Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
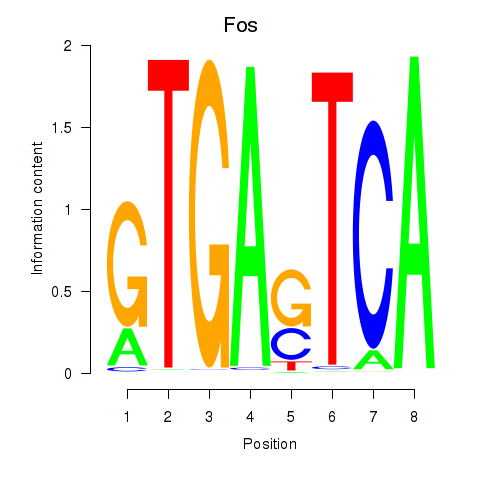
Results for Fos
Z-value: 1.22
Transcription factors associated with Fos
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Fos
|
ENSRNOG00000008015 | FBJ osteosarcoma oncogene |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Fos | rn6_v1_chr6_+_109300433_109300433 | 0.48 | 2.9e-20 | Click! |
Activity profile of Fos motif
Sorted Z-values of Fos motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_-_70626252 | 72.32 |
ENSRNOT00000036947
|
Lamc2
|
laminin subunit gamma 2 |
| chr13_-_70625842 | 67.61 |
ENSRNOT00000092499
|
Lamc2
|
laminin subunit gamma 2 |
| chr1_+_79989019 | 57.47 |
ENSRNOT00000020428
|
Dmpk
|
dystrophia myotonica-protein kinase |
| chr16_-_18766174 | 37.49 |
ENSRNOT00000084813
|
Sftpd
|
surfactant protein D |
| chr10_-_88122233 | 32.80 |
ENSRNOT00000083895
ENSRNOT00000005285 |
Krt14
|
keratin 14 |
| chr3_-_92749121 | 32.65 |
ENSRNOT00000008760
|
Cd44
|
CD44 molecule (Indian blood group) |
| chr16_-_10941414 | 29.96 |
ENSRNOT00000086627
ENSRNOT00000085414 ENSRNOT00000081631 ENSRNOT00000087521 ENSRNOT00000083623 |
Ldb3
|
LIM domain binding 3 |
| chr5_+_164796185 | 29.29 |
ENSRNOT00000010779
|
Nppb
|
natriuretic peptide B |
| chr1_+_199555722 | 28.74 |
ENSRNOT00000054983
|
Itgax
|
integrin subunit alpha X |
| chr18_+_3861539 | 28.13 |
ENSRNOT00000015363
|
Lama3
|
laminin subunit alpha 3 |
| chr4_+_169161585 | 27.94 |
ENSRNOT00000079785
|
Emp1
|
epithelial membrane protein 1 |
| chr4_+_169147243 | 27.76 |
ENSRNOT00000011580
|
Emp1
|
epithelial membrane protein 1 |
| chr2_-_187863503 | 27.57 |
ENSRNOT00000093036
ENSRNOT00000026705 ENSRNOT00000082174 |
Lmna
|
lamin A/C |
| chr20_+_12658065 | 27.37 |
ENSRNOT00000072951
ENSRNOT00000001679 |
Col6a1
|
collagen type VI alpha 1 chain |
| chr8_+_5606592 | 27.04 |
ENSRNOT00000011727
|
Mmp12
|
matrix metallopeptidase 12 |
| chr4_+_78694447 | 26.91 |
ENSRNOT00000011945
|
Gpnmb
|
glycoprotein nmb |
| chr10_-_87067456 | 25.57 |
ENSRNOT00000014163
|
Ccr7
|
C-C motif chemokine receptor 7 |
| chr17_-_55300998 | 25.05 |
ENSRNOT00000064331
|
Svil
|
supervillin |
| chr10_-_88163712 | 24.67 |
ENSRNOT00000005382
ENSRNOT00000084493 |
Krt17
|
keratin 17 |
| chr2_-_227207584 | 24.41 |
ENSRNOT00000065361
ENSRNOT00000080215 |
Myoz2
|
myozenin 2 |
| chr1_-_197770669 | 24.04 |
ENSRNOT00000023563
|
Lat
|
linker for activation of T cells |
| chr3_+_55910177 | 23.73 |
ENSRNOT00000009969
|
Klhl41
|
kelch-like family member 41 |
| chr8_+_5893249 | 23.15 |
ENSRNOT00000014041
|
Mmp7
|
matrix metallopeptidase 7 |
| chr13_+_89975267 | 23.09 |
ENSRNOT00000006266
ENSRNOT00000000053 |
Cd244
|
CD244 molecule |
| chr2_+_225310624 | 22.40 |
ENSRNOT00000015836
|
F3
|
coagulation factor III, tissue factor |
| chr1_+_81230612 | 22.14 |
ENSRNOT00000026489
|
Kcnn4
|
potassium calcium-activated channel subfamily N member 4 |
| chr8_+_75687100 | 22.00 |
ENSRNOT00000038677
|
Anxa2
|
annexin A2 |
| chr1_+_81230989 | 21.60 |
ENSRNOT00000077952
|
Kcnn4
|
potassium calcium-activated channel subfamily N member 4 |
| chr7_-_11648322 | 20.75 |
ENSRNOT00000026871
|
Gadd45b
|
growth arrest and DNA-damage-inducible, beta |
| chr8_-_33661049 | 19.74 |
ENSRNOT00000068037
|
Fli1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr15_+_87704340 | 19.71 |
ENSRNOT00000034987
|
Scel
|
sciellin |
| chr1_+_91152635 | 19.08 |
ENSRNOT00000073438
|
LOC100912070
|
dermokine-like |
| chr20_+_295250 | 18.87 |
ENSRNOT00000000955
|
Clic2
|
chloride intracellular channel 2 |
| chr1_-_198232344 | 18.81 |
ENSRNOT00000080988
|
Aldoa
|
aldolase, fructose-bisphosphate A |
| chr17_-_55346279 | 18.53 |
ENSRNOT00000025037
|
Svil
|
supervillin |
| chr3_+_37545238 | 18.36 |
ENSRNOT00000070792
|
Tnfaip6
|
TNF alpha induced protein 6 |
| chr20_+_5040337 | 18.27 |
ENSRNOT00000068435
|
Clic1
|
chloride intracellular channel 1 |
| chr10_-_88152064 | 17.97 |
ENSRNOT00000019477
|
Krt16
|
keratin 16 |
| chr14_-_85484275 | 17.91 |
ENSRNOT00000083770
|
Kremen1
|
kringle containing transmembrane protein 1 |
| chr4_+_71740532 | 17.80 |
ENSRNOT00000023537
|
Zyx
|
zyxin |
| chr4_-_131694755 | 17.76 |
ENSRNOT00000013271
|
Foxp1
|
forkhead box P1 |
| chr18_-_68551558 | 17.74 |
ENSRNOT00000016369
|
Rab27b
|
RAB27B, member RAS oncogene family |
| chr5_-_159293673 | 17.11 |
ENSRNOT00000009081
|
Padi4
|
peptidyl arginine deiminase 4 |
| chrX_+_51286737 | 16.96 |
ENSRNOT00000035692
|
Dmd
|
dystrophin |
| chr1_+_72420352 | 16.86 |
ENSRNOT00000066307
|
Sbk3
|
SH3 domain binding kinase family, member 3 |
| chr1_+_140601791 | 16.82 |
ENSRNOT00000091588
|
Isg20
|
interferon stimulated exonuclease gene 20 |
| chr10_-_88050622 | 16.43 |
ENSRNOT00000019037
|
Krt15
|
keratin 15 |
| chr14_+_44889287 | 16.06 |
ENSRNOT00000091312
ENSRNOT00000032273 |
Tmem156
|
transmembrane protein 156 |
| chr9_-_100306194 | 16.02 |
ENSRNOT00000087584
|
RGD1563692
|
similar to hypothetical protein FLJ22671 |
| chr3_+_55623634 | 16.00 |
ENSRNOT00000080525
|
Dhrs9
|
dehydrogenase/reductase 9 |
| chr13_+_27449934 | 15.81 |
ENSRNOT00000003409
|
Serpinb2
|
serpin family B member 2 |
| chr2_-_186480278 | 15.54 |
ENSRNOT00000031743
|
Kirrel
|
kin of IRRE like (Drosophila) |
| chrX_+_28593405 | 15.49 |
ENSRNOT00000071708
|
Tmsb4x
|
thymosin beta 4, X-linked |
| chr7_-_143863186 | 15.48 |
ENSRNOT00000017096
|
Rarg
|
retinoic acid receptor, gamma |
| chr1_-_252461461 | 15.33 |
ENSRNOT00000026093
|
Ankrd22
|
ankyrin repeat domain 22 |
| chr13_-_100928811 | 14.61 |
ENSRNOT00000045326
|
Capn2
|
calpain 2 |
| chr1_-_89474252 | 14.34 |
ENSRNOT00000028597
|
Fxyd5
|
FXYD domain-containing ion transport regulator 5 |
| chr20_+_31102476 | 14.03 |
ENSRNOT00000078719
|
Lrrc20
|
leucine rich repeat containing 20 |
| chr1_-_89473904 | 13.92 |
ENSRNOT00000089474
|
Fxyd5
|
FXYD domain-containing ion transport regulator 5 |
| chr10_+_83655460 | 13.91 |
ENSRNOT00000008011
|
Gngt2
|
G protein subunit gamma transducin 2 |
