Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
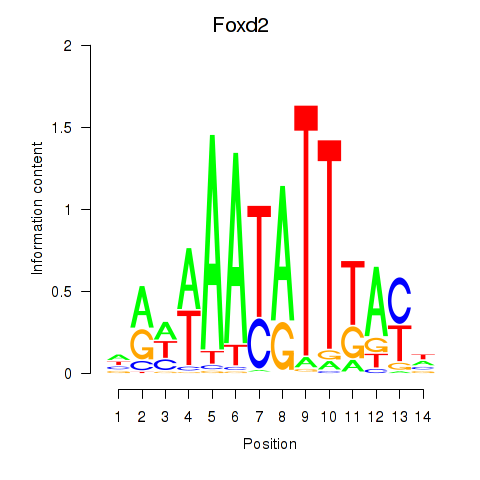
Results for Foxd2
Z-value: 0.95
Transcription factors associated with Foxd2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Foxd2
|
ENSRNOG00000007759 | forkhead box D2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Foxd2 | rn6_v1_chr5_-_133709712_133709712 | 0.30 | 2.6e-08 | Click! |
Activity profile of Foxd2 motif
Sorted Z-values of Foxd2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_+_95501778 | 64.03 |
ENSRNOT00000086805
|
Spp2
|
secreted phosphoprotein 2 |
| chr19_+_15081590 | 35.17 |
ENSRNOT00000024187
|
Ces1f
|
carboxylesterase 1F |
| chr19_+_15081158 | 34.37 |
ENSRNOT00000074070
|
Ces1f
|
carboxylesterase 1F |
| chr7_-_30105132 | 34.28 |
ENSRNOT00000091227
|
Nr1h4
|
nuclear receptor subfamily 1, group H, member 4 |
| chr4_-_176294997 | 33.91 |
ENSRNOT00000015112
ENSRNOT00000051461 ENSRNOT00000000026 ENSRNOT00000039877 ENSRNOT00000049303 |
Slc21a4
|
kidney specific organic anion transporter |
| chr14_+_85113578 | 32.79 |
ENSRNOT00000011158
|
Nipsnap1
|
nipsnap homolog 1 |
| chr1_-_253000760 | 30.47 |
ENSRNOT00000030024
|
Slc16a12
|
solute carrier family 16, member 12 |
| chr6_+_80188943 | 24.64 |
ENSRNOT00000059335
|
Mia2
|
melanoma inhibitory activity 2 |
| chr6_-_127620296 | 21.70 |
ENSRNOT00000012577
|
Serpina1
|
serpin family A member 1 |
| chr10_+_3411380 | 20.70 |
ENSRNOT00000004346
|
RGD1305733
|
similar to RIKEN cDNA 2900011O08 |
| chr2_+_235264219 | 20.69 |
ENSRNOT00000086245
|
Cfi
|
complement factor I |
| chr6_-_127508452 | 19.29 |
ENSRNOT00000073709
|
LOC100909524
|
protein Z-dependent protease inhibitor-like |
| chr2_-_187706300 | 18.54 |
ENSRNOT00000092349
ENSRNOT00000026414 |
Tmem79
|
transmembrane protein 79 |
| chr16_+_50181316 | 18.19 |
ENSRNOT00000077662
|
F11
|
coagulation factor XI |
| chr1_-_76780230 | 18.18 |
ENSRNOT00000002046
|
LOC100912485
|
alcohol sulfotransferase-like |
| chr7_+_34326087 | 17.65 |
ENSRNOT00000006971
|
Hal
|
histidine ammonia lyase |
| chr7_-_143793970 | 16.02 |
ENSRNOT00000016205
|
Csad
|
cysteine sulfinic acid decarboxylase |
| chr8_-_21901829 | 15.92 |
ENSRNOT00000027963
|
Angptl6
|
angiopoietin-like 6 |
| chr3_-_134696654 | 15.71 |
ENSRNOT00000006454
|
Flrt3
|
fibronectin leucine rich transmembrane protein 3 |
| chr13_+_91080341 | 15.27 |
ENSRNOT00000000058
|
Crp
|
C-reactive protein |
| chr7_-_2677199 | 14.40 |
ENSRNOT00000043908
|
Apon
|
apolipoprotein N |
| chr15_-_44627765 | 14.15 |
ENSRNOT00000058887
|
Dock5
|
dedicator of cytokinesis 5 |
| chr2_-_63166509 | 14.08 |
ENSRNOT00000018246
|
Cdh6
|
cadherin 6 |
| chr1_-_189181901 | 13.74 |
ENSRNOT00000092022
|
Gp2
|
glycoprotein 2 |
| chr15_-_77736892 | 13.42 |
ENSRNOT00000057924
|
Pcdh9
|
protocadherin 9 |
| chr3_-_2453933 | 13.26 |
ENSRNOT00000014060
|
Slc34a3
|
solute carrier family 34 member 3 |
| chr15_-_52320385 | 12.39 |
ENSRNOT00000067776
|
Dmtn
|
dematin actin binding protein |
| chr7_-_143523457 | 12.26 |
ENSRNOT00000012943
ENSRNOT00000082264 |
Krt4
|
keratin 4 |
| chr20_+_20519031 | 12.09 |
ENSRNOT00000000780
|
AABR07044783.1
|
|
| chr8_-_127900463 | 11.90 |
ENSRNOT00000078303
|
Slc22a13
|
solute carrier family 22 member 13 |
| chrX_-_63807810 | 11.44 |
ENSRNOT00000084632
|
Maged1
|
MAGE family member D1 |
| chr2_+_54466280 | 11.34 |
ENSRNOT00000033112
|
C6
|
complement C6 |
| chr4_-_28437676 | 11.34 |
ENSRNOT00000012995
|
Hepacam2
|
HEPACAM family member 2 |
| chr18_+_3900714 | 10.98 |
ENSRNOT00000092814
|
Lama3
|
laminin subunit alpha 3 |
| chr3_-_12155098 | 10.85 |
ENSRNOT00000082696
|
Garnl3
|
GTPase activating Rap/RanGAP domain-like 3 |
| chr3_-_66885085 | 10.48 |
ENSRNOT00000084299
ENSRNOT00000090547 |
Pde1a
|
phosphodiesterase 1A |
| chr10_-_70788309 | 10.47 |
ENSRNOT00000029184
|
Ccl9
|
chemokine (C-C motif) ligand 9 |
| chr2_+_27905535 | 9.32 |
ENSRNOT00000022120
|
Fam169a
|
family with sequence similarity 169, member A |
| chr7_+_73273985 | 9.26 |
ENSRNOT00000077730
|
Pop1
|
POP1 homolog, ribonuclease P/MRP subunit |
| chr7_+_76092503 | 9.00 |
ENSRNOT00000079692
|
Grhl2
|
grainyhead-like transcription factor 2 |
| chr8_-_54990604 | 8.90 |
ENSRNOT00000059192
|
Tex12
|
testis expressed 12 |
| chr20_+_27592379 | 8.87 |
ENSRNOT00000047000
|
Trappc3l
|
trafficking protein particle complex 3-like |
| chr4_-_85915099 | 8.72 |
ENSRNOT00000016182
|
Neurod6
|
