A |
| chr1_-_169230360 | 1.15 |
ENSRNOT00000022740
|
Olr142
|
olfactory receptor 142 |
| chr1_-_156327352 | 1.06 |
ENSRNOT00000074282
|
LOC100911776
|
coiled-coil domain-containing protein 89-like |
| chr8_+_116715755 | 1.06 |
ENSRNOT00000090239
|
Camkv
|
CaM kinase-like vesicle-associated |
| chr2_+_24546536 | 1.03 |
ENSRNOT00000014036
|
Otp
|
orthopedia homeobox |
| chr1_+_201429771 | 0.95 |
ENSRNOT00000027836
|
Plekha1
|
pleckstrin homology domain containing A1 |
| chr3_-_76279104 | 0.92 |
ENSRNOT00000007796
|
Olr608
|
olfactory receptor 608 |
| chr4_-_167130996 | 0.92 |
ENSRNOT00000040860
|
Tas2r110
|
taste receptor, type 2, member 110 |
| chr9_-_74048244 | 0.90 |
ENSRNOT00000018293
|
Lancl1
|
LanC like 1 |
| chr1_-_129776276 | 0.87 |
ENSRNOT00000051402
|
Arrdc4
|
arrestin domain containing 4 |
| chr19_-_26053762 | 0.80 |
ENSRNOT00000004646
|
MAST1
|
microtubule associated serine/threonine kinase 1 |
| chr1_+_154606490 | 0.75 |
ENSRNOT00000024095
|
Ccdc89
|
coiled-coil domain containing 89 |
| chr7_-_107223047 | 0.65 |
ENSRNOT00000007250
ENSRNOT00000084875 |
Lrrc6
|
leucine rich repeat containing 6 |
| chr13_+_98311827 | 0.62 |
ENSRNOT00000082844
|
Cdc42bpa
|
CDC42 binding protein kinase alpha |
| chr5_-_69517277 | 0.47 |
ENSRNOT00000079848
|
Olr850
|
olfactory receptor 850 |
| chr9_+_37593032 | 0.45 |
ENSRNOT00000051890
|
AABR07067274.1
|
|
| chr9_-_88748866 | 0.35 |
ENSRNOT00000071906
|
Rpl30l1
|
ribosomal protein L30-like 1 |
| chr8_+_90304148 | 0.30 |
ENSRNOT00000001195
|
AABR07070892.1
|
|
| chr3_-_77605962 | 0.27 |
ENSRNOT00000090062
|
Olr665
|
olfactory receptor 665 |
| chr2_-_155381839 | 0.25 |
ENSRNOT00000057657
|
Vom2r47
|
vomeronasal 2 receptor, 47 |
| chr1_+_169074475 | 0.25 |
ENSRNOT00000050248
|
Olr132
|
olfactory receptor 132 |
| chr10_-_29450644 | 0.24 |
ENSRNOT00000087937
|
Adra1b
|
adrenoceptor alpha 1B |
| chr5_-_12199283 | 0.12 |
ENSRNOT00000007769
|
Pcmtd1
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase domain containing 1 |
| chr4_-_82160240 | 0.08 |
ENSRNOT00000038775
ENSRNOT00000058985 |
Hoxa4
|
homeo box A4 |
| chr10_+_62566732 | 0.05 |
ENSRNOT00000021368
|
Taok1
|
TAO kinase 1 |
| chr2_+_46140482 | 0.05 |
ENSRNOT00000072858
|
Olr1262
|
olfactory receptor 1262 |
| chr2_-_118882562 | 0.03 |
ENSRNOT00000058860
|
Kcnmb3
|
potassium calcium-activated channel subfamily M regulatory beta subunit 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hoxa4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 10.5 | 62.9 | GO:1903059 | regulation of protein lipidation(GO:1903059) |
| 9.1 | 118.5 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 8.9 | 35.7 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 8.6 | 51.6 | GO:1901843 | membrane depolarization during bundle of His cell action potential(GO:0086048) positive regulation of high voltage-gated calcium channel activity(GO:1901843) |
| 8.2 | 24.6 | GO:0097325 | melanocyte proliferation(GO:0097325) |
| 8.0 | 56.3 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 7.0 | 21.1 | GO:0009249 | protein lipoylation(GO:0009249) |
| 6.7 | 79.9 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 6.1 | 54.7 | GO:0030049 | muscle filament sliding(GO:0030049) |
| 6.1 | 42.4 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) negative regulation of skeletal muscle cell differentiation(GO:2001015) |
| 5.0 | 20.1 | GO:0070093 | hormone-mediated apoptotic signaling pathway(GO:0008628) positive regulation of cortisol secretion(GO:0051464) negative regulation of glucagon secretion(GO:0070093) |
| 4.8 | 14.5 | GO:1902167 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902167) |
| 4.7 | 33.0 | GO:0035360 | positive regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035360) |
| 4.4 | 13.2 | GO:0097156 | fasciculation of motor neuron axon(GO:0097156) |
| 4.3 | 34.0 | GO:0033133 | positive regulation of glucokinase activity(GO:0033133) |
| 4.1 | 20.4 | GO:0050828 | regulation of liquid surface tension(GO:0050828) |
| 4.1 | 12.2 | GO:0006114 | glycerol biosynthetic process(GO:0006114) |
| 3.1 | 28.1 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 2.8 | 8.4 | GO:0060988 | lipid tube assembly(GO:0060988) |
| 2.7 | 13.5 | GO:0060298 | atrial cardiac muscle tissue development(GO:0003228) atrial cardiac muscle tissue morphogenesis(GO:0055009) positive regulation of sarcomere organization(GO:0060298) |
| 2.5 | 43.0 | GO:2001014 | regulation of skeletal muscle cell differentiation(GO:2001014) |
| 2.3 | 11.6 | GO:0002036 | regulation of L-glutamate transport(GO:0002036) |
| 2.2 | 38.1 | GO:0051386 | regulation of neurotrophin TRK receptor signaling pathway(GO:0051386) |
| 2.2 | 6.6 | GO:1904937 | pancreatic A cell development(GO:0003322) forebrain-midbrain boundary formation(GO:0021905) somatic motor neuron fate commitment(GO:0021917) regulation of transcription from RNA polymerase II promoter involved in somatic motor neuron fate commitment(GO:0021918) sensory neuron migration(GO:1904937) |
