Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
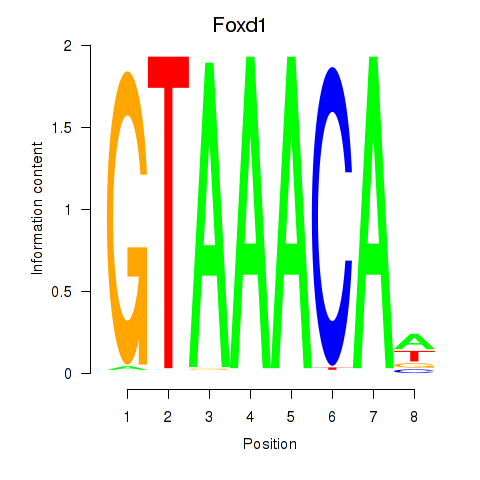
Results for Foxd1
Z-value: 0.97
Transcription factors associated with Foxd1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Foxd1
|
ENSRNOG00000043332 | forkhead box D1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Foxd1 | rn6_v1_chr2_+_28460068_28460068 | -0.11 | 5.4e-02 | Click! |
Activity profile of Foxd1 motif
Sorted Z-values of Foxd1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_58814805 | 39.11 |
ENSRNOT00000005909
|
Tspan8
|
tetraspanin 8 |
| chr5_+_152533349 | 38.89 |
ENSRNOT00000067524
|
Trim63
|
tripartite motif containing 63 |
| chr2_+_198683159 | 33.96 |
ENSRNOT00000028793
|
Txnip
|
thioredoxin interacting protein |
| chr1_+_225184939 | 27.91 |
ENSRNOT00000079456
|
Ahnak
|
AHNAK nucleoprotein |
| chr17_+_81922329 | 26.78 |
ENSRNOT00000031542
|
Cacnb2
|
calcium voltage-gated channel auxiliary subunit beta 2 |
| chr14_+_83560541 | 24.62 |
ENSRNOT00000057738
ENSRNOT00000085228 |
Pik3ip1
|
phosphoinositide-3-kinase interacting protein 1 |
| chr4_+_169161585 | 23.15 |
ENSRNOT00000079785
|
Emp1
|
epithelial membrane protein 1 |
| chr16_-_61091169 | 22.97 |
ENSRNOT00000016328
|
Dusp4
|
dual specificity phosphatase 4 |
| chr4_+_169147243 | 22.96 |
ENSRNOT00000011580
|
Emp1
|
epithelial membrane protein 1 |
| chr20_-_4863011 | 22.10 |
ENSRNOT00000079503
|
Ltb
|
lymphotoxin beta |
| chr20_-_4863198 | 21.70 |
ENSRNOT00000001108
|
Ltb
|
lymphotoxin beta |
| chr1_+_68436593 | 21.67 |
ENSRNOT00000080325
|
LOC108348118
|
leucyl-cystinyl aminopeptidase |
| chr1_-_31055453 | 21.54 |
ENSRNOT00000031083
|
Soga3
|
SOGA family member 3 |
| chr1_-_59347472 | 21.35 |
ENSRNOT00000017718
|
Lnpep
|
leucyl and cystinyl aminopeptidase |
| chr20_+_5184515 | 21.33 |
ENSRNOT00000089411
|
LOC103694381
|
lymphotoxin-beta |
| chr1_+_68436917 | 20.84 |
ENSRNOT00000088586
|
LOC108348118
|
leucyl-cystinyl aminopeptidase |
| chr7_+_120580743 | 20.62 |
ENSRNOT00000017181
|
Maff
|
MAF bZIP transcription factor F |
| chr1_-_215536980 | 20.19 |
ENSRNOT00000027344
|
Ifitm10
|
interferon induced transmembrane protein 10 |
| chr3_-_72219246 | 19.37 |
ENSRNOT00000009903
|
Smtnl1
|
smoothelin-like 1 |
| chr1_-_254735548 | 19.30 |
ENSRNOT00000025258
|
Ankrd1
|
ankyrin repeat domain 1 |
| chr20_+_29655226 | 18.34 |
ENSRNOT00000089059
|
Spock2
|
SPARC/osteonectin, cwcv and kazal like domains proteoglycan 2 |
| chr7_-_98197087 | 17.84 |
ENSRNOT00000010484
ENSRNOT00000079961 |
Klhl38
|
kelch-like family member 38 |
| chr13_+_97838361 | 17.41 |
ENSRNOT00000003641
|
Cnst
|
consortin, connexin sorting protein |
| chr8_+_104040934 | 17.27 |
ENSRNOT00000081204
|
Tfdp2
|
transcription factor Dp-2 |
| chr8_+_65733400 | 16.73 |
ENSRNOT00000089126
|
Uaca
|
uveal autoantigen with coiled-coil domains and ankyrin repeats |
| chr7_+_121841855 | 16.50 |
ENSRNOT00000024673
|
Grap2
|
GRB2-related adaptor protein 2 |
| chr1_-_90520012 | 15.96 |
ENSRNOT00000028698
|
Kctd15
|
potassium channel tetramerization domain containing 15 |
| chr18_-_5314511 | 15.23 |
ENSRNOT00000022637
ENSRNOT00000079682 |
Zfp521
|
zinc finger protein 521 |
| chr8_-_109560747 | 15.16 |
ENSRNOT00000087334
|
Ppp2r3a
|
protein phosphatase 2, regulatory subunit B'', alpha |
| chr18_+_65155685 | 14.50 |
ENSRNOT00000081797
|
Tcf4
|
transcription factor 4 |
| chr11_+_82194657 | 14.35 |
ENSRNOT00000002445
|
Etv5
|
ets variant 5 |
| chr10_+_105393072 | 14.00 |
ENSRNOT00000013359
|
Ubald2
|
UBA-like domain containing 2 |
| chr5_+_113725717 | 13.96 |
ENSRNOT00000032248
|
Tek
|
TEK receptor tyrosine kinase |
| chr1_-_90520344 | 13.55 |
ENSRNOT00000078598
|
Kctd15
|
potassium channel tetramerization domain containing 15 |
| chr7_-_139318455 | 13.20 |
ENSRNOT00000092029
|
Hdac7
|
histone deacetylase 7 |
| chr1_+_198210525 | 12.64 |
ENSRNOT00000026755
|
Ypel3
|
yippee-like 3 |
| chr4_-_30556814 | 12.14 |
ENSRNOT00000012760
|
Pdk4
|
pyruvate dehydrogenase kinase 4 |
| chr18_+_3887419 | 11.56 |
ENSRNOT00000093089
|
Lama3
|
laminin subunit alpha 3 |
| chr13_-_1946508 | 11.53 |
ENSRNOT00000043890
|
Dsel
|
dermatan sulfate epimerase-like |
| chr9_-_38196273 | 11.40 |
ENSRNOT00000044452
|
Dst
|
dystonin |
| chr8_-_104593625 | 11.32 |
ENSRNOT00000016625
|
Zbtb38
|
zinc finger and BTB domain containing 38 |
| chr10_+_18996523 | 10.96 |
ENSRNOT00000046135
|
Lcp2
|
lymphocyte cytosolic protein 2 |
| chr15_-_95514259 | 10.29 |
ENSRNOT00000038433
|
Slitrk6
|
SLIT and NTRK-like family, member 6 |
| chr2_+_41467064 | 10.05 |
ENSRNOT00000073231
|
AABR07008066.2
|
|
| chrX_+_71155601 | 10.02 |
ENSRNOT00000076453
ENSRNOT00000048521 |
Foxo4
|
forkhead box O4 |
| chr6_-_124735741 | 9.85 |
ENSRNOT00000064716
ENSRNOT00000091693 |
Rps6ka5
|
ribosomal protein S6 kinase A5 |
| chr19_-_26094756 | 9.63 |
ENSRNOT00000067780
|
Junb
|
JunB proto-oncogene, AP-1 transcription factor subunit |