| chr8_+_119228612 | 13.49 |
ENSRNOT00000078439
ENSRNOT00000043737 |
Lrrc2
|
leucine rich repeat containing 2 |
| chr6_+_50528823 | 13.48 |
ENSRNOT00000008321
|
Lamb1
|
laminin subunit beta 1 |
| chr8_+_55603968 | 13.14 |
ENSRNOT00000066848
|
Pou2af1
|
POU class 2 associating factor 1 |
| chr3_-_14019204 | 13.02 |
ENSRNOT00000072400
ENSRNOT00000092918 |
Traf1
|
TNF receptor-associated factor 1 |
| chr8_+_48571323 | 12.44 |
ENSRNOT00000059776
|
Ccdc153
|
coiled-coil domain containing 153 |
| chr1_+_218466289 | 11.91 |
ENSRNOT00000017948
|
Mrgprf
|
MAS related GPR family member F |
| chr18_+_30826260 | 11.87 |
ENSRNOT00000065235
|
Pcdhgb1
|
protocadherin gamma subfamily B, 1 |
| chr2_-_192671059 | 11.73 |
ENSRNOT00000012174
|
Sprr1a
|
small proline-rich protein 1A |
| chr5_-_144274981 | 11.70 |
ENSRNOT00000065871
|
Map7d1
|
MAP7 domain containing 1 |
| chr5_-_151824633 | 11.61 |
ENSRNOT00000043959
|
Sfn
|
stratifin |
| chr3_-_2411544 | 11.31 |
ENSRNOT00000012406
|
Tor4a
|
torsin family 4, member A |
| chr2_+_150756185 | 11.09 |
ENSRNOT00000088461
ENSRNOT00000036808 |
Mbnl1
|
muscleblind-like splicing regulator 1 |
| chr12_+_39553903 | 10.94 |
ENSRNOT00000001738
|
Atp2a2
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 2 |
| chr9_+_15166118 | 10.93 |
ENSRNOT00000020432
|
Mdfi
|
MyoD family inhibitor |
| chr7_-_93280009 | 10.78 |
ENSRNOT00000064877
|
Samd12
|
sterile alpha motif domain containing 12 |
| chr11_-_33003021 | 10.76 |
ENSRNOT00000084134
|
Runx1
|
runt-related transcription factor 1 |
| chr9_-_17835240 | 10.71 |
ENSRNOT00000026988
|
Nfkbie
|
NFKB inhibitor epsilon |
| chr18_+_14756684 | 10.62 |
ENSRNOT00000076085
ENSRNOT00000076129 |
Mapre2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr18_+_56364620 | 10.58 |
ENSRNOT00000068535
ENSRNOT00000086033 |
Pdgfrb
|
platelet derived growth factor receptor beta |
| chr5_+_139394794 | 10.24 |
ENSRNOT00000045954
|
Scmh1
|
sex comb on midleg homolog 1 (Drosophila) |
| chr1_-_124803363 | 10.15 |
ENSRNOT00000066380
|
Klf13
|
Kruppel-like factor 13 |
| chr2_-_187863349 | 10.01 |
ENSRNOT00000084455
|
Lmna
|
lamin A/C |
| chr5_+_59008933 | 9.62 |
ENSRNOT00000023060
|
Car9
|
carbonic anhydrase 9 |
| chr1_-_19376301 | 9.60 |
ENSRNOT00000015547
|
Arhgap18
|
Rho GTPase activating protein 18 |
| chr7_+_54213319 | 9.52 |
ENSRNOT00000005286
|
Nap1l1
|
nucleosome assembly protein 1-like 1 |
| chr1_+_89215266 | 9.41 |
ENSRNOT00000093612
ENSRNOT00000084799 |
Dmkn
|
dermokine |
| chr11_+_85430400 | 9.37 |
ENSRNOT00000083198
|
AABR07034729.1
|
|
| chr10_-_64657089 | 9.18 |
ENSRNOT00000080703
|
Abr
|
active BCR-related |
| chr4_+_83713666 | 9.16 |
ENSRNOT00000086473
|
Creb5
|
cAMP responsive element binding protein 5 |
| chr6_+_129438158 | 9.04 |
ENSRNOT00000005953
|
Bdkrb1
|
bradykinin receptor B1 |
| chr12_-_17519954 | 8.94 |
ENSRNOT00000089418
|
Sun1
|
Sad1 and UNC84 domain containing 1 |
| chr12_+_24761210 | 8.69 |
ENSRNOT00000002003
|
Cldn4
|
claudin 4 |
| chr8_-_128249538 | 8.63 |
ENSRNOT00000082892
|
Scn5a
|
sodium voltage-gated channel alpha subunit 5 |
| chr19_+_41631715 | 8.62 |
ENSRNOT00000022749
|
Chst4
|
carbohydrate sulfotransferase 4 |
| chr10_+_11338306 | 8.60 |
ENSRNOT00000033695
|
LOC100910875
|
protein FAM100A-like |
| chr5_+_169181418 | 8.48 |
ENSRNOT00000004508
|
Klhl21
|
kelch-like family member 21 |
| chr10_+_105499569 | 8.44 |
ENSRNOT00000088457
|
Sphk1
|
sphingosine kinase 1 |
| chr8_-_82492243 | 8.33 |
ENSRNOT00000013852
|
Tmod3
|
tropomodulin 3 |
| chr18_+_36371041 | 8.01 |
ENSRNOT00000025408
|
Sh3rf2
|
SH3 domain containing ring finger 2 |
| chr10_-_82229140 | 7.97 |
ENSRNOT00000004400
|
Epn3
|
epsin 3 |
| chr13_-_95348913 | 7.72 |
ENSRNOT00000057879
|
Akt3
|
AKT serine/threonine kinase 3 |
| chrX_+_110007214 | 7.67 |
ENSRNOT00000093539
|
Nrk
|
Nik related kinase |
| chrX_+_105537602 | 7.60 |
ENSRNOT00000029833
|
Armcx1
|
armadillo repeat containing, X-linked 1 |
| chr10_+_10889488 | 7.59 |
ENSRNOT00000071746
|
Ubald1
|
UBA-like domain containing 1 |
| chr8_+_49342067 | 7.43 |
ENSRNOT00000021693
|
Mpzl2
|
myelin protein zero-like 2 |
| chr1_+_89202527 | 7.42 |
ENSRNOT00000028526
|
Sbsn
|
suprabasin |
| chr8_-_128665988 | 7.17 |
ENSRNOT00000050543
|
Csrnp1
|
cysteine and serine rich nuclear protein 1 |
| chr9_-_17206994 | 7.00 |
ENSRNOT00000026195
|
Gtpbp2
|
GTP binding protein 2 |
| chrX_-_115073890 | 6.92 |
ENSRNOT00000006638
|
Capn6
|
calpain 6 |
| chr11_+_27364916 | 6.89 |
ENSRNOT00000002151
|
Bach1
|
BTB domain and CNC homolog 1 |
| chr1_+_267483145 | 6.88 |
ENSRNOT00000088962
|
Sfr1
|
SWI5-dependent homologous recombination repair protein 1 |
| chr3_-_166994286 | 6.82 |
ENSRNOT00000081593
|
Zfp217
|
zinc finger protein 217 |
| chr3_-_166993940 | 6.63 |
ENSRNOT00000034669
|
Zfp217
|
zinc finger protein 217 |
| chr5_+_58636083 | 6.52 |
ENSRNOT00000067281
|
Unc13b
|
unc-13 homolog B |
| chr3_+_164665532 | 6.50 |
ENSRNOT00000014309
|
Ptpn1
|
protein tyrosine phosphatase, non-receptor type 1 |
| chr10_-_71382058 | 6.45 |
ENSRNOT00000043148
|
Dusp14
|
dual specificity phosphatase 14 |
| chr1_+_220826560 | 6.45 |
ENSRNOT00000027891
|
Fosl1
|
FOS like 1, AP-1 transcription factor subunit |
| chr1_+_14224393 | 6.43 |
ENSRNOT00000016037
|
Perp
|
PERP, TP53 apoptosis effector |
| chr9_+_18564927 | 6.31 |
ENSRNOT00000061014
|
Runx2
|
runt-related transcription factor 2 |
| chr8_-_120446455 | 6.11 |
ENSRNOT00000085161
ENSRNOT00000042854 ENSRNOT00000037199 |
Arpp21
|
cAMP regulated phosphoprotein 21 |
| chr11_+_87435185 | 5.97 |
ENSRNOT00000002558
|
P2rx6
|
purinergic receptor P2X 6 |
| chr5_-_153924896 | 5.94 |
ENSRNOT00000065247
|
Grhl3
|
grainyhead-like transcription factor 3 |
| chr2_+_198040536 | 5.89 |
ENSRNOT00000028744
|
Anp32e
|
acidic nuclear phosphoprotein 32 family member E |
| chr1_-_170318935 | 5.83 |
ENSRNOT00000024119
|
Prkcdbp
|
protein kinase C, delta binding protein |
| chr7_-_115963046 | 5.80 |
ENSRNOT00000007923