| 2.0 | 32.6 | GO:1901663 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 2.0 | 9.9 | GO:0061727 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 1.9 | 13.2 | GO:0046882 | negative regulation of follicle-stimulating hormone secretion(GO:0046882) |
| 1.9 | 18.7 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 1.8 | 12.6 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 1.6 | 16.5 | GO:0021540 | corpus callosum morphogenesis(GO:0021540) |
| 1.5 | 18.0 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 1.5 | 17.8 | GO:1902004 | positive regulation of beta-amyloid formation(GO:1902004) positive regulation of amyloid precursor protein catabolic process(GO:1902993) |
| 1.4 | 8.2 | GO:0002904 | positive regulation of B cell apoptotic process(GO:0002904) |
| 1.3 | 6.4 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 1.1 | 8.0 | GO:0035166 | post-embryonic hemopoiesis(GO:0035166) |
| 1.1 | 7.4 | GO:2000675 | negative regulation of type B pancreatic cell apoptotic process(GO:2000675) |
| 1.0 | 7.7 | GO:0090394 | negative regulation of excitatory postsynaptic potential(GO:0090394) regulation of retina development in camera-type eye(GO:1902866) |
| 0.9 | 13.2 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.9 | 2.8 | GO:1990790 | response to glial cell derived neurotrophic factor(GO:1990790) cellular response to glial cell derived neurotrophic factor(GO:1990792) |
| 0.8 | 17.9 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.8 | 10.3 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.7 | 4.5 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) |
| 0.7 | 18.6 | GO:0010758 | regulation of macrophage chemotaxis(GO:0010758) |
| 0.7 | 12.1 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.7 | 8.4 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.7 | 8.7 | GO:0034285 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.6 | 2.6 | GO:0033131 | regulation of glucokinase activity(GO:0033131) |
| 0.6 | 26.1 | GO:0048741 | skeletal muscle fiber development(GO:0048741) |
| 0.6 | 1.8 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) regulation of telomeric DNA binding(GO:1904742) |
| 0.5 | 6.0 | GO:0021559 | trigeminal nerve development(GO:0021559) |
| 0.4 | 5.4 | GO:0097623 | potassium ion export across plasma membrane(GO:0097623) |
| 0.4 | 5.3 | GO:0003356 | regulation of cilium movement(GO:0003352) regulation of cilium beat frequency(GO:0003356) |
| 0.4 | 11.9 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.4 | 7.2 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.4 | 23.6 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.4 | 1.6 | GO:1900377 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.4 | 55.6 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.4 | 9.0 | GO:0044126 | regulation of growth of symbiont in host(GO:0044126) modulation of growth of symbiont involved in interaction with host(GO:0044144) |
| 0.3 | 2.4 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 0.3 | 10.0 | GO:0015991 | ATP hydrolysis coupled proton transport(GO:0015991) |
| 0.3 | 3.7 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.3 | 7.6 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.2 | 9.3 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.2 | 0.2 | GO:0001982 | baroreceptor response to decreased systemic arterial blood pressure(GO:0001982) |
| 0.2 | 3.0 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.2 | 6.8 | GO:0002053 | positive regulation of mesenchymal cell proliferation(GO:0002053) |
| 0.2 | 10.1 | GO:0030521 | androgen receptor signaling pathway(GO:0030521) |
| 0.2 | 14.5 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.2 | 1.0 | GO:0021979 | hypothalamus cell differentiation(GO:0021979) |
| 0.2 | 8.6 | GO:0060292 | long term synaptic depression(GO:0060292) |
| 0.2 | 5.0 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.1 | 8.6 | GO:0006406 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.1 | 15.0 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.1 | 0.6 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.1 | 34.8 | GO:0042692 | muscle cell differentiation(GO:0042692) |
| 0.1 | 4.7 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.1 | 0.6 | GO:0007097 | nuclear migration(GO:0007097) |
| 0.1 | 3.0 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 3.8 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.1 | 0.8 | GO:0050912 | detection of chemical stimulus involved in sensory perception of taste(GO:0050912) |
| 0.0 | 1.8 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.0 | 0.9 | GO:0051443 | positive regulation of ubiquitin-protein transferase activity(GO:0051443) |
| 0.0 | 6.1 | GO:0007411 | axon guidance(GO:0007411) |
| 0.0 | 6.5 | GO:0050900 | leukocyte migration(GO:0050900) |
| 0.0 | 32.6 | GO:0050907 | detection of chemical stimulus involved in sensory perception(GO:0050907) detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 2.8 | GO:0050684 | regulation of mRNA processing(GO:0050684) |
| 0.0 | 1.3 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.0 | 2.0 | GO:0006626 | protein targeting to mitochondrion(GO:0006626) |