| chr20_-_4390436 | 9.41 |
ENSRNOT00000000497
ENSRNOT00000077655 |
Ppt2
|
palmitoyl-protein thioesterase 2 |
| chr17_-_22143324 | 8.97 |
ENSRNOT00000019361
|
Edn1
|
endothelin 1 |
| chr6_-_77421286 | 8.93 |
ENSRNOT00000011453
|
Nkx2-1
|
NK2 homeobox 1 |
| chr2_-_250232295 | 8.85 |
ENSRNOT00000082132
|
Lmo4
|
LIM domain only 4 |
| chr1_+_137014272 | 8.83 |
ENSRNOT00000014802
|
Akap13
|
A-kinase anchoring protein 13 |
| chr15_-_37983882 | 8.79 |
ENSRNOT00000087978
|
Lats2
|
large tumor suppressor kinase 2 |
| chr6_+_93462852 | 8.79 |
ENSRNOT00000089473
|
Arid4a
|
AT-rich interaction domain 4A |
| chr15_+_8730871 | 8.70 |
ENSRNOT00000081845
|
Nr1d2
|
nuclear receptor subfamily 1, group D, member 2 |
| chr4_-_157433467 | 8.65 |
ENSRNOT00000028965
|
Lag3
|
lymphocyte activating 3 |
| chr1_+_274049533 | 8.46 |
ENSRNOT00000076779
ENSRNOT00000042822 |
Mxi1
|
MAX interactor 1, dimerization protein |
| chr1_-_206282575 | 8.38 |
ENSRNOT00000024974
|
Adam12
|
ADAM metallopeptidase domain 12 |
| chr5_-_50075454 | 8.25 |
ENSRNOT00000011085
|
Orc3
|
origin recognition complex, subunit 3 |
| chr4_-_40136061 | 8.09 |
ENSRNOT00000009752
|
Bmt2
|
base methyltransferase of 25S rRNA 2 homolog |
| chr10_-_90307658 | 7.75 |
ENSRNOT00000092102
|
Slc4a1
|
solute carrier family 4 member 1 |
| chr3_+_177351518 | 7.73 |
ENSRNOT00000023989
|
Pcmtd2
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase domain containing 2 |
| chr6_+_27768943 | 7.56 |
ENSRNOT00000015820
|
Kif3c
|
kinesin family member 3C |
| chr2_-_250241590 | 7.49 |
ENSRNOT00000077221
ENSRNOT00000067502 |
Lmo4
|
LIM domain only 4 |
| chr8_+_11931767 | 7.36 |
ENSRNOT00000087963
|
Maml2
|
mastermind-like transcriptional coactivator 2 |
| chr2_-_250235435 | 7.30 |
ENSRNOT00000088618
|
Lmo4
|
LIM domain only 4 |
| chr13_-_113872097 | 7.20 |
ENSRNOT00000010799
ENSRNOT00000084320 |
Cr1l
|
complement C3b/C4b receptor 1 like |
| chr1_-_100845392 | 7.00 |
ENSRNOT00000027436
|
Tbc1d17
|
TBC1 domain family, member 17 |
| chr1_-_134870255 | 6.99 |
ENSRNOT00000055829
|
Chd2
|
chromodomain helicase DNA binding protein 2 |
| chr8_+_77107536 | 6.82 |
ENSRNOT00000083255
|
Adam10
|
ADAM metallopeptidase domain 10 |
| chrX_-_15627235 | 6.76 |
ENSRNOT00000013369
|
Wdr45
|
WD repeat domain 45 |
| chr17_-_9721542 | 6.54 |
ENSRNOT00000047958
ENSRNOT00000079063 |
Grk6
|
G protein-coupled receptor kinase 6 |
| chr15_+_108956448 | 6.54 |
ENSRNOT00000046324
|
AC123185.1
|
|
| chr13_-_113871842 | 6.50 |
ENSRNOT00000079549
|
Cr1l
|
complement C3b/C4b receptor 1 like |
| chr10_-_72556564 | 6.40 |
ENSRNOT00000048373
|
AABR07030162.1
|
|
| chr10_-_40296470 | 6.33 |
ENSRNOT00000086456
|
Tnip1
|
TNFAIP3 interacting protein 1 |
| chr3_-_2453933 | 6.30 |
ENSRNOT00000014060
|
Slc34a3
|
solute carrier family 34 member 3 |
| chr1_+_100845563 | 6.17 |
ENSRNOT00000027478
|
Akt1s1
|
AKT1 substrate 1 |
| chr8_-_116469915 | 6.10 |
ENSRNOT00000024193
|
Sema3f
|
semaphorin 3F |
| chr2_-_235951275 | 6.05 |
ENSRNOT00000050291
|
AABR07013410.1
|
|
| chr1_+_240908483 | 5.96 |
ENSRNOT00000019367
|
Klf9
|
Kruppel-like factor 9 |
| chr3_-_46051096 | 5.74 |
ENSRNOT00000081302
|
Baz2b
|
bromodomain adjacent to zinc finger domain, 2B |
| chr4_-_17594598 | 5.72 |
ENSRNOT00000008936
|
Sema3e
|
semaphorin 3E |
| chr7_+_99954492 | 5.72 |
ENSRNOT00000005885
|
Trib1
|
tribbles pseudokinase 1 |
| chr4_-_115239723 | 5.68 |
ENSRNOT00000042699
|
Actg2
|
actin, gamma 2, smooth muscle, enteric |
| chr8_-_36438730 | 5.56 |
ENSRNOT00000015366
|
Fam118b
|
family with sequence similarity 118, member B |
| chr16_-_20807070 | 5.51 |
ENSRNOT00000072536
|
Comp
|
cartilage oligomeric matrix protein |
| chr4_-_61502778 | 5.15 |
ENSRNOT00000051232
|
AABR07060190.1
|
|
| chr3_-_58009514 | 5.10 |
ENSRNOT00000048356
|
AABR07052498.1
|
|
| chr13_+_51240911 | 5.09 |
ENSRNOT00000005551
|
Adipor1
|
adiponectin receptor 1 |
| chr1_+_254974789 | 4.94 |
ENSRNOT00000025020
|
Pcgf5
|
polycomb group ring finger 5 |
| chr3_+_120726906 | 4.93 |
ENSRNOT00000051069
|
Bcl2l11
|
BCL2 like 11 |
| chr2_+_54897424 | 4.92 |
ENSRNOT00000017665
|
Ttc33
|
tetratricopeptide repeat domain 33 |
| chr4_+_157594436 | 4.89 |
ENSRNOT00000029053
|
Lpar5
|
lysophosphatidic acid receptor 5 |
| chr18_-_69671199 | 4.79 |
ENSRNOT00000082484
|
Smad4
|
SMAD family member 4 |
| chr1_+_260798239 | 4.75 |
ENSRNOT00000036791
|
Lcor
|
ligand dependent nuclear receptor corepressor |
| chr17_-_74679696 | 4.73 |
ENSRNOT00000048420
|
AABR07028558.1
|
|
| chr16_-_32299542 | 4.66 |
ENSRNOT00000072321
|
AABR07025299.1
|
|
| chr17_-_84705052 | 4.54 |
ENSRNOT00000050803
|
AABR07028749.1
|
|
| chr2_-_208225888 | 4.45 |
ENSRNOT00000054860
|
AABR07012775.1
|
|
| chr14_-_3351553 | 4.40 |
ENSRNOT00000061556
|
Btbd8
|
BTB (POZ) domain containing 8 |
| chr4_+_41364441 | 4.36 |
ENSRNOT00000087146
|
Foxp2
|
forkhead box P2 |