neuronal differentiation 6 |
| chr3_-_48831417 | 8.46 |
ENSRNOT00000009920
ENSRNOT00000085246 |
Kcnh7
|
potassium voltage-gated channel subfamily H member 7 |
| chr14_-_42560174 | 8.40 |
ENSRNOT00000003128
|
Tmem33
|
transmembrane protein 33 |
| chr17_-_77527894 | 8.06 |
ENSRNOT00000032173
|
Bend7
|
BEN domain containing 7 |
| chr19_+_38669230 | 7.88 |
ENSRNOT00000027273
|
Cdh3
|
cadherin 3 |
| chr1_+_185356975 | 7.70 |
ENSRNOT00000086681
|
Plekha7
|
pleckstrin homology domain containing A7 |
| chr17_-_51912496 | 7.57 |
ENSRNOT00000019272
|
Inhba
|
inhibin beta A subunit |
| chr18_-_77579969 | 7.46 |
ENSRNOT00000034896
|
Sall3
|
spalt-like transcription factor 3 |
| chr5_+_116421894 | 7.26 |
ENSRNOT00000080577
ENSRNOT00000086628 ENSRNOT00000004017 |
Nfia
|
nuclear factor I/A |
| chr10_+_71546977 | 7.11 |
ENSRNOT00000041140
|
Acaca
|
acetyl-CoA carboxylase alpha |
| chr5_-_28131133 | 6.96 |
ENSRNOT00000067331
|
Slc26a7
|
solute carrier family 26 member 7 |
| chr2_+_143475323 | 6.94 |
ENSRNOT00000044028
ENSRNOT00000015437 |
Trpc4
|
transient receptor potential cation channel, subfamily C, member 4 |
| chr19_-_44101365 | 6.65 |
ENSRNOT00000082182
|
Tmem170a
|
transmembrane protein 170A |
| chr2_+_204427608 | 6.61 |
ENSRNOT00000083374
|
Nhlh2
|
nescient helix loop helix 2 |
| chr20_+_18833481 | 6.32 |
ENSRNOT00000080846
|
Bicc1
|
BicC family RNA binding protein 1 |
| chr17_-_21705773 | 6.25 |
ENSRNOT00000078010
|
Gcnt2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme |
| chr5_-_33664435 | 6.23 |
ENSRNOT00000009047
|
Wwp1
|
WW domain containing E3 ubiquitin protein ligase 1 |
| chr10_+_83154423 | 6.08 |
ENSRNOT00000005939
|
Fam117a
|
family with sequence similarity 117, member A |
| chr4_+_108301129 | 5.98 |
ENSRNOT00000007993
|
LRRTM1
|
leucine rich repeat transmembrane neuronal 1 |
| chr4_+_35279063 | 5.89 |
ENSRNOT00000011833
|
Nxph1
|
neurexophilin 1 |
| chr5_+_103251986 | 5.81 |
ENSRNOT00000008757
|
Cntln
|
centlein |
| chr1_-_67065797 | 5.72 |
ENSRNOT00000048152
|
Vom1r46
|
vomeronasal 1 receptor 46 |
| chr2_-_191294374 | 5.70 |
ENSRNOT00000067469
|
RGD1562234
|
similar to S100 calcium-binding protein, ventral prostate |
| chr8_+_14060394 | 5.57 |
ENSRNOT00000014827
|
Smco4
|
single-pass membrane protein with coiled-coil domains 4 |
| chr4_-_2201749 | 5.37 |
ENSRNOT00000089327
|
Lmbr1
|
limb development membrane protein 1 |
| chr17_-_10622226 | 5.29 |
ENSRNOT00000044559
|
Simc1
|
SUMO-interacting motifs containing 1 |
| chr15_-_60752106 | 5.13 |
ENSRNOT00000058148
|
Akap11
|
A-kinase anchoring protein 11 |
| chr17_+_81455955 | 5.11 |
ENSRNOT00000044313
|
Slc39a12
|
solute carrier family 39 member 12 |
| chr15_+_62406873 | 5.06 |
ENSRNOT00000047572
|
Olfm4
|
olfactomedin 4 |
| chr9_+_73493027 | 5.00 |
ENSRNOT00000074427
ENSRNOT00000089478 |
Unc80
|
unc-80 homolog, NALCN activator |
| chr4_+_57034675 | 4.97 |
ENSRNOT00000080223
|
Smo
|
smoothened, frizzled class receptor |
| chr11_-_1983513 | 4.89 |
ENSRNOT00000000907
|
Htr1f
|
5-hydroxytryptamine receptor 1F |
| chr1_+_243276403 | 4.86 |
ENSRNOT00000021496
|
Kank1
|
KN motif and ankyrin repeat domains 1 |
| chr17_-_66295014 | 4.84 |
ENSRNOT00000023974
|
Mtr
|
5-methyltetrahydrofolate-homocysteine methyltransferase |
| chr9_-_28732919 | 4.66 |
ENSRNOT00000083915
|
Rims1
|
regulating synaptic membrane exocytosis 1 |
| chr15_+_87886783 | 4.58 |
ENSRNOT00000065710
|
Slain1
|
SLAIN motif family, member 1 |
| chr2_-_186232292 | 4.56 |
ENSRNOT00000087088
|
Dclk2
|
doublecortin-like kinase 2 |
| chr8_+_82038967 | 4.40 |
ENSRNOT00000079535
|
Myo5a
|
myosin VA |
| chr4_+_31333970 | 4.26 |
ENSRNOT00000064866
|
LOC100911994
|
coiled-coil domain-containing protein 132-like |
| chr8_+_61671513 | 4.18 |
ENSRNOT00000091128
|
Ptpn9
|
protein tyrosine phosphatase, non-receptor type 9 |
| chr18_+_30581530 | 4.16 |
ENSRNOT00000048166
|
Pcdhb20
|
protocadherin beta 20 |
| chr3_-_78066123 | 4.15 |
ENSRNOT00000073165
|
Olr690
|
olfactory receptor 690 |
| chr7_-_55604403 | 4.12 |
ENSRNOT00000088732
|
Atxn7l3b
|
ataxin 7-like 3B |
| chr7_-_26983172 | 3.93 |
ENSRNOT00000089545
|
Txnrd1
|
thioredoxin reductase 1 |
| chr5_-_128333805 | 3.84 |
ENSRNOT00000037523
|
Zfyve9
|
zinc finger FYVE-type containing 9 |
| chr7_+_51794173 | 3.82 |
ENSRNOT00000043774
|
Otogl
|
otogelin-like |
| chr2_+_110306363 | 3.67 |
ENSRNOT00000040463
|
LOC499584
|
LRRGT00202 |
| chr15_-_86142672 | 3.63 |
ENSRNOT00000057830
|
Commd6
|
COMM domain containing 6 |
| chr1_+_53874860 | 3.60 |
ENSRNOT00000090486
|
Tcte2
|
t-complex-associated testis expressed 2 |
| chr5_+_71742911 | 3.51 |
ENSRNOT00000047225
|
Zfp462
|
zinc finger protein 462 |
| chr10_-_56465393 | 3.36 |
ENSRNOT00000056860
|
Tnk1
|
tyrosine kinase, non-receptor, 1 |
| chr14_-_114649173 | 3.12 |
ENSRNOT00000083528
|
Sptbn1
|
spectrin, beta, non-erythrocytic 1 |
| chr9_-_30844199 | 3.01 |
ENSRNOT00000017169
|
Col19a1
|
collagen type XIX alpha 1 chain |
| chr9_+_73418607 | 2.93 |
ENSRNOT00000092547
|
Map2
|
microtubule-associated protein 2 |
| chr18_+_15856801 | 2.92 |
ENSRNOT00000071548
|
Zfp397
|
zinc finger protein 397 |
| chr17_+_45199178 | 2.92 |
ENSRNOT00000080047
|
Zscan26
|
zinc finger and SCAN domain containing 26 |
| chr6_+_43234526 | 2.91 |
ENSRNOT00000086808
|
Asap2
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 2 |
| chr14_-_5006594 | 2.91 |
ENSRNOT00000076571
|
Zfp326
|
zinc finger protein 326 |
| chr5_+_16463597 | 2.90 |
ENSRNOT00000091550
|
AABR07047036.1
|
|
| chr5_+_60850852 | 2.90 |
ENSRNOT00000016720
|
Trmt10b
|
tRNA methyltransferase 10B |
| chr2_-_250923744 | 2.88 |
ENSRNOT00000084996
|
Clca1
|
chloride channel accessory 1 |
| chr3_+_119354904 | 2.74 |
ENSRNOT00000046977
|
LOC100363112
|
Aa2-296-like |
| chr14_+_34455934 | 2.68 |
ENSRNOT00000085991
ENSRNOT00000002981 |
Clock
|
clock circadian regulator |
| chrX_-_111179152 | 2.56 |
ENSRNOT00000089115
|
Morc4
|
MORC family CW-type zinc finger 4 |
| chr4_+_67359531 | 2.46 |
ENSRNOT00000013277
ENSRNOT00000082241 |
Adck2
|
aarF domain containing kinase 2 |
| chr4_-_29778039 | 2.12 |
ENSRNOT00000074177
|
Sgce
|
sarcoglycan, epsilon |
| chr1_+_167719947 | 2.08 |
ENSRNOT00000024971
|
Olr43
|
olfactory receptor 43 |
| chr1_-_275882444 | 2.05 |
ENSRNOT00000083215
|
Gpam
|
glycerol-3-phosphate acyltransferase, mitochondrial |
| chrX_+_40460047 | 2.05 |
ENSRNOT00000010970
|
Phex
|
phosphate regulating endopeptidase homolog, X-linked |
| chr18_+_30587872 | 1.98 |
ENSRNOT00000040016
|
Pcdhb21
|
protocadherin beta 21 |
| chr19_-_41161765 | 1.95 |
ENSRNOT00000023117
|
Hydin
|
Hydin, axonemal central pair apparatus protein |
| chr7_+_117773948 | 1.91 |
ENSRNOT00000021673
|
Lrrc14
|
leucine rich repeat containing 14 |
| chrX_-_153878806 | 1.79 |
ENSRNOT00000087990
|
Aff2
|
AF4/FMR2 family, member 2 |
| chr17_-_53688966 | 1.75 |
ENSRNOT00000078643
|
LOC100912163
|
AT-rich interactive domain-containing protein 4B-like |
| chr5_+_5616483 | 1.74 |
ENSRNOT00000011026
|
Ncoa2
|
nuclear receptor coactivator 2 |
| chr8_-_21453190 | 1.65 |
ENSRNOT00000078192
|
Zfp26
|
zinc finger protein 26 |
| chr8_-_19527513 | 1.63 |
ENSRNOT00000008594
|
Olr1147
|
olfactory receptor 1147 |
| chr13_+_68707776 | 1.61 |
ENSRNOT00000003558
ENSRNOT00000087997 |
Ivns1abp
|
influenza virus NS1A binding protein |
| chr1_+_203524426 | 1.60 |
ENSRNOT00000028020
|
Bub3
|
BUB3 mitotic checkpoint protein |
| chr1_+_150225373 | 1.55 |
ENSRNOT00000051266
|
Olr30
|
olfactory receptor 30 |
| chrX_+_105419285 | 1.52 |
ENSRNOT00000015518
|
Hnrnph2
|
heterogeneous nuclear ribonucleoprotein H2 (H') |
| chr19_-_44211208 | 1.25 |
ENSRNOT00000026309
|
Adat1
|
adenosine deaminase, tRNA-specific 1 |
| chr3_+_75987796 | 1.24 |
ENSRNOT00000091187
|
Olr594
|
olfactory receptor 594 |
| chr17_-_34945317 | 1.23 |
ENSRNOT00000090457
|
Vma21
|
VMA21 vacuolar H+-ATPase homolog (S. cerevisiae) |
| chr1_+_225120061 | 1.23 |
ENSRNOT00000026875
|
B3gat3
|
beta-1,3-glucuronyltransferase 3 |
| chr7_-_91756865 | 1.21 |
ENSRNOT00000041268
|
LOC100363319
|
high mobility group nucleosomal binding domain 2 |
| chr14_+_45033309 | 1.21 |
ENSRNOT00000002933
|
Tlr6
|
toll-like receptor 6 |
| chr6_-_125853461 | 1.17 |
ENSRNOT00000007505
|
Atxn3
|
ataxin 3 |
| chr14_-_34172893 | 1.15 |
ENSRNOT00000090470
|
Exoc1
|
exocyst complex component 1 |
| chr10_-_37311625 | 1.15 |
ENSRNOT00000043343
|
Jade2
|
jade family PHD finger 2 |
| chr10_-_11005952 | 1.13 |
ENSRNOT00000005031
|
Hmox2
|
heme oxygenase 2 |
| chr6_+_100337226 | 1.09 |
ENSRNOT00000011220
|
Fut8
|
fucosyltransferase 8 |
| chr11_-_43561883 | 1.03 |
ENSRNOT00000060877
|
Olr1553
|
olfactory receptor 1553 |
| chr10_-_89130339 | 0.96 |
ENSRNOT00000027640
|
Ezh1
|
enhancer of zeste 1 polycomb repressive complex 2 subunit |
| chr8_-_68275720 | 0.91 |
ENSRNOT00000079122
|
Map2k5
|
mitogen activated protein kinase kinase 5 |
| chr14_-_2610929 | 0.86 |
ENSRNOT00000036442
|
Ccdc18
|
coiled-coil domain containing 18 |
| chr3_+_149668102 | 0.81 |
ENSRNOT00000055342
|
Bpifb1
|
BPI fold containing family B, member 1 |
| chr3_+_154910626 | 0.81 |
ENSRNOT00000079515
|
Ralgapb
|
Ral GTPase activating protein non-catalytic beta subunit |
| chr1_-_170005597 | 0.78 |
ENSRNOT00000023325
|
Olr192
|
olfactory receptor 192 |
| chr12_-_51965779 | 0.71 |
ENSRNOT00000056733
|
LOC100362927
|
replication protein A3-like |
| chrX_-_143274180 | 0.68 |
ENSRNOT00000051550
|
Mcf2
|
MCF.2 cell line derived transforming sequence |
| chr8_+_5734348 | 0.68 |
ENSRNOT00000013119
|
Mmp10
|
matrix metallopeptidase 10 |
| chr4_-_80395502 | 0.67 |
ENSRNOT00000014437
|
Npvf
|
neuropeptide VF precursor |
| chr15_+_35761388 | 0.56 |
ENSRNOT00000086118
|
Olr1286
|
olfactory receptor 1286 |
| chr18_-_52017734 | 0.50 |
ENSRNOT00000081020
|
March3
|
membrane associated ring-CH-type finger 3 |
| chr3_+_77018186 | 0.49 |
ENSRNOT00000074443
|
Olr646
|
olfactory receptor 646 |
| chr3_-_78271875 | 0.48 |
ENSRNOT00000008475
|
Olr701
|
olfactory receptor 701 |
| chr16_-_3765917 | 0.44 |
ENSRNOT00000088284
|
Duxbl1
|
double homeobox B-like 1 |
| chr12_+_16899025 | 0.41 |
ENSRNOT00000001716
|
Psmg3
|
proteasome assembly chaperone 3 |
| chr1_+_172348583 | 0.36 |
ENSRNOT00000041144
|
Olr250
|
olfactory receptor 250 |
| chr19_+_11450760 | 0.27 |
ENSRNOT00000026297
|
Nudt21
|
nudix hydrolase 21 |
| chr3_+_146981984 | 0.26 |
ENSRNOT00000090211
|
Nsfl1c
|
NSFL1 cofactor |
| chr2_+_185846232 | 0.25 |
ENSRNOT00000023418
|
Lrba
|
LPS responsive beige-like anchor protein |
| chr17_+_45467015 | 0.24 |
ENSRNOT00000081408
|
Gpx5
|
glutathione peroxidase 5 |
| chr7_+_18120715 | 0.21 |
ENSRNOT00000068151
|
Vom1r108
|
vomeronasal 1 receptor 108 |
| chr15_-_32888095 | 0.16 |
ENSRNOT00000012233
|
Dad1
|
defender against cell death 1 |
| chr10_+_86564928 | 0.12 |
ENSRNOT00000039892
|
Lrrc3c
|
leucine rich repeat containing 3C |
| chr1_+_203526850 | 0.07 |
ENSRNOT00000087065
|
Bub3
|
BUB3 mitotic checkpoint protein |
| chr3_+_102841615 | 0.01 |
ENSRNOT00000075178
|
Olr770
|
olfactory receptor 770 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Foxd2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 11.4 | 34.3 | GO:0034971 | histone H3-R17 methylation(GO:0034971) |
| 8.5 | 33.9 | GO:0051958 | methotrexate transport(GO:0051958) |
| 7.2 | 21.7 | GO:0033986 | response to methanol(GO:0033986) |
| 6.2 | 18.5 | GO:0042335 | cuticle development(GO:0042335) cornification(GO:0070268) |
| 5.9 | 17.7 | GO:0015942 | formate metabolic process(GO:0015942) histidine catabolic process to glutamate and formamide(GO:0019556) formamide metabolic process(GO:0043606) |
| 5.3 | 16.0 | GO:0042412 | taurine biosynthetic process(GO:0042412) |
| 5.2 | 20.7 | GO:1903976 | negative regulation of glial cell migration(GO:1903976) |
| 4.7 | 14.1 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 4.1 | 24.6 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 3.8 | 11.3 | GO:0001969 | activation of membrane attack complex(GO:0001905) regulation of activation of membrane attack complex(GO:0001969) |
| 3.4 | 13.7 | GO:0002386 | immune response in mucosal-associated lymphoid tissue(GO:0002386) |
| 3.1 | 12.4 | GO:0070560 | protein secretion by platelet(GO:0070560) |
| 2.9 | 20.4 | GO:0051799 | negative regulation of hair follicle development(GO:0051799) |
| 2.6 | 18.2 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 2.5 | 15.3 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 2.3 | 9.3 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 2.3 | 9.0 | GO:0060671 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 2.1 | 8.4 | GO:1903371 | regulation of endoplasmic reticulum tubular network organization(GO:1903371) |
| 1.9 | 13.3 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 1.8 | 7.1 | GO:0044268 | multicellular organismal protein metabolic process(GO:0044268) malonyl-CoA biosynthetic process(GO:2001295) |
| 1.6 | 15.7 | GO:0003344 | pericardium morphogenesis(GO:0003344) |
| 1.6 | 4.7 | GO:0031632 | positive regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031632) |
| 1.5 | 7.7 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 1.3 | 8.9 | GO:0000711 | meiotic DNA repair synthesis(GO:0000711) |
| 1.2 | 4.9 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 1.1 | 4.4 | GO:0051643 | endoplasmic reticulum localization(GO:0051643) |
| 1.1 | 11.9 | GO:0015747 | urate transport(GO:0015747) |
| 1.1 | 30.5 | GO:0072337 | modified amino acid transport(GO:0072337) |
| 1.0 | 6.2 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 1.0 | 11.4 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 1.0 | 4.8 | GO:1904640 | response to methionine(GO:1904640) |
| 0.8 | 10.5 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.7 | 6.0 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 0.7 | 14.1 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.7 | 20.7 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.7 | 2.0 | GO:1904383 | response to sodium phosphate(GO:1904383) |
| 0.6 | 5.1 | GO:2000389 | regulation of neutrophil extravasation(GO:2000389) |
| 0.6 | 11.0 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.6 | 3.9 | GO:0016259 | selenocysteine metabolic process(GO:0016259) |
| 0.5 | 5.8 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.5 | 6.7 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 0.5 | 53.8 | GO:0046849 | bone remodeling(GO:0046849) |
| 0.5 | 2.0 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.5 | 7.5 | GO:0021891 | olfactory bulb interneuron development(GO:0021891) |
| 0.5 | 8.7 | GO:0021542 | dentate gyrus development(GO:0021542) |
| 0.4 | 2.7 | GO:2000323 | negative regulation of glucocorticoid receptor signaling pathway(GO:2000323) |
| 0.4 | 7.0 | GO:0019532 | oxalate transport(GO:0019532) sulfate transmembrane transport(GO:1902358) |
| 0.4 | 7.3 | GO:0072189 | ureter development(GO:0072189) |
| 0.4 | 10.5 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.4 | 1.2 | GO:0070340 | detection of diacyl bacterial lipopeptide(GO:0042496) detection of bacterial lipopeptide(GO:0070340) |
| 0.4 | 3.1 | GO:0007182 | common-partner SMAD protein phosphorylation(GO:0007182) |
| 0.4 | 1.1 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.4 | 1.8 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.3 | 6.9 | GO:0006828 | manganese ion transport(GO:0006828) gamma-aminobutyric acid secretion(GO:0014051) |
| 0.3 | 3.8 | GO:0019321 | pentose metabolic process(GO:0019321) |
| 0.3 | 0.9 | GO:2000342 | negative regulation of chemokine (C-X-C motif) ligand 2 production(GO:2000342) |
| 0.3 | 3.8 | GO:0007183 | SMAD protein complex assembly(GO:0007183) |
| 0.3 | 1.7 | GO:0045925 | positive regulation of female receptivity(GO:0045925) |
| 0.3 | 1.1 | GO:0046368 | GDP-L-fucose metabolic process(GO:0046368) |
| 0.2 | 1.2 | GO:0050651 | dermatan sulfate proteoglycan biosynthetic process(GO:0050651) |
| 0.2 | 4.6 | GO:0021860 | pyramidal neuron development(GO:0021860) |
| 0.2 | 2.0 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.2 | 6.6 | GO:0044705 | mating behavior(GO:0007617) multi-organism reproductive behavior(GO:0044705) |
| 0.2 | 0.8 | GO:0060178 | regulation of exocyst localization(GO:0060178) |
| 0.2 | 27.6 | GO:0019233 | sensory perception of pain(GO:0019233) |
| 0.1 | 1.2 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.1 | 2.9 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.1 | 1.1 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.1 | 5.1 | GO:0031113 | regulation of microtubule polymerization(GO:0031113) |
| 0.1 | 19.3 | GO:0007596 | blood coagulation(GO:0007596) |
| 0.1 | 1.2 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) positive regulation of ERAD pathway(GO:1904294) |
| 0.1 | 1.0 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.1 | 1.2 | GO:0070070 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.1 | 0.8 | GO:0034144 | negative regulation of toll-like receptor 4 signaling pathway(GO:0034144) |
| 0.1 | 4.0 | GO:0007193 | adenylate cyclase-inhibiting G-protein coupled receptor signaling pathway(GO:0007193) |
| 0.1 | 5.4 | GO:0042733 | embryonic digit morphogenesis(GO:0042733) |
| 0.1 | 6.3 | GO:0090090 | negative regulation of canonical Wnt signaling pathway(GO:0090090) |
| 0.1 | 3.4 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.1 | 0.4 | GO:0070317 | negative regulation of G0 to G1 transition(GO:0070317) |
| 0.1 | 2.9 | GO:0032784 | regulation of DNA-templated transcription, elongation(GO:0032784) |
| 0.1 | 3.6 | GO:0032088 | negative regulation of NF-kappaB transcription factor activity(GO:0032088) |
| 0.1 | 2.9 | GO:0032570 | response to progesterone(GO:0032570) |
| 0.0 | 8.5 | GO:0051291 | protein heterooligomerization(GO:0051291) |
| 0.0 | 6.1 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 8.9 | GO:0048193 | Golgi vesicle transport(GO:0048193) |
| 0.0 | 1.5 | GO:0043392 | negative regulation of DNA binding(GO:0043392) |
| 0.0 | 0.7 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.0 | 2.7 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 5.1 | GO:0007519 | skeletal muscle tissue development(GO:0007519) |
| 0.0 | 0.3 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.0 | 1.9 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 1.6 | GO:2001243 | negative regulation of intrinsic apoptotic signaling pathway(GO:2001243) |
| 0.0 | 1.2 | GO:0008033 | tRNA processing(GO:0008033) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.1 | 12.4 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 3.1 | 9.3 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 2.5 | 7.6 | GO:0043511 | inhibin complex(GO:0043511) |
| 1.8 | 11.0 | GO:0005608 | laminin-3 complex(GO:0005608) |
| 1.6 | 11.3 | GO:0005579 | membrane attack complex(GO:0005579) |
| 1.5 | 4.4 | GO:0042642 | actomyosin, myosin complex part(GO:0042642) |
| 1.2 | 24.6 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 1.0 | 34.3 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 1.0 | 2.9 | GO:0044609 | DBIRD complex(GO:0044609) |
| 0.8 | 69.5 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.7 | 8.9 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.7 | 7.7 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.7 | 8.9 | GO:0000801 | central element(GO:0000801) |
| 0.7 | 2.7 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.7 | 18.5 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.6 | 4.7 | GO:0098831 | presynaptic active zone cytoplasmic component(GO:0098831) |
| 0.4 | 20.7 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.4 | 1.2 | GO:0035355 | Toll-like receptor 2-Toll-like receptor 6 protein complex(GO:0035355) |
| 0.4 | 2.0 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 0.4 | 2.1 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.3 | 2.9 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.3 | 3.1 | GO:0008091 | spectrin(GO:0008091) cuticular plate(GO:0032437) |
| 0.3 | 5.1 | GO:0042581 | specific granule(GO:0042581) |
| 0.2 | 1.2 | GO:0098592 | cytoplasmic side of apical plasma membrane(GO:0098592) |
| 0.2 | 1.2 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.2 | 12.3 | GO:0045095 | keratin filament(GO:0045095) |
| 0.2 | 30.1 | GO:0030426 | growth cone(GO:0030426) |
| 0.1 | 13.7 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 8.4 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.1 | 6.9 | GO:0034704 | calcium channel complex(GO:0034704) |
| 0.1 | 32.6 | GO:0097060 | synaptic membrane(GO:0097060) |
| 0.1 | 1.7 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.1 | 0.3 | GO:1990730 | VCP-NSFL1C complex(GO:1990730) |
| 0.1 | 7.9 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.1 | 11.0 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.1 | 4.3 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.1 | 4.9 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.1 | 68.2 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.1 | 11.7 | GO:0030141 | secretory granule(GO:0030141) |
| 0.1 | 1.2 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.1 | 1.0 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.3 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.2 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 55.3 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 1.2 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 3.4 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 4.1 | GO:0030054 | cell junction(GO:0030054) |
| 0.0 | 10.7 | GO:0000785 | chromatin(GO:0000785) |
| 0.0 | 2.0 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 0.4 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.0 | 1.1 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 2.2 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 5.1 | GO:0005911 | cell-cell junction(GO:0005911) |
| 0.0 | 1.7 | GO:0000776 | kinetochore(GO:0000776) |
| 0.0 | 10.0 | GO:0031410 | cytoplasmic vesicle(GO:0031410) |
| 0.0 | 30.9 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.0 | 1.5 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 7.3 | GO:0005794 | Golgi apparatus(GO:0005794) |
| 0.0 | 23.5 | GO:0005829 | cytosol(GO:0005829) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.6 | 34.3 | GO:0038181 | bile acid receptor activity(GO:0038181) |
| 8.5 | 33.9 | GO:0015350 | methotrexate transporter activity(GO:0015350) |
| 4.1 | 24.6 | GO:0042954 | lipoprotein transporter activity(GO:0042954) |
| 3.5 | 17.7 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 3.1 | 9.3 | GO:0000171 | ribonuclease MRP activity(GO:0000171) |
| 2.7 | 13.3 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 2.6 | 10.5 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 2.2 | 15.3 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 1.9 | 7.6 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 1.8 | 7.1 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 1.6 | 6.2 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 1.5 | 30.5 | GO:0051184 | cofactor transporter activity(GO:0051184) modified amino acid transmembrane transporter activity(GO:0072349) |
| 1.5 | 18.2 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 1.1 | 11.4 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 1.1 | 15.7 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 1.0 | 32.8 | GO:0042165 | neurotransmitter binding(GO:0042165) |
| 0.9 | 10.7 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) |
| 0.8 | 11.9 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.7 | 5.3 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.6 | 4.8 | GO:0008172 | S-methyltransferase activity(GO:0008172) cobalamin binding(GO:0031419) |
| 0.6 | 7.7 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.6 | 2.9 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.6 | 3.9 | GO:0045340 | mercury ion binding(GO:0045340) |
| 0.6 | 5.0 | GO:0005113 | patched binding(GO:0005113) |
| 0.5 | 6.9 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.5 | 7.0 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.4 | 8.4 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.4 | 1.2 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.4 | 1.2 | GO:0035663 | Toll-like receptor 2 binding(GO:0035663) diacyl lipopeptide binding(GO:0042498) |
| 0.4 | 1.1 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.4 | 8.5 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.4 | 16.0 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.3 | 4.9 | GO:0051378 | serotonin binding(GO:0051378) |
| 0.3 | 2.0 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.3 | 54.3 | GO:0052689 | carboxylic ester hydrolase activity(GO:0052689) |
| 0.3 | 10.5 | GO:0048020 | chemokine activity(GO:0008009) CCR chemokine receptor binding(GO:0048020) |
| 0.3 | 4.4 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.3 | 12.4 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.3 | 12.9 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.2 | 1.8 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.2 | 5.1 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.2 | 1.2 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.2 | 21.2 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.2 | 3.8 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 6.6 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.1 | 13.7 | GO:0003823 | antigen binding(GO:0003823) |
| 0.1 | 2.9 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.1 | 1.0 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.1 | 2.9 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.1 | 2.9 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.1 | 1.2 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.1 | 3.6 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.1 | 1.1 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.1 | 3.1 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.1 | 0.9 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.1 | 6.1 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.1 | 2.7 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 5.9 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.1 | 1.2 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.1 | 4.9 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 11.0 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 12.9 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.0 | 28.8 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 3.8 | GO:0004553 | hydrolase activity, hydrolyzing O-glycosyl compounds(GO:0004553) |
| 0.0 | 3.8 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 4.7 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 6.2 | GO:0016874 | ligase activity(GO:0016874) |
| 0.0 | 2.0 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 1.9 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 5.1 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.2 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.2 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.3 | GO:0017091 | AU-rich element binding(GO:0017091) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 34.3 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.6 | 11.0 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.5 | 29.0 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.3 | 5.0 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.3 | 15.3 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.2 | 7.6 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.2 | 13.2 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.2 | 11.4 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.2 | 7.7 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.1 | 2.7 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.1 | 1.7 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 4.1 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.1 | 3.0 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 0.9 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 2.9 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.8 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 12.0 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 1.7 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 2.0 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.7 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 20.7 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 1.4 | 15.3 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 1.1 | 11.3 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.9 | 18.2 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.8 | 11.4 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.8 | 22.0 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.8 | 7.6 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.7 | 20.9 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.6 | 6.9 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.4 | 10.5 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.3 | 4.9 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.3 | 4.7 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.3 | 7.1 | REACTOME FATTY ACYL COA BIOSYNTHESIS | Genes involved in Fatty Acyl-CoA Biosynthesis |
| 0.3 | 5.5 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.2 | 21.7 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.2 | 3.9 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.2 | 4.4 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.2 | 8.5 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.2 | 18.3 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.2 | 8.9 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.2 | 11.0 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.1 | 3.8 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.1 | 3.1 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.1 | 17.7 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.1 | 1.7 | REACTOME INHIBITION OF THE PROTEOLYTIC ACTIVITY OF APC C REQUIRED FOR THE ONSET OF ANAPHASE BY MITOTIC SPINDLE CHECKPOINT COMPONENTS | Genes involved in Inhibition of the proteolytic activity of APC/C required for the onset of anaphase by mitotic spindle checkpoint components |
| 0.1 | 9.6 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.1 | 2.0 | REACTOME TRIGLYCERIDE BIOSYNTHESIS | Genes involved in Triglyceride Biosynthesis |
| 0.1 | 1.1 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 6.2 | REACTOME SIGNALING BY ERBB4 | Genes involved in Signaling by ERBB4 |
| 0.1 | 3.0 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 1.1 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.0 | 1.2 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.0 | 0.7 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 1.8 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.9 | REACTOME NGF SIGNALLING VIA TRKA FROM THE PLASMA MEMBRANE | Genes involved in NGF signalling via TRKA from the plasma membrane |