| 0.0 | 1.1 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 13.2 | 118.5 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 8.7 | 78.1 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 3.9 | 54.7 | GO:0032982 | myosin filament(GO:0032982) |
| 3.5 | 24.6 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 2.5 | 29.5 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 1.7 | 56.3 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 1.5 | 20.4 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 1.1 | 18.0 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 1.0 | 20.1 | GO:0043196 | varicosity(GO:0043196) |
| 0.8 | 10.0 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.8 | 39.0 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.6 | 88.7 | GO:0030018 | Z disc(GO:0030018) |
| 0.6 | 6.7 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.4 | 14.5 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.3 | 12.5 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.2 | 3.0 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.2 | 28.4 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.2 | 43.0 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.2 | 10.3 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.2 | 6.4 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.2 | 5.3 | GO:0097546 | ciliary base(GO:0097546) |
| 0.1 | 1.6 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 30.4 | GO:0030427 | site of polarized growth(GO:0030427) |
| 0.1 | 8.7 | GO:0044291 | cell-cell contact zone(GO:0044291) |
| 0.1 | 18.7 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.1 | 3.6 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.1 | 18.8 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.1 | 7.9 | GO:0005902 | microvillus(GO:0005902) |
| 0.1 | 4.2 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.1 | 5.0 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.1 | 5.4 | GO:0005901 | caveola(GO:0005901) |
| 0.1 | 11.5 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 6.8 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 2.6 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 13.6 | GO:0044445 | cytosolic part(GO:0044445) |
| 0.0 | 20.0 | GO:0005794 | Golgi apparatus(GO:0005794) |
| 0.0 | 1.6 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 5.0 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 249.0 | GO:0016021 | integral component of membrane(GO:0016021) |
| 0.0 | 1.5 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 28.5 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 1.0 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.6 | GO:0042641 | actomyosin(GO:0042641) |
| 0.0 | 1.1 | GO:0030659 | cytoplasmic vesicle membrane(GO:0030659) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 17.2 | 51.6 | GO:0086057 | voltage-gated calcium channel activity involved in bundle of His cell action potential(GO:0086057) |
| 7.0 | 21.1 | GO:0031177 | phosphopantetheine binding(GO:0031177) |
| 5.0 | 20.1 | GO:0051431 | corticotropin-releasing hormone receptor 2 binding(GO:0051431) |
| 4.9 | 34.0 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 4.7 | 28.1 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 3.7 | 18.7 | GO:0070739 | protein-glutamic acid ligase activity(GO:0070739) |
| 3.6 | 14.5 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 3.4 | 10.3 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 2.9 | 8.7 | GO:0001605 | adrenomedullin receptor activity(GO:0001605) |
| 2.5 | 54.7 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 2.5 | 24.6 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 2.1 | 24.8 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 2.0 | 12.2 | GO:0098519 | nucleotide phosphatase activity, acting on free nucleotides(GO:0098519) |
| 2.0 | 9.9 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 1.9 | 13.5 | GO:0031433 | telethonin binding(GO:0031433) |
| 1.8 | 16.5 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 1.8 | 34.2 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 1.4 | 38.5 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 1.3 | 13.2 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 1.3 | 6.4 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 1.2 | 8.6 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 1.2 | 8.2 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 1.1 | 8.0 | GO:0015119 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 1.1 | 10.1 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 1.1 | 13.2 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 1.1 | 12.1 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 1.1 | 5.4 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 1.1 | 13.7 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 1.1 | 8.4 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 1.0 | 13.2 | GO:0048185 | activin binding(GO:0048185) |
| 0.9 | 12.2 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.8 | 44.2 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.8 | 10.2 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.8 | 32.6 | GO:0043531 | ADP binding(GO:0043531) |