|
Slurp1
|
secreted Ly6/Plaur domain containing 1 |
| chr13_-_83425641 | 5.76 |
ENSRNOT00000063870
|
Tbx19
|
T-box 19 |
| chr10_-_35005287 | 5.73 |
ENSRNOT00000032386
|
LOC103689949
|
nedd4 binding protein 3 |
| chr6_+_42092467 | 5.47 |
ENSRNOT00000060499
|
E2f6
|
E2F transcription factor 6 |
| chr15_+_2733114 | 5.39 |
ENSRNOT00000074545
|
LOC108348179
|
dual specificity protein phosphatase 13 |
| chr13_+_92264231 | 5.30 |
ENSRNOT00000066509
ENSRNOT00000004716 |
Spta1
|
spectrin, alpha, erythrocytic 1 |
| chr16_-_2226121 | 5.28 |
ENSRNOT00000091513
|
Slmap
|
sarcolemma associated protein |
| chr1_-_24191908 | 5.26 |
ENSRNOT00000061157
|
Sgk1
|
serum/glucocorticoid regulated kinase 1 |
| chr4_+_129574264 | 5.05 |
ENSRNOT00000010185
|
Arl6ip5
|
ADP-ribosylation factor like GTPase 6 interacting protein 5 |
| chr2_-_60657712 | 5.03 |
ENSRNOT00000040348
|
Rai14
|
retinoic acid induced 14 |
| chr5_+_161889342 | 4.88 |
ENSRNOT00000040481
|
LOC100362684
|
ribosomal protein S20-like |
| chr7_+_11152038 | 4.88 |
ENSRNOT00000006168
|
Nfic
|
nuclear factor I/C |
| chr20_+_6205903 | 4.79 |
ENSRNOT00000092333
ENSRNOT00000092655 |
Kctd20
|
potassium channel tetramerization domain containing 20 |
| chr9_+_117737611 | 4.75 |
ENSRNOT00000022396
|
Zfp161
|
zinc finger protein 161 |
| chr1_+_220428481 | 4.74 |
ENSRNOT00000027335
|
LOC108348044
|
ras and Rab interactor 1 |
| chr7_+_23854846 | 4.63 |
ENSRNOT00000037290
|
Bpifc
|
BPI fold containing family C |
| chr1_-_78180216 | 4.61 |
ENSRNOT00000071576
|
C5ar2
|
complement component 5a receptor 2 |
| chr8_-_115179191 | 4.50 |
ENSRNOT00000017224
|
Parp3
|
poly (ADP-ribose) polymerase family, member 3 |
| chr10_-_37099004 | 4.46 |
ENSRNOT00000075465
|
N4bp3
|
Nedd4 binding protein 3 |
| chr3_+_22964230 | 4.44 |
ENSRNOT00000041813
|
LOC100362149
|
ribosomal protein S20-like |
| chr16_+_1749191 | 4.39 |
ENSRNOT00000014004
|
Zmiz1
|
zinc finger, MIZ-type containing 1 |
| chr1_+_87066289 | 4.35 |
ENSRNOT00000027645
|
Capn12
|
calpain 12 |
| chr1_+_84293102 | 4.22 |
ENSRNOT00000028468
|
Sertad1
|
SERTA domain containing 1 |
| chr5_-_19559244 | 4.20 |
ENSRNOT00000014289
ENSRNOT00000089666 |
Nsmaf
|
neutral sphingomyelinase activation associated factor |
| chr1_+_198199622 | 4.18 |
ENSRNOT00000026688
|
Gdpd3
|
glycerophosphodiester phosphodiesterase domain containing 3 |
| chr4_-_155401480 | 4.15 |
ENSRNOT00000020735
|
Apobec1
|
apolipoprotein B mRNA editing enzyme catalytic subunit 1 |
| chr2_+_203200427 | 4.14 |
ENSRNOT00000020566
|
Vtcn1
|
V-set domain containing T cell activation inhibitor 1 |
| chr4_-_39102807 | 4.14 |
ENSRNOT00000052063
|
Thsd7a
|
thrombospondin type 1 domain containing 7A |
| chrX_-_15627235 | 4.09 |
ENSRNOT00000013369
|
Wdr45
|
WD repeat domain 45 |
| chr3_-_81911197 | 3.98 |
ENSRNOT00000066526
|
Prdm11
|
PR/SET domain 11 |
| chr1_+_220335254 | 3.97 |
ENSRNOT00000072261
|
Rin1
|
Ras and Rab interactor 1 |
| chr19_+_55094585 | 3.81 |
ENSRNOT00000068452
|
Zfpm1
|
zinc finger protein, multitype 1 |
| chr4_+_9882904 | 3.79 |
ENSRNOT00000016909
|
Dnajc2
|
DnaJ heat shock protein family (Hsp40) member C2 |
| chr17_-_84488480 | 3.69 |
ENSRNOT00000000158
ENSRNOT00000075983 |
Nebl
|
nebulette |
| chr18_+_27923572 | 3.65 |
ENSRNOT00000008041
|
Ctnna1
|
catenin alpha 1 |
| chr1_+_197999336 | 3.63 |
ENSRNOT00000023555
|
Apobr
|
apolipoprotein B receptor |
| chr13_+_88606894 | 3.63 |
ENSRNOT00000048692
|
Sh2d1b
|
SH2 domain containing 1B |
| chr1_+_197999037 | 3.57 |
ENSRNOT00000091065
|
Apobr
|
apolipoprotein B receptor |
| chr13_-_47440682 | 3.56 |
ENSRNOT00000037679
ENSRNOT00000005729 ENSRNOT00000050354 ENSRNOT00000050859 |
Pfkfb2
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2 |
| chrX_-_71127237 | 3.56 |
ENSRNOT00000076403
ENSRNOT00000076635 ENSRNOT00000068098 |
Snx12
|
sorting nexin 12 |
| chr1_+_137799185 | 3.53 |
ENSRNOT00000083590
ENSRNOT00000092778 |
Agbl1
|
ATP/GTP binding protein-like 1 |
| chr8_+_43628182 | 3.47 |
ENSRNOT00000090359
|
Olr1320
|
olfactory receptor 1320 |
| chr15_+_2526368 | 3.17 |
ENSRNOT00000048713
ENSRNOT00000074803 |
Dusp13
Dusp13
|
dual specificity phosphatase 13 dual specificity phosphatase 13 |
| chr10_-_56491715 | 3.07 |
ENSRNOT00000020970
|
Kctd11
|
potassium channel tetramerization domain containing 11 |
| chr6_-_99400822 | 3.03 |
ENSRNOT00000089594
|
Zbtb25
|
zinc finger and BTB domain containing 25 |
| chr3_+_113976687 | 2.97 |
ENSRNOT00000022437
|
Eif3j
|
eukaryotic translation initiation factor 3, subunit J |
| chr15_-_27968358 | 2.94 |
ENSRNOT00000040616
|
Rnase9
|
ribonuclease A family member 9 |
| chr2_-_123972356 | 2.91 |
ENSRNOT00000023348
|
Il21
|
interleukin 21 |
| chr3_-_164055561 | 2.90 |
ENSRNOT00000064849
|
B4galt5
|
beta-1,4-galactosyltransferase 5 |
| chr20_+_5008508 | 2.84 |
ENSRNOT00000001153
|
Vwa7
|
von Willebrand factor A domain containing 7 |
| chr12_+_16988136 | 2.81 |
ENSRNOT00000078925
ENSRNOT00000036744 |
Micall2
|
MICAL-like 2 |
| chr14_+_104250617 | 2.75 |
ENSRNOT00000079874
|
Spred2
|
sprouty-related, EVH1 domain containing 2 |
| chr10_+_104582955 | 2.74 |
ENSRNOT00000009733
|
Unk
|
unkempt family zinc finger |
| chr5_+_135687538 | 2.74 |
ENSRNOT00000091664
|
AC126292.2
|
|
| chr1_+_199941161 | 2.71 |
ENSRNOT00000027616
|
Bag3
|
Bcl2-associated athanogene 3 |
| chr20_-_3419831 | 2.69 |
ENSRNOT00000046798
|
Mdc1
|
mediator of DNA damage checkpoint 1 |
| chr16_+_90325304 | 2.67 |
ENSRNOT00000057310
|
Slc10a2
|
solute carrier family 10 member 2 |
| chr8_-_85840818 | 2.57 |
ENSRNOT00000013608
|
Eef1a1
|
eukaryotic translation elongation factor 1 alpha 1 |
| chr11_-_24641820 | 2.48 |
ENSRNOT00000044081
ENSRNOT00000048854 |
App
|
amyloid beta precursor protein |
| chr13_+_51384389 | 2.46 |
ENSRNOT00000087025
|
Kdm5b
|
lysine demethylase 5B |
| chr18_+_36596585 | 2.38 |
ENSRNOT00000036613
|
Rbm27
|
RNA binding motif protein 27 |
| chr13_-_67206688 | 2.37 |
ENSRNOT00000003630
ENSRNOT00000090693 |
Pla2g4a
|
phospholipase A2 group IVA |
| chr2_+_188748359 | 2.28 |
ENSRNOT00000028038
|
Shc1
|
SHC adaptor protein 1 |
| chr7_-_2712723 | 2.21 |
ENSRNOT00000004363
|
Il23a
|
interleukin 23 subunit alpha |
| chr7_+_99954492 | 2.02 |
ENSRNOT00000005885
|
Trib1
|
tribbles pseudokinase 1 |
| chr7_+_3332788 | 1.97 |
ENSRNOT00000010180
|
Cd63
|
Cd63 molecule |
| chrX_+_69730242 | 1.96 |
ENSRNOT00000075980
ENSRNOT00000076425 |
Eda
|
ectodysplasin-A |
| chr9_+_20951260 | 1.95 |
ENSRNOT00000016906
|
Adgrf4
|
adhesion G protein-coupled receptor F4 |
| chr6_-_107678156 | 1.87 |
ENSRNOT00000014158
|
Elmsan1
|
ELM2 and Myb/SANT domain containing 1 |
| chr4_-_99546905 | 1.82 |
ENSRNOT00000077447
|
Kdm3a
|
lysine demethylase 3A |
| chr3_+_8643936 | 1.60 |
ENSRNOT00000077797
|
Set
|
SET nuclear proto-oncogene |
| chr3_+_1452644 | 1.54 |
ENSRNOT00000007949
|
Il1rn
|
interleukin 1 receptor antagonist |
| chr10_-_90312386 | 1.41 |
ENSRNOT00000028445
|
Slc4a1
|
solute carrier family 4 member 1 |
| chr7_+_143122269 | 1.38 |
ENSRNOT00000082542
ENSRNOT00000045495 ENSRNOT00000081386 ENSRNOT00000067422 |
Krt86
|
keratin 86 |
| chr4_+_7314369 | 1.28 |
ENSRNOT00000029724
|
Atg9b
|
autophagy related 9B |
| chr1_+_240908483 | 1.26 |
ENSRNOT00000019367
|
Klf9
|
Kruppel-like factor 9 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Fos
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 14.9 | 74.4 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) |
| 12.9 | 38.6 | GO:1904178 | sterol regulatory element binding protein import into nucleus(GO:0035105) negative regulation of adipose tissue development(GO:1904178) |
| 9.0 | 27.0 | GO:0072642 | type I interferon secretion(GO:0072641) interferon-alpha secretion(GO:0072642) regulation of interferon-alpha secretion(GO:1902739) positive regulation of interferon-alpha secretion(GO:1902741) |
| 8.2 | 32.7 | GO:1900623 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 7.7 | 23.1 | GO:0002780 | antimicrobial peptide biosynthetic process(GO:0002777) antibacterial peptide biosynthetic process(GO:0002780) |
| 7.7 | 23.1 | GO:0071613 | granzyme B production(GO:0071613) regulation of granzyme B production(GO:0071661) positive regulation of granzyme B production(GO:0071663) |
| 7.5 | 37.5 | GO:0050828 | regulation of liquid surface tension(GO:0050828) |
| 7.5 | 22.4 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 7.1 | 28.5 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 6.7 | 139.9 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 6.4 | 25.6 | GO:2000525 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 5.9 | 17.8 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 5.6 | 16.8 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 5.5 | 32.8 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 5.2 | 15.5 | GO:0003430 | growth plate cartilage chondrocyte growth(GO:0003430) |
| 5.1 | 15.4 | GO:2000645 | negative regulation of receptor catabolic process(GO:2000645) |
| 4.5 | 13.5 | GO:0021812 | neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 4.4 | 43.7 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 4.2 | 29.3 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 3.6 | 10.9 | GO:1903233 | positive regulation of endoplasmic reticulum calcium ion concentration(GO:0032470) regulation of calcium ion-dependent exocytosis of neurotransmitter(GO:1903233) calcium ion import into sarcoplasmic reticulum(GO:1990036) |
| 3.6 | 10.8 | GO:2000872 | positive regulation of progesterone secretion(GO:2000872) |
| 3.4 | 17.1 | GO:0036414 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 3.1 | 66.0 | GO:0031424 | keratinization(GO:0031424) |
| 3.1 | 55.7 | GO:0032060 | bleb assembly(GO:0032060) |
| 3.1 | 9.2 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) |
| 3.0 | 8.9 | GO:0021817 | nucleokinesis involved in cell motility in cerebral cortex radial glia guided migration(GO:0021817) nuclear migration along microtubule(GO:0030473) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 2.7 | 10.8 | GO:0007296 | vitellogenesis(GO:0007296) |
| 2.7 | 18.9 | GO:0060315 | negative regulation of ryanodine-sensitive calcium-release channel activity(GO:0060315) |
| 2.6 | 10.6 | GO:0038086 | cell migration involved in vasculogenesis(GO:0035441) VEGF-activated platelet-derived growth factor receptor signaling pathway(GO:0038086) positive regulation of cell proliferation by VEGF-activated platelet derived growth factor receptor signaling pathway(GO:0038091) metanephric glomerulus morphogenesis(GO:0072275) metanephric glomerulus vasculature morphogenesis(GO:0072276) metanephric glomerular capillary formation(GO:0072277) |
| 2.6 | 15.5 | GO:2000483 | negative regulation of interleukin-8 secretion(GO:2000483) |
| 2.2 | 8.6 | GO:0042339 | keratan sulfate metabolic process(GO:0042339) |
| 2.0 | 16.0 | GO:0042904 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 2.0 | 23.7 | GO:0031272 | regulation of pseudopodium assembly(GO:0031272) |
| 1.8 | 17.7 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 1.7 | 8.4 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 1.7 | 28.1 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 1.6 | 6.5 | GO:1903898 | regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) negative regulation of PERK-mediated unfolded protein response(GO:1903898) |
| 1.5 | 4.6 | GO:0071623 | negative regulation of granulocyte chemotaxis(GO:0071623) negative regulation of neutrophil chemotaxis(GO:0090024) |
| 1.5 | 10.7 | GO:0042940 | D-amino acid transport(GO:0042940) |
| 1.5 | 4.5 | GO:1990166 | protein localization to site of double-strand break(GO:1990166) |
| 1.4 | 8.6 | GO:0086047 | membrane depolarization during Purkinje myocyte cell action potential(GO:0086047) membrane depolarization during bundle of His cell action potential(GO:0086048) membrane depolarization during atrial cardiac muscle cell action potential(GO:0098912) |
| 1.4 | 7.0 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
| 1.4 | 4.1 | GO:0090310 | negative regulation of methylation-dependent chromatin silencing(GO:0090310) |
| 1.3 | 18.8 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 1.3 | 3.8 | GO:0071733 | mitral valve formation(GO:0003192) transcriptional activation by promoter-enhancer looping(GO:0071733) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 1.3 | 5.0 | GO:0002036 | regulation of L-glutamate transport(GO:0002036) |
| 1.2 | 27.4 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 1.2 | 3.7 | GO:0071691 | cardiac muscle thin filament assembly(GO:0071691) |
| 1.2 | 20.7 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 1.2 | 3.6 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 1.1 | 2.2 | GO:0043382 | positive regulation of memory T cell differentiation(GO:0043382) |
| 1.1 | 5.3 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 1.1 | 6.3 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 1.0 | 14.6 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 1.0 | 8.3 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 1.0 | 10.5 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.9 | 25.4 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.9 | 19.7 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.9 | 54.4 | GO:0030239 | myofibril assembly(GO:0030239) |
| 0.9 | 5.3 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.8 | 12.4 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.8 | 2.3 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.7 | 14.6 | GO:0061436 | establishment of skin barrier(GO:0061436) |
| 0.7 | 10.2 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.7 | 6.4 | GO:0002934 | desmosome organization(GO:0002934) |
| 0.7 | 3.6 | GO:1903336 | negative regulation of vacuolar transport(GO:1903336) |
| 0.7 | 7.1 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.7 | 4.1 | GO:0072602 | interleukin-4 secretion(GO:0072602) |
| 0.7 | 2.0 | GO:0061153 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.7 | 68.3 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.6 | 9.5 | GO:0014010 | Schwann cell proliferation(GO:0014010) |
| 0.6 | 7.7 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.6 | 3.5 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) |
| 0.5 | 10.9 | GO:0060707 | trophoblast giant cell differentiation(GO:0060707) |
| 0.5 | 26.4 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.5 | 2.0 | GO:0045658 | regulation of neutrophil differentiation(GO:0045658) |
| 0.5 | 2.5 | GO:0051124 | synaptic growth at neuromuscular junction(GO:0051124) |
| 0.5 | 2.9 | GO:0002729 | positive regulation of natural killer cell cytokine production(GO:0002729) |
| 0.5 | 2.4 | GO:2001280 | positive regulation of prostaglandin biosynthetic process(GO:0031394) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.4 | 3.6 | GO:0033133 | positive regulation of glucokinase activity(GO:0033133) positive regulation of hexokinase activity(GO:1903301) |
| 0.4 | 7.7 | GO:0010765 | positive regulation of sodium ion transport(GO:0010765) |
| 0.4 | 18.3 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.4 | 11.1 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.4 | 14.0 | GO:0030728 | ovulation(GO:0030728) |
| 0.4 | 15.7 | GO:0045103 | intermediate filament-based process(GO:0045103) |
| 0.3 | 1.4 | GO:0035937 | estrogen secretion(GO:0035937) estradiol secretion(GO:0035938) regulation of estrogen secretion(GO:2000861) regulation of estradiol secretion(GO:2000864) |
| 0.3 | 1.3 | GO:1902534 | single-organism membrane invagination(GO:1902534) |
| 0.3 | 1.2 | GO:0000414 | regulation of histone H3-K36 methylation(GO:0000414) |
| 0.3 | 5.9 | GO:0043486 | histone exchange(GO:0043486) |
| 0.3 | 1.8 | GO:0046292 | formaldehyde metabolic process(GO:0046292) |
| 0.3 | 0.6 | GO:0072126 | positive regulation of glomerular mesangial cell proliferation(GO:0072126) |
| 0.3 | 1.4 | GO:0010037 | response to carbon dioxide(GO:0010037) |
| 0.3 | 9.0 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.3 | 1.1 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.3 | 19.5 | GO:0030216 | keratinocyte differentiation(GO:0030216) |
| 0.3 | 9.6 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.3 | 12.9 | GO:0007588 | excretion(GO:0007588) |
| 0.2 | 8.5 | GO:0034629 | cellular protein complex localization(GO:0034629) |
| 0.2 | 17.9 | GO:0030279 | negative regulation of ossification(GO:0030279) |
| 0.2 | 6.0 | GO:0033198 | response to ATP(GO:0033198) |
| 0.2 | 7.2 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.2 | 3.1 | GO:0007406 | negative regulation of neuroblast proliferation(GO:0007406) |
| 0.2 | 2.7 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.2 | 34.6 | GO:0007519 | skeletal muscle tissue development(GO:0007519) |
| 0.2 | 1.2 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.2 | 9.2 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.2 | 5.3 | GO:0006779 | porphyrin-containing compound biosynthetic process(GO:0006779) |
| 0.2 | 2.7 | GO:0097201 | negative regulation of transcription from RNA polymerase II promoter in response to stress(GO:0097201) |
| 0.2 | 4.8 | GO:0003170 | heart valve development(GO:0003170) |
| 0.2 | 5.8 | GO:0021983 | pituitary gland development(GO:0021983) |
| 0.1 | 0.4 | GO:2000015 | regulation of determination of dorsal identity(GO:2000015) |
| 0.1 | 0.4 | GO:1903660 | vacuolar transmembrane transport(GO:0034486) chaperone-mediated protein transport involved in chaperone-mediated autophagy(GO:0061741) negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.1 | 2.6 | GO:1903427 | negative regulation of reactive oxygen species biosynthetic process(GO:1903427) |
| 0.1 | 2.7 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.1 | 1.2 | GO:0007614 | short-term memory(GO:0007614) |
| 0.1 | 6.9 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.1 | 7.4 | GO:0071594 | T cell differentiation in thymus(GO:0033077) thymocyte aggregation(GO:0071594) |
| 0.1 | 0.4 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.1 | 3.8 | GO:2000279 | negative regulation of DNA biosynthetic process(GO:2000279) |
| 0.1 | 10.4 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.1 | 2.8 | GO:0070830 | bicellular tight junction assembly(GO:0070830) |
| 0.1 | 2.3 | GO:0045907 | positive regulation of vasoconstriction(GO:0045907) |
| 0.1 | 4.9 | GO:0042475 | odontogenesis of dentin-containing tooth(GO:0042475) |
| 0.1 | 9.6 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.1 | 6.9 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.1 | 4.2 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.1 | 0.4 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 0.0 | 1.2 | GO:0008608 | attachment of spindle microtubules to kinetochore(GO:0008608) |
| 0.0 | 0.4 | GO:0032825 | positive regulation of natural killer cell differentiation(GO:0032825) |
| 0.0 | 9.9 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 6.1 | GO:2001236 | regulation of extrinsic apoptotic signaling pathway(GO:2001236) |
| 0.0 | 0.8 | GO:2000272 | negative regulation of receptor activity(GO:2000272) |
| 0.0 | 1.5 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 7.2 | GO:0006898 | receptor-mediated endocytosis(GO:0006898) |
| 0.0 | 4.0 | GO:0009952 | anterior/posterior pattern specification(GO:0009952) |
| 0.0 | 0.3 | GO:0030225 | macrophage differentiation(GO:0030225) |
| 0.0 | 4.1 | GO:0090305 | nucleic acid phosphodiester bond hydrolysis(GO:0090305) |
| 0.0 | 1.6 | GO:0043524 | negative regulation of neuron apoptotic process(GO:0043524) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 38.4 | 153.4 | GO:0005607 | laminin-2 complex(GO:0005607) |
| 10.9 | 32.7 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 4.8 | 38.6 | GO:0005638 | lamin filament(GO:0005638) |
| 4.7 | 28.1 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 3.6 | 10.9 | GO:0014801 | longitudinal sarcoplasmic reticulum(GO:0014801) calcium ion-transporting ATPase complex(GO:0090534) |
| 3.3 | 19.7 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 2.9 | 38.3 | GO:0031143 | pseudopodium(GO:0031143) |
| 2.7 | 22.0 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 2.3 | 6.9 | GO:0032798 | Swi5-Sfr1 complex(GO:0032798) |
| 2.1 | 57.5 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 1.4 | 60.5 | GO:0043034 | costamere(GO:0043034) |
| 1.3 | 8.9 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 1.2 | 8.5 | GO:0005827 | polar microtubule(GO:0005827) |
| 1.1 | 18.8 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 1.1 | 37.5 | GO:0005771 | multivesicular body(GO:0005771) |
| 1.0 | 74.2 | GO:0045095 | keratin filament(GO:0045095) |
| 0.9 | 5.3 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.9 | 24.0 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.8 | 28.8 | GO:0008305 | integrin complex(GO:0008305) |
| 0.8 | 6.5 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.8 | 6.5 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.8 | 10.6 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.7 | 37.1 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.6 | 2.5 | GO:1990761 | growth cone lamellipodium(GO:1990761) growth cone filopodium(GO:1990812) |
| 0.5 | 70.8 | GO:0030018 | Z disc(GO:0030018) |
| 0.5 | 11.5 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.5 | 5.9 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.5 | 10.2 | GO:0010369 | chromocenter(GO:0010369) |
| 0.4 | 8.7 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.4 | 3.6 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.4 | 10.6 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.4 | 36.8 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.4 | 6.0 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.3 | 13.9 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.3 | 8.3 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.3 | 16.8 | GO:0015030 | Cajal body(GO:0015030) |
| 0.3 | 16.8 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.2 | 13.4 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.2 | 0.7 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.2 | 28.3 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.2 | 15.5 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.2 | 5.5 | GO:0044665 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.2 | 4.6 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.2 | 48.1 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.1 | 2.8 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.1 | 0.4 | GO:0036488 | CHOP-C/EBP complex(GO:0036488) CHOP-ATF3 complex(GO:1990622) |
| 0.1 | 8.3 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.1 | 6.9 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.1 | 1.2 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.1 | 4.5 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.1 | 2.4 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.1 | 32.2 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.1 | 10.8 | GO:0005604 | basement membrane(GO:0005604) |
| 0.1 | 5.0 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.1 | 0.9 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.1 | 24.4 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.1 | 1.2 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.1 | 11.7 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.1 | 6.4 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.1 | 9.0 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.1 | 15.5 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.1 | 2.6 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.1 | 2.7 | GO:0005844 | polysome(GO:0005844) |
| 0.1 | 1.3 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.1 | 30.7 | GO:0016604 | nuclear body(GO:0016604) |
| 0.1 | 7.4 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 12.1 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 5.3 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 4.0 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 2.0 | GO:0045177 | apical part of cell(GO:0045177) |
| 0.0 | 1.2 | GO:0000776 | kinetochore(GO:0000776) |
| 0.0 | 0.1 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.6 | 32.8 | GO:1990254 | keratin filament binding(GO:1990254) |
| 6.2 | 43.7 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 4.9 | 24.4 | GO:0051373 | FATZ binding(GO:0051373) |
| 4.4 | 22.0 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 4.3 | 17.1 | GO:0034618 | arginine binding(GO:0034618) |
| 4.1 | 24.7 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 3.7 | 11.1 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 3.5 | 10.6 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 3.4 | 26.9 | GO:0045545 | syndecan binding(GO:0045545) |
| 3.4 | 16.8 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 3.3 | 32.7 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 2.8 | 8.4 | GO:0008481 | sphinganine kinase activity(GO:0008481) |
| 2.7 | 27.4 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 2.7 | 18.8 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 2.6 | 25.6 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 2.3 | 16.0 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 2.0 | 5.9 | GO:0019212 | phosphatase inhibitor activity(GO:0019212) |
| 1.9 | 30.0 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 1.7 | 8.6 | GO:0086062 | voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 1.7 | 46.6 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 1.5 | 4.6 | GO:0001847 | opsonin receptor activity(GO:0001847) |
| 1.5 | 37.5 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 1.5 | 13.5 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 1.4 | 7.2 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 1.4 | 8.6 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 1.4 | 4.2 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 1.3 | 25.9 | GO:0005521 | lamin binding(GO:0005521) |
| 1.3 | 15.5 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 1.2 | 29.3 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 1.0 | 166.7 | GO:0008201 | heparin binding(GO:0008201) |
| 1.0 | 18.3 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.9 | 24.0 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.9 | 23.1 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.9 | 2.7 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 0.9 | 58.6 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.9 | 6.0 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.8 | 10.9 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.8 | 5.3 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.8 | 12.0 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.7 | 9.6 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.7 | 18.4 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.7 | 2.9 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.7 | 6.5 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.7 | 17.7 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.7 | 4.1 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.7 | 12.2 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.6 | 13.7 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.6 | 2.3 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.6 | 15.5 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.5 | 19.6 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.5 | 4.5 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.5 | 6.9 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.5 | 18.9 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.4 | 2.7 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.4 | 16.6 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.4 | 6.4 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.4 | 2.7 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.4 | 15.5 | GO:0017022 | myosin binding(GO:0017022) |
| 0.3 | 6.6 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.3 | 10.8 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.3 | 3.6 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.3 | 50.0 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.3 | 6.5 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.2 | 22.4 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.2 | 21.4 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.2 | 15.6 | GO:0005518 | collagen binding(GO:0005518) |
| 0.2 | 13.1 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.2 | 1.4 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.2 | 1.5 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.2 | 6.3 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.2 | 0.7 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.2 | 6.9 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.1 | 11.6 | GO:0001158 | enhancer sequence-specific DNA binding(GO:0001158) |
| 0.1 | 3.5 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 17.3 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.1 | 0.4 | GO:0002134 | UTP binding(GO:0002134) sulfonylurea receptor binding(GO:0017098) pyrimidine ribonucleoside binding(GO:0032551) |
| 0.1 | 2.6 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 2.0 | GO:0055103 | ligase regulator activity(GO:0055103) ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.1 | 0.4 | GO:0004082 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.1 | 2.0 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.1 | 4.2 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.1 | 9.2 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.1 | 5.9 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.1 | 3.8 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.1 | 2.7 | GO:0030291 | protein serine/threonine kinase inhibitor activity(GO:0030291) |
| 0.1 | 1.4 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.1 | 0.6 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.1 | 38.2 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.1 | 0.4 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 0.8 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.0 | 3.0 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 10.8 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.0 | 10.4 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.4 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.0 | 7.7 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.0 | 3.4 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 7.7 | GO:0042277 | peptide binding(GO:0042277) |
| 0.0 | 1.1 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 2.9 | GO:0004540 | ribonuclease activity(GO:0004540) |
| 0.0 | 2.2 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 12.0 | GO:0003700 | nucleic acid binding transcription factor activity(GO:0001071) transcription factor activity, sequence-specific DNA binding(GO:0003700) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 10.7 | 181.5 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 1.8 | 32.7 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 1.7 | 24.0 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 1.1 | 28.8 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 1.0 | 61.6 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 1.0 | 44.8 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.9 | 10.7 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.9 | 13.0 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.8 | 21.4 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.8 | 19.0 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.8 | 42.4 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.8 | 20.7 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.8 | 49.8 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.7 | 10.0 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.6 | 36.7 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.6 | 17.9 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.6 | 15.5 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.5 | 9.2 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.5 | 13.9 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.3 | 6.5 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.3 | 14.6 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.3 | 7.8 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.2 | 7.6 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.2 | 14.8 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.2 | 9.2 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.2 | 4.2 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.2 | 12.2 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.2 | 6.3 | PID FGF PATHWAY | FGF signaling pathway |
| 0.2 | 4.0 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.1 | 22.0 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 20.4 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 2.4 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.1 | 2.2 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.1 | 1.4 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.1 | 2.3 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.1 | 2.7 | PID ATM PATHWAY | ATM pathway |
| 0.1 | 3.8 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.1 | 11.5 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 1.1 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 1.2 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 0.4 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.4 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 0.4 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 32.7 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 1.8 | 180.4 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 1.0 | 7.7 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 1.0 | 24.0 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.8 | 37.9 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.7 | 22.4 | REACTOME FORMATION OF FIBRIN CLOT CLOTTING CASCADE | Genes involved in Formation of Fibrin Clot (Clotting Cascade) |
| 0.6 | 27.4 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.6 | 44.4 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.6 | 13.9 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.5 | 20.6 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.5 | 25.6 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.4 | 10.9 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.4 | 13.9 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.4 | 17.0 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.4 | 43.7 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.3 | 2.4 | REACTOME ADP SIGNALLING THROUGH P2RY1 | Genes involved in ADP signalling through P2Y purinoceptor 1 |
| 0.3 | 16.8 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.3 | 6.5 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.3 | 8.4 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.2 | 2.9 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.2 | 6.3 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.2 | 2.7 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.1 | 7.7 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.1 | 0.7 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.1 | 10.6 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.1 | 2.7 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.1 | 9.6 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 1.5 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.1 | 5.9 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 9.6 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 2.6 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.4 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 2.6 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 4.1 | REACTOME MRNA PROCESSING | Genes involved in mRNA Processing |