| chr7_+_139271698 | 4.29 |
ENSRNOT00000079388
|
Slc48a1
|
solute carrier family 48 member 1 |
| chr8_+_111209908 | 4.29 |
ENSRNOT00000090221
|
Amotl2
|
angiomotin like 2 |
| chr13_-_30800451 | 4.28 |
ENSRNOT00000046791
|
Rpl21
|
ribosomal protein L21 |
| chr3_-_148932878 | 4.23 |
ENSRNOT00000013881
|
Nol4l
|
nucleolar protein 4-like |
| chr9_+_43093138 | 4.09 |
ENSRNOT00000021592
|
Cnnm3
|
cyclin and CBS domain divalent metal cation transport mediator 3 |
| chrX_-_20359945 | 4.08 |
ENSRNOT00000049138
|
AABR07037412.1
|
|
| chr6_+_73553210 | 4.03 |
ENSRNOT00000006562
|
Akap6
|
A-kinase anchoring protein 6 |
| chr18_+_27558089 | 3.98 |
ENSRNOT00000027499
|
Fam53c
|
family with sequence similarity 53, member C |
| chr9_+_61720583 | 3.96 |
ENSRNOT00000020536
|
Mob4
|
MOB family member 4, phocein |
| chr4_+_83713666 | 3.95 |
ENSRNOT00000086473
|
Creb5
|
cAMP responsive element binding protein 5 |
| chr20_+_3823596 | 3.92 |
ENSRNOT00000087670
|
Rxrb
|
retinoid X receptor beta |
| chr10_-_90501819 | 3.86 |
ENSRNOT00000050474
|
Gpatch8
|
G patch domain containing 8 |
| chr7_+_70753101 | 3.82 |
ENSRNOT00000090001
|
R3hdm2
|
R3H domain containing 2 |
| chrX_-_111887906 | 3.61 |
ENSRNOT00000085118
|
Tsc22d3
|
TSC22 domain family, member 3 |
| chr2_+_31378743 | 3.56 |
ENSRNOT00000050384
|
AABR07007853.1
|
|
| chr1_-_13395370 | 3.56 |
ENSRNOT00000085519
|
Ccdc28a
|
coiled-coil domain containing 28A |
| chr6_+_8220228 | 3.55 |
ENSRNOT00000079279
ENSRNOT00000048656 |
Ppm1b
|
protein phosphatase, Mg2+/Mn2+ dependent, 1B |
| chrX_+_128493614 | 3.43 |
ENSRNOT00000044240
|
Stag2
|
stromal antigen 2 |
| chr20_+_5262946 | 3.38 |
ENSRNOT00000082900
|
Brd2
|
bromodomain containing 2 |
| chr19_-_52206310 | 3.34 |
ENSRNOT00000087886
|
Mbtps1
|
membrane-bound transcription factor peptidase, site 1 |
| chr9_-_86103158 | 3.30 |
ENSRNOT00000021528
|
Cul3
|
cullin 3 |
| chr18_+_29972808 | 3.30 |
ENSRNOT00000074051
|
Pcdha4
|
protocadherin alpha 4 |
| chr6_+_108167716 | 3.22 |
ENSRNOT00000064426
|
Lin52
|
lin-52 DREAM MuvB core complex component |
| chrX_+_159513800 | 3.17 |
ENSRNOT00000065636
|
Htatsf1
|
HIV-1 Tat specific factor 1 |
| chr1_-_222590112 | 3.13 |
ENSRNOT00000028763
ENSRNOT00000091990 |
Mark2
|
microtubule affinity regulating kinase 2 |
| chr5_-_157165767 | 3.11 |
ENSRNOT00000000167
|
Ubxn10
|
UBX domain protein 10 |
| chr1_-_246110218 | 3.11 |
ENSRNOT00000077544
|
Rfx3
|
regulatory factor X3 |
| chr7_-_12457513 | 3.06 |
ENSRNOT00000060683
|
Stk11
|
serine/threonine kinase 11 |
| chr7_-_137856485 | 3.05 |
ENSRNOT00000007003
|
LOC688906
|
similar to splicing factor, arginine/serine-rich 2, interacting protein |
| chr1_-_24302298 | 2.93 |
ENSRNOT00000083452
|
Sgk1
|
serum/glucocorticoid regulated kinase 1 |
| chr13_-_98480419 | 2.89 |
ENSRNOT00000086306
|
Coq8a
|
coenzyme Q8A |
| chr4_-_183426439 | 2.78 |
ENSRNOT00000083310
|
Fam60a
|
family with sequence similarity 60, member A |
| chr19_+_33130303 | 2.70 |
ENSRNOT00000048673
|
LOC102550668
|
60S ribosomal protein L21-like |
| chr13_-_101697684 | 2.66 |
ENSRNOT00000078834
|
Brox
|
BRO1 domain and CAAX motif containing |
| chr18_-_410098 | 2.65 |
ENSRNOT00000084138
|
LOC102546764
|
cx9C motif-containing protein 4-like |
| chr8_-_117446019 | 2.65 |
ENSRNOT00000042577
|
Arih2
|
ariadne RBR E3 ubiquitin protein ligase 2 |
| chr10_-_109267500 | 2.64 |
ENSRNOT00000005977
|
Cep131
|
centrosomal protein 131 |
| chr13_-_68360664 | 2.61 |
ENSRNOT00000030971
|
Hmcn1
|
hemicentin 1 |
| chr7_+_74047814 | 2.55 |
ENSRNOT00000014879
|
Osr2
|
odd-skipped related transciption factor 2 |
| chr10_-_87136026 | 2.52 |
ENSRNOT00000014230
ENSRNOT00000083233 |
Smarce1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily e, member 1 |
| chr2_-_98610368 | 2.48 |
ENSRNOT00000011641
|
Zfhx4
|
zinc finger homeobox 4 |
| chr10_+_29029590 | 2.41 |
ENSRNOT00000005132
|
Slu7
|
SLU7 homolog, splicing factor |
| chr3_-_147865393 | 2.36 |
ENSRNOT00000009852
|
Sox12
|
SRY box 12 |
| chr4_-_27331508 | 2.20 |
ENSRNOT00000090751
ENSRNOT00000010118 |
Akap9
|
A-kinase anchoring protein 9 |
| chr14_-_23002011 | 2.01 |
ENSRNOT00000002736
|
Ythdc1
|
YTH domain containing 1 |
| chrX_+_157374305 | 2.01 |
ENSRNOT00000081973
|
Trex2
|
three prime repair exonuclease 2 |
| chr5_-_60559533 | 2.00 |
ENSRNOT00000092899
|
Zbtb5
|
zinc finger and BTB domain containing 5 |
| chr19_-_25345790 | 1.99 |
ENSRNOT00000010050
|
Zswim4
|
zinc finger, SWIM-type containing 4 |
| chr9_+_42315682 | 1.93 |
ENSRNOT00000071995
|
AABR07067388.1
|
|
| chr4_-_67520356 | 1.90 |
ENSRNOT00000014604
|
Braf
|
B-Raf proto-oncogene, serine/threonine kinase |
| chr5_+_48374096 | 1.90 |
ENSRNOT00000010172
|
Gabrr1
|
gamma-aminobutyric acid type A receptor rho 1 subunit |
| chrX_+_10430847 | 1.89 |
ENSRNOT00000047936
|
Rpl21
|
ribosomal protein L21 |
| chr2_+_257633425 | 1.88 |
ENSRNOT00000071770
|
Zzz3
|
zinc finger, ZZ-type containing 3 |
| chr16_-_69280109 | 1.88 |
ENSRNOT00000058595
|
AABR07026240.1
|
|
| chr4_-_115516296 | 1.88 |
ENSRNOT00000019399
|
Paip2b
|
poly(A) binding protein interacting protein 2B |
| chr1_-_264706476 | 1.86 |
ENSRNOT00000045446
|
AC121209.1
|
|
| chr9_+_94702129 | 1.85 |
ENSRNOT00000080930
|
Neu2
|
neuraminidase 2 |
| chr13_+_90533365 | 1.78 |
ENSRNOT00000082469
|
Dcaf8
|
DDB1 and CUL4 associated factor 8 |
| chr15_-_61876430 | 1.73 |
ENSRNOT00000049690
|
AC123280.1
|
|
| chr16_+_53957858 | 1.64 |
ENSRNOT00000013078
|
Frg1
|
FSHD region gene 1 |
| chr8_+_128087345 | 1.59 |
ENSRNOT00000019777
|
Acvr2b
|
activin A receptor type 2B |
| chr6_-_50786967 | 1.48 |
ENSRNOT00000009566
|
Cbll1
|
Cbl proto-oncogene like 1 |
| chr10_+_89645973 | 1.43 |
ENSRNOT00000086892
|
Dhx8
|
DEAH-box helicase 8 |
| chr6_-_26486695 | 1.41 |
ENSRNOT00000073236
|
Krtcap3
|
keratinocyte associated protein 3 |
| chr16_-_19583386 | 1.21 |
ENSRNOT00000090131
|
Zfp617
|
zinc finger protein 617 |
| chr7_+_24534615 | 1.15 |
ENSRNOT00000009124
|
Cry1
|
cryptochrome circadian clock 1 |
| chr4_+_168599331 | 1.11 |
ENSRNOT00000086719
|
Crebl2
|
cAMP responsive element binding protein-like 2 |
| chr15_+_32894938 | 1.04 |
ENSRNOT00000012837
|
Abhd4
|
abhydrolase domain containing 4 |
| chr10_+_89646195 | 1.02 |
ENSRNOT00000048140
|
Dhx8
|
DEAH-box helicase 8 |
| chr7_-_120780108 | 0.88 |
ENSRNOT00000018185
|
Ddx17
|
DEAD-box helicase 17 |
| chr9_-_37144015 | 0.87 |
ENSRNOT00000015638
|
Phf3
|
PHD finger protein 3 |
| chr9_-_71651512 | 0.82 |
ENSRNOT00000032782
|
Plekhm3
|
pleckstrin homology domain containing M3 |
| chr9_-_61528882 | 0.69 |
ENSRNOT00000015432
|
Ankrd44
|
ankyrin repeat domain 44 |
| chr13_-_97838228 | 0.61 |
ENSRNOT00000003618
|
Tfb2m
|
transcription factor B2, mitochondrial |
| chr7_-_91538673 | 0.59 |
ENSRNOT00000006209
|
Rad21
|
RAD21 cohesin complex component |
| chr2_+_185590986 | 0.56 |
ENSRNOT00000088807
ENSRNOT00000088188 |
Lrba
|
LPS responsive beige-like anchor protein |
| chr2_+_51672722 | 0.56 |
ENSRNOT00000016485
|
Fgf10
|
fibroblast growth factor 10 |
| chr11_-_78861716 | 0.28 |
ENSRNOT00000002634
|
Tprg1
|
tumor protein p63 regulated 1 |
| chr17_+_86408151 | 0.17 |
ENSRNOT00000022734
|
Otud1
|
OTU deubiquitinase 1 |
| chr3_-_160038078 | 0.16 |
ENSRNOT00000013445
|
Serinc3
|
serine incorporator 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Foxd1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.9 | 26.8 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 7.8 | 38.9 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
| 7.1 | 21.4 | GO:0010813 | neuropeptide catabolic process(GO:0010813) |
| 5.1 | 15.2 | GO:0090249 | cell motility involved in somitogenic axis elongation(GO:0090247) regulation of cell motility involved in somitogenic axis elongation(GO:0090249) |
| 4.7 | 14.0 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 4.4 | 17.4 | GO:0042998 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) |
| 4.1 | 24.6 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 3.0 | 21.3 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 3.0 | 12.1 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 3.0 | 9.0 | GO:0031583 | phospholipase D-activating G-protein coupled receptor signaling pathway(GO:0031583) |
| 2.9 | 8.8 | GO:0001827 | inner cell mass cell fate commitment(GO:0001827) inner cell mass cellular morphogenesis(GO:0001828) |
| 2.9 | 14.3 | GO:0042078 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 2.7 | 8.2 | GO:0006267 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 2.7 | 42.5 | GO:0038166 | angiotensin-activated signaling pathway(GO:0038166) |
| 2.6 | 46.1 | GO:0032060 | bleb assembly(GO:0032060) |
| 2.5 | 9.8 | GO:0043988 | histone H3-S10 phosphorylation(GO:0043987) histone H3-S28 phosphorylation(GO:0043988) histone H2A phosphorylation(GO:1990164) |
| 2.3 | 11.5 | GO:0030205 | dermatan sulfate metabolic process(GO:0030205) |
| 2.3 | 18.3 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 2.3 | 6.8 | GO:0051088 | PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 2.2 | 8.8 | GO:0071883 | activation of MAPK activity by adrenergic receptor signaling pathway(GO:0071883) |
| 2.1 | 10.3 | GO:0060005 | vestibular reflex(GO:0060005) |
| 2.0 | 18.3 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 2.0 | 6.0 | GO:0071387 | cellular response to cortisol stimulus(GO:0071387) |
| 1.8 | 26.5 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 1.7 | 8.6 | GO:0045085 | negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 1.6 | 4.8 | GO:0035262 | left ventricular cardiac muscle tissue morphogenesis(GO:0003220) cell proliferation involved in heart valve morphogenesis(GO:0003249) regulation of cell proliferation involved in heart valve morphogenesis(GO:0003250) gonad morphogenesis(GO:0035262) nephrogenic mesenchyme morphogenesis(GO:0072134) |
| 1.5 | 7.7 | GO:0010037 | response to carbon dioxide(GO:0010037) |
| 1.5 | 8.9 | GO:0021759 | globus pallidus development(GO:0021759) |
| 1.5 | 34.0 | GO:0002347 | response to tumor cell(GO:0002347) |
| 1.4 | 10.0 | GO:0070317 | negative regulation of G0 to G1 transition(GO:0070317) |
| 1.4 | 5.7 | GO:0045658 | regulation of neutrophil differentiation(GO:0045658) |
| 1.4 | 12.6 | GO:2000774 | positive regulation of cellular senescence(GO:2000774) |
| 1.4 | 5.6 | GO:0030576 | Cajal body organization(GO:0030576) |
| 1.3 | 8.8 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) |
| 1.2 | 13.7 | GO:0045916 | negative regulation of complement activation(GO:0045916) negative regulation of protein activation cascade(GO:2000258) |
| 1.2 | 23.6 | GO:0021527 | spinal cord association neuron differentiation(GO:0021527) |
| 1.2 | 4.9 | GO:1902263 | apoptotic process involved in embryonic digit morphogenesis(GO:1902263) |
| 1.2 | 7.4 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 1.2 | 23.0 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 1.2 | 14.5 | GO:1900746 | regulation of vascular endothelial growth factor signaling pathway(GO:1900746) |
| 1.2 | 9.4 | GO:0098734 | macromolecule depalmitoylation(GO:0098734) |
| 1.1 | 5.7 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 1.1 | 3.3 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 1.1 | 17.6 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 1.1 | 6.5 | GO:0010360 | negative regulation of anion channel activity(GO:0010360) |
| 1.1 | 9.6 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 1.0 | 3.1 | GO:0002071 | glandular epithelial cell maturation(GO:0002071) positive regulation of type B pancreatic cell development(GO:2000078) |
| 1.0 | 6.1 | GO:0097490 | trunk segmentation(GO:0035290) trunk neural crest cell migration(GO:0036484) ventral trunk neural crest cell migration(GO:0036486) sympathetic neuron projection extension(GO:0097490) sympathetic neuron projection guidance(GO:0097491) |
| 1.0 | 4.0 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.9 | 16.7 | GO:0042347 | negative regulation of NF-kappaB import into nucleus(GO:0042347) |
| 0.9 | 6.3 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.9 | 2.6 | GO:0036023 | embryonic skeletal limb joint morphogenesis(GO:0036023) |
| 0.6 | 4.4 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.6 | 2.4 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.6 | 8.7 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.6 | 0.6 | GO:0071338 | submandibular salivary gland formation(GO:0060661) positive regulation of hair follicle cell proliferation(GO:0071338) |
| 0.5 | 22.8 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.5 | 4.3 | GO:0015886 | heme transport(GO:0015886) |
| 0.5 | 1.9 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.5 | 1.8 | GO:0051692 | cellular oligosaccharide catabolic process(GO:0051692) |
| 0.4 | 6.2 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.4 | 8.7 | GO:2000505 | regulation of energy homeostasis(GO:2000505) |
| 0.4 | 19.4 | GO:0045907 | positive regulation of vasoconstriction(GO:0045907) |
| 0.4 | 3.4 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.4 | 2.6 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.4 | 6.3 | GO:0044068 | modulation by symbiont of host cellular process(GO:0044068) |
| 0.4 | 7.0 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
| 0.4 | 2.9 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.4 | 3.6 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.3 | 16.1 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.3 | 3.1 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.3 | 2.2 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.2 | 4.6 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.2 | 1.1 | GO:2000847 | DNA damage induced protein phosphorylation(GO:0006975) negative regulation of corticosteroid hormone secretion(GO:2000847) negative regulation of glucocorticoid secretion(GO:2000850) |
| 0.2 | 1.6 | GO:0060836 | lymphatic endothelial cell differentiation(GO:0060836) |
| 0.2 | 2.5 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.2 | 2.9 | GO:1901663 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.2 | 5.7 | GO:0001953 | negative regulation of cell-matrix adhesion(GO:0001953) |
| 0.2 | 3.4 | GO:0003417 | growth plate cartilage development(GO:0003417) |
| 0.2 | 3.6 | GO:0055012 | ventricular cardiac muscle cell differentiation(GO:0055012) |
| 0.1 | 11.0 | GO:0045576 | mast cell activation(GO:0045576) |
| 0.1 | 8.5 | GO:0048663 | neuron fate commitment(GO:0048663) |
| 0.1 | 1.9 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.1 | 2.7 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.1 | 7.6 | GO:0072384 | organelle transport along microtubule(GO:0072384) |
| 0.1 | 7.0 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.1 | 21.5 | GO:0010506 | regulation of autophagy(GO:0010506) |
| 0.1 | 2.0 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.1 | 28.6 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.1 | 4.0 | GO:0006900 | membrane budding(GO:0006900) |
| 0.1 | 3.3 | GO:0080171 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.1 | 0.6 | GO:0045876 | positive regulation of sister chromatid cohesion(GO:0045876) |
| 0.1 | 9.5 | GO:0006275 | regulation of DNA replication(GO:0006275) |
| 0.1 | 0.7 | GO:0033148 | positive regulation of intracellular estrogen receptor signaling pathway(GO:0033148) |
| 0.0 | 7.7 | GO:0006479 | protein methylation(GO:0006479) protein alkylation(GO:0008213) |
| 0.0 | 0.2 | GO:0009597 | detection of virus(GO:0009597) |
| 0.0 | 3.4 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 1.1 | GO:0046326 | positive regulation of glucose import(GO:0046326) |
| 0.0 | 2.4 | GO:0065004 | protein-DNA complex assembly(GO:0065004) |
| 0.0 | 3.1 | GO:0030010 | establishment of cell polarity(GO:0030010) |
| 0.0 | 1.9 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 10.7 | GO:0000278 | mitotic cell cycle(GO:0000278) |
| 0.0 | 6.9 | GO:0007283 | spermatogenesis(GO:0007283) |
| 0.0 | 0.2 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.5 | 14.0 | GO:0036398 | TCR signalosome(GO:0036398) |
| 3.3 | 16.7 | GO:0043293 | apoptosome(GO:0043293) |
| 2.7 | 8.2 | GO:0005656 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 2.3 | 11.4 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 2.2 | 26.8 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 1.9 | 9.6 | GO:0035976 | AP1 complex(GO:0035976) |
| 1.9 | 11.6 | GO:0005608 | laminin-3 complex(GO:0005608) |
| 1.8 | 58.3 | GO:0031430 | M band(GO:0031430) |
| 1.6 | 21.4 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 1.5 | 9.0 | GO:0048237 | Weibel-Palade body(GO:0033093) rough endoplasmic reticulum lumen(GO:0048237) |
| 1.3 | 4.0 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.9 | 6.8 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.8 | 4.8 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.8 | 3.1 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.8 | 27.9 | GO:0043034 | costamere(GO:0043034) |
| 0.8 | 6.2 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.7 | 4.9 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.6 | 15.2 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.5 | 2.2 | GO:0044307 | dendritic branch(GO:0044307) |
| 0.5 | 5.7 | GO:0032982 | myosin filament(GO:0032982) |
| 0.5 | 3.3 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.5 | 36.6 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.5 | 3.2 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.3 | 2.7 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.3 | 14.0 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.3 | 2.5 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.2 | 2.6 | GO:0002177 | manchette(GO:0002177) |
| 0.2 | 16.0 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.2 | 27.0 | GO:0031674 | I band(GO:0031674) |
| 0.2 | 14.9 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.2 | 2.5 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.2 | 3.2 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.1 | 8.8 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.1 | 41.2 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.1 | 5.6 | GO:0015030 | Cajal body(GO:0015030) |
| 0.1 | 1.9 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.1 | 1.9 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.1 | 26.3 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.1 | 0.6 | GO:0000798 | nuclear cohesin complex(GO:0000798) |
| 0.1 | 6.3 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.1 | 22.3 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.1 | 9.8 | GO:0005840 | ribosome(GO:0005840) |
| 0.1 | 7.7 | GO:0000922 | spindle pole(GO:0000922) |
| 0.1 | 6.2 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 4.0 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.1 | 9.5 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.1 | 29.0 | GO:0009986 | cell surface(GO:0009986) |
| 0.0 | 2.0 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 4.3 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 12.8 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 2.4 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 2.6 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 59.7 | GO:0005654 | nucleoplasm(GO:0005654) |
| 0.0 | 4.3 | GO:0010008 | endosome membrane(GO:0010008) |
| 0.0 | 6.9 | GO:0005768 | endosome(GO:0005768) |
| 0.0 | 3.1 | GO:0000790 | nuclear chromatin(GO:0000790) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.8 | 43.8 | GO:0043120 | tumor necrosis factor binding(GO:0043120) |
| 7.7 | 23.0 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 4.8 | 14.5 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 4.5 | 26.8 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 4.2 | 58.2 | GO:0031432 | titin binding(GO:0031432) |
| 3.8 | 11.5 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 3.1 | 21.4 | GO:0001595 | angiotensin receptor activity(GO:0001595) |
| 3.0 | 12.1 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 3.0 | 9.0 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 2.2 | 8.8 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 2.2 | 17.4 | GO:0071253 | connexin binding(GO:0071253) |
| 2.2 | 8.7 | GO:0016433 | rRNA (adenine) methyltransferase activity(GO:0016433) |
| 2.1 | 27.9 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 1.9 | 42.5 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 1.9 | 7.7 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 1.4 | 8.6 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 1.3 | 12.1 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 1.3 | 9.4 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 1.3 | 5.1 | GO:0055100 | adiponectin binding(GO:0055100) |
| 1.3 | 6.3 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 1.1 | 3.3 | GO:0031208 | POZ domain binding(GO:0031208) |
| 1.1 | 6.5 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 1.1 | 19.4 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 1.0 | 13.2 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.9 | 5.5 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.8 | 2.4 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.8 | 8.7 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.7 | 8.9 | GO:0044213 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.6 | 3.6 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.6 | 4.8 | GO:0043199 | sulfate binding(GO:0043199) |
| 0.6 | 8.2 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.5 | 21.3 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.5 | 11.4 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.5 | 3.6 | GO:0043426 | MRF binding(GO:0043426) |
| 0.5 | 11.3 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.5 | 2.9 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.5 | 4.3 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.5 | 6.1 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.5 | 1.8 | GO:0052796 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.4 | 4.0 | GO:0043495 | protein anchor(GO:0043495) |
| 0.4 | 5.7 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.4 | 20.6 | GO:0043548 | phosphatidylinositol 3-kinase binding(GO:0043548) |
| 0.4 | 7.6 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.4 | 18.3 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.4 | 7.7 | GO:0015106 | bicarbonate transmembrane transporter activity(GO:0015106) |
| 0.3 | 39.1 | GO:0005178 | integrin binding(GO:0005178) |
| 0.3 | 3.1 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.3 | 8.3 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.3 | 2.0 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.2 | 1.6 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.2 | 2.0 | GO:0008296 | 3'-5'-exodeoxyribonuclease activity(GO:0008296) |
| 0.2 | 6.3 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.1 | 4.4 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.1 | 3.4 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.1 | 2.5 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.1 | 7.6 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.1 | 3.1 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.1 | 1.1 | GO:0009881 | photoreceptor activity(GO:0009881) |
| 0.1 | 15.2 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.1 | 1.9 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.1 | 15.2 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.1 | 0.6 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.1 | 2.2 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.1 | 10.0 | GO:0001158 | enhancer sequence-specific DNA binding(GO:0001158) |
| 0.1 | 31.9 | GO:0003712 | transcription cofactor activity(GO:0003712) |
| 0.1 | 8.8 | GO:0000975 | regulatory region DNA binding(GO:0000975) transcription regulatory region DNA binding(GO:0044212) |
| 0.1 | 2.7 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.1 | 1.9 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.1 | 22.8 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.1 | 2.9 | GO:0043531 | ADP binding(GO:0043531) |
| 0.1 | 3.6 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.1 | 7.5 | GO:0004386 | helicase activity(GO:0004386) |
| 0.1 | 9.8 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.0 | 7.0 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 12.1 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 28.2 | GO:0001071 | nucleic acid binding transcription factor activity(GO:0001071) transcription factor activity, sequence-specific DNA binding(GO:0003700) |
| 0.0 | 6.2 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 7.9 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |
| 0.0 | 10.4 | GO:0003677 | DNA binding(GO:0003677) |
| 0.0 | 1.6 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.2 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 27.5 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.7 | 11.6 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.6 | 19.5 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.6 | 24.3 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.6 | 33.3 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.5 | 9.8 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.5 | 14.3 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.4 | 10.0 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.4 | 18.3 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.4 | 8.8 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.4 | 4.9 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.4 | 23.3 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.4 | 19.4 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.4 | 25.7 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.3 | 4.8 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.3 | 16.6 | PID E2F PATHWAY | E2F transcription factor network |
| 0.2 | 10.0 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.2 | 9.0 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.2 | 9.0 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.2 | 58.4 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.2 | 5.7 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 6.5 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.1 | 1.6 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.1 | 2.9 | PID FOXO PATHWAY | FoxO family signaling |
| 0.1 | 5.4 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.1 | 3.3 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 2.6 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.1 | 3.0 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 1.5 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 5.5 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.1 | 21.4 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 2.8 | 34.0 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 1.6 | 23.0 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 1.0 | 27.5 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.9 | 9.8 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.9 | 26.8 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.8 | 12.1 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.8 | 6.8 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.7 | 8.2 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.6 | 7.4 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.6 | 13.1 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.6 | 14.4 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.5 | 11.2 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.5 | 14.5 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.4 | 6.2 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.4 | 1.9 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.3 | 13.2 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.3 | 5.7 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.3 | 5.7 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.3 | 4.9 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.2 | 28.2 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.2 | 3.3 | REACTOME ACTIVATION OF CHAPERONES BY ATF6 ALPHA | Genes involved in Activation of Chaperones by ATF6-alpha |
| 0.2 | 5.7 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.2 | 3.2 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.2 | 3.1 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.2 | 41.0 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.2 | 12.3 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.2 | 2.2 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.2 | 8.4 | REACTOME SIGNALING BY EGFR IN CANCER | Genes involved in Signaling by EGFR in Cancer |
| 0.2 | 8.4 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.1 | 11.6 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.1 | 1.6 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 4.0 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.1 | 1.9 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.1 | 6.2 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.1 | 9.0 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |
| 0.1 | 3.6 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 1.8 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.6 | REACTOME FGFR1 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR1 ligand binding and activation |