| 0.8 | 3.8 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.7 | 29.1 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.7 | 6.6 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.6 | 77.9 | GO:0003774 | motor activity(GO:0003774) |
| 0.6 | 33.7 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.6 | 18.0 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.6 | 12.6 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.5 | 10.0 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.4 | 3.7 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.4 | 1.5 | GO:0004027 | alcohol sulfotransferase activity(GO:0004027) |
| 0.3 | 76.4 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.3 | 132.9 | GO:0003779 | actin binding(GO:0003779) |
| 0.3 | 9.0 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.3 | 28.3 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.3 | 1.6 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 0.2 | 6.8 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.2 | 3.6 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.2 | 3.0 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.2 | 1.7 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 0.1 | 34.9 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.1 | 8.8 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.1 | 2.8 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.1 | 15.2 | GO:0042626 | ATPase activity, coupled to transmembrane movement of substances(GO:0042626) |
| 0.1 | 4.5 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.1 | 4.5 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.1 | 0.2 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.1 | 1.8 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.1 | 25.1 | GO:0004857 | enzyme inhibitor activity(GO:0004857) |
| 0.1 | 2.3 | GO:0099589 | G-protein coupled serotonin receptor activity(GO:0004993) serotonin receptor activity(GO:0099589) |
| 0.1 | 0.8 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.1 | 1.0 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.1 | 0.8 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.1 | 2.6 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.1 | 0.9 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.0 | 22.2 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 4.9 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 1.1 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.1 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.0 | 5.4 | GO:0001664 | G-protein coupled receptor binding(GO:0001664) |
| 0.0 | 3.3 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 30.1 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 8.4 | GO:0004930 | G-protein coupled receptor activity(GO:0004930) |
| 0.0 | 2.6 | GO:0008022 | protein C-terminus binding(GO:0008022) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 118.5 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.7 | 29.1 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.3 | 8.4 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.3 | 11.3 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.3 | 15.0 | PID BMP PATHWAY | BMP receptor signaling |
| 0.2 | 65.6 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.2 | 18.7 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.2 | 3.0 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.1 | 31.3 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 18.4 | NABA MATRISOME ASSOCIATED | Ensemble of genes encoding ECM-associated proteins including ECM-affilaited proteins, ECM regulators and secreted factors |
| 0.1 | 6.8 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.1 | 10.3 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.1 | 5.6 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 1.0 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 54.7 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 1.3 | 56.3 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.9 | 34.0 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.8 | 24.5 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.8 | 15.2 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.6 | 18.7 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.5 | 4.5 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.4 | 12.5 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.4 | 6.6 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.3 | 21.1 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.3 | 28.8 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.2 | 9.0 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.2 | 6.4 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.2 | 8.0 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.1 | 3.0 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.1 | 5.4 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 8.6 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.1 | 4.2 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.1 | 1.2 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 4.1 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |