Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
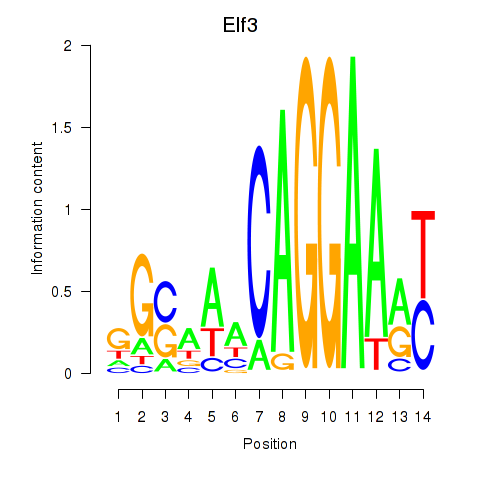
Results for Elf3
Z-value: 1.36
Transcription factors associated with Elf3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Elf3
|
ENSRNOG00000006330 | E74-like factor 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Elf3 | rn6_v1_chr13_-_52088780_52088780 | 0.52 | 9.7e-24 | Click! |
Activity profile of Elf3 motif
Sorted Z-values of Elf3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_+_47930633 | 57.00 |
ENSRNOT00000003515
|
Grap
|
GRB2-related adaptor protein |
| chr14_-_86796378 | 51.78 |
ENSRNOT00000092021
|
Myo1g
|
myosin IG |
| chr13_+_50164563 | 49.42 |
ENSRNOT00000029533
|
Lax1
|
lymphocyte transmembrane adaptor 1 |
| chrX_+_134979646 | 48.02 |
ENSRNOT00000006035
|
Sash3
|
SAM and SH3 domain containing 3 |
| chr13_-_111917587 | 40.87 |
ENSRNOT00000007649
|
Traf3ip3
|
TRAF3 interacting protein 3 |
| chr9_+_43331155 | 38.82 |
ENSRNOT00000023036
|
Zap70
|
zeta chain of T cell receptor associated protein kinase 70 |
| chr6_-_138508753 | 36.43 |
ENSRNOT00000006888
|
Ighm
|
immunoglobulin heavy constant mu |
| chr4_-_78342863 | 34.67 |
ENSRNOT00000049038
|
Gimap6
|
GTPase, IMAP family member 6 |
| chr7_+_12782491 | 33.24 |
ENSRNOT00000065093
|
Cnn2
|
calponin 2 |
| chr3_+_161018511 | 33.12 |
ENSRNOT00000019804
ENSRNOT00000039664 |
Wfdc2
|
WAP four-disulfide core domain 2 |
| chr10_-_19574094 | 31.69 |
ENSRNOT00000059810
|
Dock2
|
dedicator of cytokinesis 2 |
| chr9_-_54484533 | 31.34 |
ENSRNOT00000083514
|
Stat1
|
signal transducer and activator of transcription 1 |
| chr14_-_43992587 | 29.70 |
ENSRNOT00000003425
|
Rhoh
|
ras homolog family member H |
| chr4_+_78371121 | 29.64 |
ENSRNOT00000059157
|
Gimap1
|
GTPase, IMAP family member 1 |
| chr2_+_183674522 | 29.44 |
ENSRNOT00000014433
|
Tmem154
|
transmembrane protein 154 |
| chr7_-_60341264 | 29.40 |
ENSRNOT00000007747
|
Lyz2
|
lysozyme 2 |
| chr1_+_64114721 | 28.66 |
ENSRNOT00000080466
|
Tmc4
|
transmembrane channel-like 4 |
| chr20_+_9948908 | 28.65 |
ENSRNOT00000001541
|
Ubash3a
|
ubiquitin associated and SH3 domain containing, A |
| chrX_+_15155230 | 28.47 |
ENSRNOT00000073289
ENSRNOT00000051439 |
Was
|
Wiskott-Aldrich syndrome |
| chr6_-_137664133 | 27.23 |
ENSRNOT00000018613
|
Gpr132
|
G protein-coupled receptor 132 |
| chr14_-_82347679 | 27.04 |
ENSRNOT00000032972
|
Tacc3
|
transforming, acidic coiled-coil containing protein 3 |
| chr3_+_20303979 | 26.65 |
ENSRNOT00000058331
|
AABR07051730.1
|
|
| chr19_+_19395655 | 25.70 |
ENSRNOT00000019130
|
Snx20
|
sorting nexin 20 |
| chr2_-_196323881 | 22.94 |
ENSRNOT00000028642
|
Tnfaip8l2
|
TNF alpha induced protein 8 like 2 |
| chr17_+_44794130 | 22.91 |
ENSRNOT00000077571
|
Hist1h2ac
|
histone cluster 1, H2ac |
| chr15_+_52451161 | 22.25 |
ENSRNOT00000018725
|
Dok2
|
docking protein 2 |
| chr16_+_18716019 | 21.73 |
ENSRNOT00000047870
|
Sftpa1
|
surfactant protein A1 |
| chr4_+_109467272 | 21.51 |
ENSRNOT00000008212
|
Reg3b
|
regenerating family member 3 beta |
| chr1_+_223214132 | 21.32 |
ENSRNOT00000066559
|
AABR07006142.1
|
|
| chr7_+_11077411 | 20.96 |
ENSRNOT00000007117
|
S1pr4
|
sphingosine-1-phosphate receptor 4 |
| chr13_+_91954138 | 20.92 |
ENSRNOT00000074608
ENSRNOT00000037655 |
Aim2
|
absent in melanoma 2 |
| chr2_+_186776644 | 20.78 |
ENSRNOT00000046778
|
Fcrl3
|
Fc receptor-like 3 |
| chr10_+_39109522 | 20.69 |
ENSRNOT00000010968
|
Irf1
|
interferon regulatory factor 1 |
| chr20_+_9743269 | 20.63 |
ENSRNOT00000001533
ENSRNOT00000083505 |
Abcg1
|
ATP binding cassette subfamily G member 1 |
| chr10_-_70802782 | 20.59 |
ENSRNOT00000045867
|
Ccl6
|
chemokine (C-C motif) ligand 6 |
| chr13_+_47602692 | 20.39 |
ENSRNOT00000038822
|
Fcmr
|
Fc fragment of IgM receptor |
| chr9_+_65620658 | 20.20 |
ENSRNOT00000084498
|
Casp8
|
caspase 8 |
| chr7_-_107616038 | 20.04 |
ENSRNOT00000088752
|
Sla
|
src-like adaptor |
| chr20_+_5184515 | 19.72 |
ENSRNOT00000089411
|
LOC103694381
|
lymphotoxin-beta |
| chr5_+_154260062 | 19.61 |
ENSRNOT00000074998
|
Cnr2
|
cannabinoid receptor 2 |
| chr20_-_4863198 | 19.47 |
ENSRNOT00000001108
|
Ltb
|
lymphotoxin beta |
| chr7_-_117680004 | 19.44 |
ENSRNOT00000040422
|
Slc39a4
|
solute carrier family 39 member 4 |
| chr19_-_52499433 | 19.36 |
ENSRNOT00000021954
|
Cotl1
|
coactosin-like F-actin binding protein 1 |
| chr7_+_70614617 | 19.36 |
ENSRNOT00000035382
|
Arhgap9
|
Rho GTPase activating protein 9 |
| chr20_+_30915213 | 19.28 |
ENSRNOT00000000681
|
Prf1
|
perforin 1 |
| chr15_+_32817343 | 19.23 |
ENSRNOT00000073853
|
AABR07017902.1
|
|
| chr8_+_133195288 | 18.94 |
ENSRNOT00000078529
|
Ccr2
|
C-C motif chemokine receptor 2 |
| chr1_+_64046377 | 18.69 |
ENSRNOT00000085010
|
Tmc4
|
transmembrane channel-like 4 |
| chr20_-_4863011 | 18.26 |
ENSRNOT00000079503
|
Ltb
|
lymphotoxin beta |
| chr11_-_66759402 | 18.19 |
ENSRNOT00000003326
|
Hcls1
|
hematopoietic cell specific Lyn substrate 1 |
| chr1_+_81643816 | 18.16 |
ENSRNOT00000027214
|
LOC103689942
|
carcinoembryonic antigen-related cell adhesion molecule 1-like |
| chr10_+_17421075 | 18.14 |
ENSRNOT00000047011
|
Stk10
|
serine/threonine kinase 10 |
| chr17_-_44793927 | 18.06 |
ENSRNOT00000086309
|
Hist1h2bo
|
histone cluster 1 H2B family member o |
| chr16_-_19349080 | 17.64 |
ENSRNOT00000038494
|
Hsh2d
|
hematopoietic SH2 domain containing |
| chr1_+_63842277 | 17.22 |
ENSRNOT00000087957
ENSRNOT00000080820 |
Lilrb3a
|
leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 3A |
| chr1_+_219397850 | 17.05 |
ENSRNOT00000029829
|
Coro1b
|
coronin 1B |
| chr7_+_118895405 | 16.75 |
ENSRNOT00000092095
|
RGD1309808
|
similar to apolipoprotein L2; apolipoprotein L-II |
| chr11_+_84745904 | 16.52 |
ENSRNOT00000002617
|
Klhl6
|
kelch-like family member 6 |
| chr20_+_5040337 | 16.44 |
ENSRNOT00000068435
|
Clic1
|
chloride intracellular channel 1 |
| chr14_+_5928737 | 16.26 |
ENSRNOT00000071877
ENSRNOT00000040985 ENSRNOT00000074889 |
Mpa2l
|
macrophage activation 2 like |
| chr13_+_88606894 | 16.16 |
ENSRNOT00000048692
|
Sh2d1b
|
SH2 domain containing 1B |
| chr1_+_91152635 | 15.97 |
ENSRNOT00000073438
|
LOC100912070
|
dermokine-like |
| chr12_+_19890749 | 15.82 |
ENSRNOT00000074970
|
RGD1559588
|
similar to cell surface receptor FDFACT |
| chr20_+_5351605 | 15.76 |
ENSRNOT00000089306
ENSRNOT00000041590 ENSRNOT00000081240 ENSRNOT00000082538 |
RT1-A1
|
RT1 class Ia, locus A1 |
| chr1_+_83003841 | 15.68 |
ENSRNOT00000057384
|
Ceacam4
|
carcinoembryonic antigen-related cell adhesion molecule 4 |
| chr9_-_45206427 | 15.19 |
ENSRNOT00000042227
|
Aff3
|
AF4/FMR2 family, member 3 |
| chr15_+_19338175 | 15.11 |
ENSRNOT00000075209
|
Ptger2
|
prostaglandin E receptor 2 |
| chr10_-_57275708 | 14.61 |
ENSRNOT00000005370
|
Pfn1
|
profilin 1 |
| chr8_+_49378644 | 14.53 |
ENSRNOT00000007588
|
Jaml
|
junction adhesion molecule like |
| chr16_-_10706073 | 14.26 |
ENSRNOT00000089114
|
Fam25a
|
family with sequence similarity 25, member A |
| chr10_+_12046541 | 14.23 |
ENSRNOT00000081191
|
Mefv
|
MEFV, pyrin innate immunity regulator |
| chr10_+_49000973 | 14.17 |
ENSRNOT00000057880
|
LOC102553715
|
microfibril-associated glycoprotein 4-like |
| chr1_-_43638161 | 13.99 |
ENSRNOT00000024460
|
Ipcef1
|
interaction protein for cytohesin exchange factors 1 |
| chr12_-_2826378 | 13.98 |
ENSRNOT00000061749
|
Clec4m
|
C-type lectin domain family 4 member M |
| chr15_+_60084918 | 13.87 |
ENSRNOT00000012632
|
Epsti1
|
epithelial stromal interaction 1 |
| chr10_+_47765432 | 13.22 |
ENSRNOT00000078231
|
LOC102553715
|
microfibril-associated glycoprotein 4-like |
| chr4_+_163162211 | 12.77 |
ENSRNOT00000082537
|
Clec1b
|
C-type lectin domain family 1, member B |
| chr8_-_103608913 | 12.66 |
ENSRNOT00000013209
|
Pls1
|
plastin 1 |
| chr4_+_90990088 | 12.53 |
ENSRNOT00000030320
|
Mmrn1
|
multimerin 1 |
| chr1_-_191007503 | 12.45 |
ENSRNOT00000023262
|
Igsf6
|
immunoglobulin superfamily, member 6 |
| chr4_-_170092848 | 12.40 |
ENSRNOT00000072147
|
Hist2h4
|
histone cluster 2, H4 |
| chr6_-_138852571 | 12.37 |
ENSRNOT00000081803
|
AABR07065656.8
|
|
| chr10_+_104952237 | 12.23 |
ENSRNOT00000085222
|
RGD1559482
|
similar to immunoglobulin superfamily, member 7 |
| chr2_-_112831476 | 12.19 |
ENSRNOT00000018055
|
Ect2
|
epithelial cell transforming 2 |
| chr3_+_55369214 | 12.04 |
ENSRNOT00000067161
|
Nostrin
|
nitric oxide synthase trafficking |
| chr9_+_65614142 | 11.78 |
ENSRNOT00000016613
|
Casp8
|
caspase 8 |
| chr12_-_20486276 | 11.70 |
ENSRNOT00000074057
|
LOC680910
|
similar to paired immunoglobin-like type 2 receptor beta |
| chr1_+_75440034 | 11.60 |
ENSRNOT00000033633
|
Pla2g4c
|
phospholipase A2, group IVC |
| chr1_-_206282575 | 11.48 |
ENSRNOT00000024974
|
Adam12
|
ADAM metallopeptidase domain 12 |
| chr15_+_31579478 | 11.30 |
ENSRNOT00000071642
|
AABR07017830.1
|
|
| chr7_+_95309928 | 10.91 |
ENSRNOT00000005887
|
Mtbp
|
MDM2 binding protein |
| chr15_-_45524582 | 10.59 |
ENSRNOT00000081912
|
Gucy1b2
|
guanylate cyclase 1 soluble subunit beta 2 |
| chr15_+_87704340 | 10.58 |
ENSRNOT00000034987
|
Scel
|
sciellin |
| chr1_+_282638017 | 10.35 |
ENSRNOT00000079722
ENSRNOT00000082254 |
Ces2c
|
carboxylesterase 2C |
| chr5_+_48303366 | 10.31 |
ENSRNOT00000009973
|
Gabrr2
|
gamma-aminobutyric acid type A receptor rho 2 subunit |
| chr8_+_48925604 | 10.27 |
ENSRNOT00000077445
|
Ddx6
|
DEAD-box helicase 6 |
| chr15_-_30323833 | 10.03 |
ENSRNOT00000071631
|
AABR07017714.1
|
|
| chr11_+_82680253 | 10.00 |
ENSRNOT00000077119
ENSRNOT00000075512 |
Liph
|
lipase H |
| chr15_+_30270740 | 9.94 |
ENSRNOT00000070991
|
AABR07017707.1
|
|
| chr9_+_47134034 | 9.90 |
ENSRNOT00000020108
|
Il1rl1
|
interleukin 1 receptor-like 1 |
| chr6_+_139523495 | 9.82 |
ENSRNOT00000075467
|
AABR07065699.4
|
|
| chr1_-_260254600 | 9.81 |
ENSRNOT00000019014
|
Blnk
|
B-cell linker |
| chr19_+_37652969 | 9.79 |
ENSRNOT00000041970
|
Carmil2
|
capping protein regulator and myosin 1 linker 2 |
| chr6_+_126170911 | 9.57 |
ENSRNOT00000077477
|
Rin3
|
Ras and Rab interactor 3 |
| chr1_-_89509343 | 9.30 |
ENSRNOT00000028637
|
Fxyd3
|
FXYD domain-containing ion transport regulator 3 |
| chr6_+_126170720 | 9.29 |
ENSRNOT00000065246
|
Rin3
|
Ras and Rab interactor 3 |
| chr1_-_221015929 | 9.24 |
ENSRNOT00000028137
|
Sipa1
|
signal-induced proliferation-associated 1 |
| chr16_+_2379480 | 9.12 |
ENSRNOT00000079215
|
Dnah12
|
dynein, axonemal, heavy chain 12 |
| chr6_+_102477036 | 9.08 |
ENSRNOT00000087132
|
Rad51b
|
RAD51 paralog B |
| chr10_+_103562725 | 9.04 |
ENSRNOT00000075199
|
AABR07030773.1
|
|
| chr15_-_33218456 | 9.02 |
ENSRNOT00000017753
|
Ajuba
|
ajuba LIM protein |
| chr15_-_42898150 | 8.98 |
ENSRNOT00000030036
|
Ptk2b
|
protein tyrosine kinase 2 beta |
| chr15_+_32160364 | 8.94 |
ENSRNOT00000072888
|
AABR07017875.1
|
|
| chr6_-_125590049 | 8.90 |
ENSRNOT00000006371
|
Tc2n
|
tandem C2 domains, nuclear |
| chr4_+_71652354 | 8.84 |
ENSRNOT00000022672
ENSRNOT00000022543 |
Casp2
|
caspase 2 |
| chr8_-_68275720 | 8.74 |
ENSRNOT00000079122
|
Map2k5
|
mitogen activated protein kinase kinase 5 |
| chr4_-_118595580 | 8.72 |
ENSRNOT00000024436
|
Anxa4
|
annexin A4 |
| chr6_-_135877883 | 8.58 |
ENSRNOT00000079092
|
AABR07065589.1
|
|
| chr10_+_104437648 | 8.52 |
ENSRNOT00000035001
|
AC130970.1
|
|
| chr3_-_165360292 | 8.34 |
ENSRNOT00000065615
|
Nfatc2
|
nuclear factor of activated T-cells 2 |
| chr1_+_282694906 | 8.26 |
ENSRNOT00000074303
|
Ces2c
|
carboxylesterase 2C |
| chr10_-_59049482 | 8.20 |
ENSRNOT00000078272
|
Spns2
|
spinster homolog 2 |
| chr7_+_15242227 | 8.19 |
ENSRNOT00000084634
|
Zfp472
|
zinc finger protein 472 |
| chr1_-_175586802 | 8.09 |
ENSRNOT00000071333
|
AABR07005027.1
|
|
| chr13_+_105684420 | 8.07 |
ENSRNOT00000040543
|
Gpatch2
|
G patch domain containing 2 |
| chrX_+_65566047 | 8.03 |
ENSRNOT00000092103
|
Heph
|
hephaestin |
| chr10_+_35133252 | 7.99 |
ENSRNOT00000051916
|
Scgb3a1
|
secretoglobin, family 3A, member 1 |
| chr8_-_72714664 | 7.78 |
ENSRNOT00000024286
|
Rab8b
|
RAB8B, member RAS oncogene family |
| chr1_+_189550354 | 7.76 |
ENSRNOT00000083153
|
Exnef
|
exonuclease NEF-sp |
| chr15_+_32188736 | 7.68 |
ENSRNOT00000081436
|
AABR07017878.1
|
|
| chr17_-_75886523 | 7.66 |
ENSRNOT00000046266
|
Usp6nl
|
USP6 N-terminal like |
| chr1_+_189549960 | 7.66 |
ENSRNOT00000019654
|
Exnef
|
exonuclease NEF-sp |
| chr10_-_56289882 | 7.47 |
ENSRNOT00000090762
ENSRNOT00000056903 |
Tnfsf13
|
tumor necrosis factor superfamily member 13 |
| chr5_+_120498883 | 7.43 |
ENSRNOT00000007859
|
Leprot
|
leptin receptor overlapping transcript |
| chr15_-_34469350 | 7.31 |
ENSRNOT00000067536
|
Adcy4
|
adenylate cyclase 4 |
| chr15_-_33766438 | 7.16 |
ENSRNOT00000033977
|
Ap1g2
|
adaptor-related protein complex 1, gamma 2 subunit |
| chr12_-_9864791 | 6.91 |
ENSRNOT00000073026
|
Gtf3a
|
general transcription factor III A |
| chr18_-_36285444 | 6.87 |
ENSRNOT00000087852
|
Prelid2
|
PRELI domain containing 2 |
| chr7_-_66909470 | 6.80 |
ENSRNOT00000066381
|
Ppm1h
|
protein phosphatase, Mg2+/Mn2+ dependent, 1H |
| chr10_+_45322248 | 6.77 |
ENSRNOT00000047268
|
Trim11
|
tripartite motif-containing 11 |
| chr1_-_100993269 | 6.66 |
ENSRNOT00000027777
|
Bcl2l12
|
BCL2 like 12 |
| chr15_-_29922522 | 6.45 |
ENSRNOT00000081778
|
AABR07017669.2
|
|
| chr5_-_169630340 | 6.41 |
ENSRNOT00000087043
|
Kcnab2
|
potassium voltage-gated channel subfamily A regulatory beta subunit 2 |
| chr17_-_13586554 | 6.40 |
ENSRNOT00000041371
|
Secisbp2
|
SECIS binding protein 2 |
| chr11_+_36736586 | 6.35 |
ENSRNOT00000082098
|
Igsf5
|
immunoglobulin superfamily, member 5 |
| chr4_+_167754525 | 6.32 |
ENSRNOT00000007889
|
Etv6
|
ets variant 6 |
| chr8_-_85840818 | 6.28 |
ENSRNOT00000013608
|
Eef1a1
|
eukaryotic translation elongation factor 1 alpha 1 |
| chr7_+_70987861 | 6.27 |
ENSRNOT00000072906
|
Nemp1
|
nuclear envelope integral membrane protein 1 |
| chr10_+_59743544 | 6.19 |
ENSRNOT00000093497
ENSRNOT00000056460 |
Tax1bp3
|
Tax1 binding protein 3 |
| chr12_+_41340815 | 6.15 |
ENSRNOT00000077542
|
Oas2
|
2'-5' oligoadenylate synthetase 2 |
| chr15_+_30735921 | 6.14 |
ENSRNOT00000078887
|
AABR07017763.2
|
|
| chr15_+_32051651 | 6.09 |
ENSRNOT00000071887
|
LOC103693854
|
uncharacterized LOC103693854 |
| chr13_+_52662996 | 6.08 |
ENSRNOT00000047682
|
Tnnt2
|
troponin T2, cardiac type |
| chr5_+_172986291 | 6.06 |
ENSRNOT00000022900
|
Nadk
|
NAD kinase |
| chr6_+_129519709 | 6.05 |
ENSRNOT00000006307
|
Gskip
|
GSK3B interacting protein |
| chr5_-_154393697 | 6.03 |
ENSRNOT00000090019
|
Rpl11
|
ribosomal protein L11 |
| chr3_+_124088157 | 6.00 |
ENSRNOT00000028876
|
Smox
|
spermine oxidase |
| chr3_-_2411544 | 5.98 |
ENSRNOT00000012406
|
Tor4a
|
torsin family 4, member A |
| chr12_-_41590244 | 5.97 |
ENSRNOT00000038736
|
Slc8b1
|
solute carrier family 8 member B1 |
| chr4_-_163214678 | 5.96 |
ENSRNOT00000091602
|
Clec1a
|
C-type lectin domain family 1, member A |
| chr1_-_13050644 | 5.72 |
ENSRNOT00000081259
|
Heca
|
hdc homolog, cell cycle regulator |
| chr9_+_69497121 | 5.64 |
ENSRNOT00000042562
|
Nrp2
|
neuropilin 2 |
| chr15_-_33358138 | 5.61 |
ENSRNOT00000019155
|
Cebpe
|
CCAAT/enhancer binding protein epsilon |
| chr7_-_12601674 | 5.30 |
ENSRNOT00000093489
|
Arid3a
|
AT-rich interaction domain 3A |
| chr15_+_27875911 | 5.29 |
ENSRNOT00000013582
|
Pnp
|
purine nucleoside phosphorylase |
| chr1_-_77844189 | 5.10 |
ENSRNOT00000017555
|
Gltscr2
|
glioma tumor suppressor candidate region gene 2 |
| chr11_+_57430166 | 5.06 |
ENSRNOT00000093201
|
Phldb2
|
pleckstrin homology-like domain, family B, member 2 |
| chr3_+_113918629 | 5.04 |
ENSRNOT00000078978
ENSRNOT00000037168 |
Ctdspl2
|
CTD small phosphatase like 2 |
| chr3_+_59819157 | 5.04 |
ENSRNOT00000040114
|
LOC103695172
|
uncharacterized LOC103695172 |
| chr5_+_60250546 | 5.01 |
ENSRNOT00000017707
|
Zcchc7
|
zinc finger CCHC-type containing 7 |
| chr4_+_45555077 | 5.00 |
ENSRNOT00000089007
|
Lsm8
|
LSM8 homolog, U6 small nuclear RNA associated |
| chr8_-_71533069 | 4.97 |
ENSRNOT00000021863
|
Trip4
|
thyroid hormone receptor interactor 4 |
| chr15_+_32012543 | 4.97 |
ENSRNOT00000078174
|
AABR07017868.5
|
|
| chr11_-_67647818 | 4.92 |
ENSRNOT00000003068
|
Wdr5b
|
WD repeat domain 5B |
| chrX_-_38196060 | 4.88 |
ENSRNOT00000006741
ENSRNOT00000006438 |
Sh3kbp1
|
SH3 domain-containing kinase-binding protein 1 |
| chr12_+_11179329 | 4.87 |
ENSRNOT00000001302
|
Zfp394
|
zinc finger protein 394 |
| chr5_+_5215196 | 4.86 |
ENSRNOT00000010481
|
Tram1
|
translocation associated membrane protein 1 |
| chrX_+_77119911 | 4.80 |
ENSRNOT00000080141
|
Atp7a
|
ATPase copper transporting alpha |
| chr6_+_12415805 | 4.74 |
ENSRNOT00000022380
|
Gtf2a1l
|
general transcription factor 2A subunit 1 like |
| chr9_+_95274707 | 4.74 |
ENSRNOT00000045163
|
Ugt1a5
|
UDP glucuronosyltransferase family 1 member A5 |
| chr7_-_122329443 | 4.68 |
ENSRNOT00000032232
|
Mkl1
|
megakaryoblastic leukemia (translocation) 1 |
| chr17_-_27602934 | 4.48 |
ENSRNOT00000079298
|
Rreb1
|
ras responsive element binding protein 1 |
| chr13_-_61306939 | 4.38 |
ENSRNOT00000086610
|
Rgs21
|
regulator of G-protein signaling 21 |
| chr11_+_36737022 | 4.35 |
ENSRNOT00000032566
|
Igsf5
|
immunoglobulin superfamily, member 5 |
| chr7_-_117068332 | 4.30 |
ENSRNOT00000082433
|
Fam83h
|
family with sequence similarity 83, member H |
| chr1_-_100231460 | 4.28 |
ENSRNOT00000032200
|
Acpt
|
acid phosphatase, testicular |
| chr7_-_120026755 | 4.19 |
ENSRNOT00000077677
|
Card10
|
caspase recruitment domain family, member 10 |
| chr13_+_88557860 | 4.14 |
ENSRNOT00000058547
|
Sh2d1b2
|
SH2 domain containing 1B2 |
| chr1_+_44019059 | 4.09 |
ENSRNOT00000065733
ENSRNOT00000083425 |
Scaf8
|
SR-related CTD-associated factor 8 |
| chr18_-_399242 | 3.97 |
ENSRNOT00000045926
|
F8
|
coagulation factor VIII |
| chr3_-_112174269 | 3.92 |
ENSRNOT00000067836
|
Tmem87a
|
transmembrane protein 87A |
| chr9_+_121802673 | 3.89 |
ENSRNOT00000086534
|
Yes1
|
YES proto-oncogene 1, Src family tyrosine kinase |
| chr1_+_252375933 | 3.88 |
ENSRNOT00000078405
|
Lipn
|
lipase, family member N |
| chr2_+_45668969 | 3.74 |
ENSRNOT00000014761
ENSRNOT00000071353 |
Arl15
|
ADP-ribosylation factor like GTPase 15 |
| chr3_-_59688692 | 3.72 |
ENSRNOT00000078752
|
Sp3
|
Sp3 transcription factor |
| chr4_-_51946715 | 3.71 |
ENSRNOT00000079130
|
Pot1
|
protection of telomeres 1 |
| chr3_-_147487170 | 3.62 |
ENSRNOT00000077580
|
Fam110a
|
family with sequence similarity 110, member A |
| chr19_-_41349681 | 3.54 |
ENSRNOT00000080694
ENSRNOT00000059147 |
Hydin
|
Hydin, axonemal central pair apparatus protein |
| chr9_+_92435896 | 3.51 |
ENSRNOT00000022901
|
Fbxo36
|
F-box protein 36 |
| chr15_+_48327461 | 3.50 |
ENSRNOT00000018071
|
Ints9
|
integrator complex subunit 9 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Elf3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 12.9 | 38.8 | GO:0043366 | beta selection(GO:0043366) |
| 10.6 | 31.7 | GO:0002277 | myeloid dendritic cell activation involved in immune response(GO:0002277) |
| 8.6 | 51.8 | GO:0038096 | immune response-regulating cell surface receptor signaling pathway involved in phagocytosis(GO:0002433) Fc-gamma receptor signaling pathway involved in phagocytosis(GO:0038096) |
| 8.4 | 33.5 | GO:0035696 | monocyte extravasation(GO:0035696) |
| 7.8 | 31.3 | GO:0034240 | negative regulation of macrophage fusion(GO:0034240) |
| 6.5 | 19.4 | GO:0034224 | cellular response to zinc ion starvation(GO:0034224) |
| 5.8 | 40.4 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 5.7 | 17.0 | GO:1902463 | protein localization to cell leading edge(GO:1902463) |
| 5.4 | 21.5 | GO:1903576 | response to L-arginine(GO:1903576) |
| 5.3 | 15.8 | GO:0002462 | tolerance induction to nonself antigen(GO:0002462) |
| 5.2 | 20.6 | GO:0034436 | glycoprotein transport(GO:0034436) |
| 4.7 | 28.5 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 4.5 | 18.2 | GO:0042509 | regulation of tyrosine phosphorylation of STAT protein(GO:0042509) positive regulation of tyrosine phosphorylation of STAT protein(GO:0042531) |
| 4.5 | 9.0 | GO:2000537 | regulation of B cell chemotaxis(GO:2000537) positive regulation of B cell chemotaxis(GO:2000538) |
| 4.0 | 16.0 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 3.7 | 48.0 | GO:0002726 | positive regulation of T cell cytokine production(GO:0002726) |
| 3.6 | 10.9 | GO:0007089 | traversing start control point of mitotic cell cycle(GO:0007089) |
| 3.6 | 14.2 | GO:0071640 | regulation of macrophage inflammatory protein 1 alpha production(GO:0071640) |
| 3.5 | 17.6 | GO:0051902 | negative regulation of mitochondrial depolarization(GO:0051902) |
| 3.3 | 9.9 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 3.2 | 12.7 | GO:1902896 | terminal web assembly(GO:1902896) |
| 3.1 | 30.8 | GO:0060546 | negative regulation of necroptotic process(GO:0060546) |
| 3.1 | 9.2 | GO:0042631 | cellular response to water deprivation(GO:0042631) |
| 3.0 | 20.9 | GO:0070269 | pyroptosis(GO:0070269) |
| 2.9 | 8.7 | GO:2000342 | negative regulation of chemokine (C-X-C motif) ligand 2 production(GO:2000342) |
| 2.7 | 27.0 | GO:1990001 | inhibition of cysteine-type endopeptidase activity involved in apoptotic process(GO:1990001) |
| 2.7 | 5.3 | GO:0002426 | immunoglobulin production in mucosal tissue(GO:0002426) |
| 2.6 | 20.8 | GO:0050859 | negative regulation of B cell receptor signaling pathway(GO:0050859) |
| 2.6 | 10.3 | GO:0019072 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 2.4 | 9.8 | GO:2000813 | negative regulation of barbed-end actin filament capping(GO:2000813) |
| 2.4 | 21.7 | GO:0008228 | opsonization(GO:0008228) |
| 2.2 | 29.2 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 2.2 | 19.6 | GO:0038171 | cannabinoid signaling pathway(GO:0038171) |
| 2.1 | 6.4 | GO:1904569 | regulation of selenocysteine incorporation(GO:1904569) |
| 2.1 | 27.0 | GO:0030953 | astral microtubule organization(GO:0030953) |
| 2.0 | 6.1 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 1.9 | 5.6 | GO:0021649 | vestibulocochlear nerve structural organization(GO:0021649) ganglion morphogenesis(GO:0061552) dorsal root ganglion morphogenesis(GO:1904835) |
| 1.9 | 7.4 | GO:0060398 | regulation of growth hormone receptor signaling pathway(GO:0060398) |
| 1.8 | 48.1 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 1.8 | 5.3 | GO:0046495 | nicotinamide riboside catabolic process(GO:0006738) urate biosynthetic process(GO:0034418) nicotinamide riboside metabolic process(GO:0046495) pyridine nucleoside metabolic process(GO:0070637) pyridine nucleoside catabolic process(GO:0070638) |
| 1.7 | 33.2 | GO:0050765 | negative regulation of phagocytosis(GO:0050765) |
| 1.6 | 14.6 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 1.6 | 28.6 | GO:0038065 | collagen-activated signaling pathway(GO:0038065) |
| 1.6 | 6.3 | GO:0007296 | vitellogenesis(GO:0007296) |
| 1.6 | 14.0 | GO:0042535 | positive regulation of tumor necrosis factor biosynthetic process(GO:0042535) |
| 1.4 | 9.5 | GO:2000435 | regulation of protein neddylation(GO:2000434) negative regulation of protein neddylation(GO:2000435) |
| 1.2 | 8.7 | GO:2000483 | negative regulation of interleukin-8 secretion(GO:2000483) |
| 1.2 | 75.5 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 1.2 | 3.7 | GO:0060383 | positive regulation of DNA strand elongation(GO:0060383) |
| 1.2 | 6.1 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 1.2 | 19.3 | GO:0001771 | immunological synapse formation(GO:0001771) |
| 1.2 | 4.8 | GO:1904959 | regulation of iron ion transmembrane transport(GO:0034759) negative regulation of iron ion transmembrane transport(GO:0034760) elastin biosynthetic process(GO:0051542) regulation of cytochrome-c oxidase activity(GO:1904959) |
| 1.2 | 1.2 | GO:0014735 | regulation of muscle atrophy(GO:0014735) |
| 1.1 | 6.8 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 1.1 | 4.5 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 1.1 | 7.8 | GO:0051461 | positive regulation of corticotropin secretion(GO:0051461) |
| 1.1 | 7.7 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 1.0 | 3.1 | GO:1904815 | negative regulation of protein localization to chromosome, telomeric region(GO:1904815) |
| 1.0 | 5.1 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 1.0 | 6.0 | GO:0046208 | spermine catabolic process(GO:0046208) |
| 1.0 | 4.0 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 1.0 | 8.8 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 1.0 | 13.4 | GO:0002467 | germinal center formation(GO:0002467) |
| 0.9 | 3.5 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.9 | 3.5 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.8 | 16.3 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.8 | 2.4 | GO:0035330 | regulation of hippo signaling(GO:0035330) |
| 0.8 | 16.4 | GO:0050832 | defense response to fungus(GO:0050832) |
| 0.8 | 6.1 | GO:0018377 | protein myristoylation(GO:0018377) |
| 0.7 | 12.4 | GO:0045653 | negative regulation of megakaryocyte differentiation(GO:0045653) |
| 0.7 | 2.1 | GO:0000105 | histidine biosynthetic process(GO:0000105) |
| 0.7 | 2.1 | GO:0001828 | inner cell mass cellular morphogenesis(GO:0001828) endodermal cell fate determination(GO:0007493) gall bladder development(GO:0061010) |
| 0.7 | 6.8 | GO:0046598 | positive regulation of viral entry into host cell(GO:0046598) |
| 0.6 | 6.4 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.6 | 38.4 | GO:0072676 | lymphocyte migration(GO:0072676) |
| 0.6 | 29.4 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.6 | 5.0 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.6 | 9.7 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.6 | 27.8 | GO:0010972 | negative regulation of G2/M transition of mitotic cell cycle(GO:0010972) |
| 0.6 | 1.8 | GO:0071922 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.6 | 4.7 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.6 | 1.7 | GO:0051409 | response to nitrosative stress(GO:0051409) |
| 0.6 | 6.2 | GO:2000009 | negative regulation of protein localization to cell surface(GO:2000009) |
| 0.6 | 15.1 | GO:0071380 | cellular response to prostaglandin E stimulus(GO:0071380) |
| 0.6 | 9.9 | GO:0010971 | positive regulation of G2/M transition of mitotic cell cycle(GO:0010971) |
| 0.5 | 4.7 | GO:0052697 | xenobiotic glucuronidation(GO:0052697) |
| 0.5 | 2.6 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.5 | 1.5 | GO:0043323 | regulation of natural killer cell degranulation(GO:0043321) positive regulation of natural killer cell degranulation(GO:0043323) |
| 0.5 | 2.0 | GO:1901314 | negative regulation of histone ubiquitination(GO:0033183) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
| 0.5 | 9.3 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.5 | 3.4 | GO:0070092 | negative regulation of glycogen biosynthetic process(GO:0045719) glucagon secretion(GO:0070091) regulation of glucagon secretion(GO:0070092) |
| 0.5 | 29.7 | GO:0045576 | mast cell activation(GO:0045576) |
| 0.5 | 3.7 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.4 | 5.6 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.4 | 6.0 | GO:0006851 | mitochondrial calcium ion transport(GO:0006851) |
| 0.4 | 16.4 | GO:1903959 | regulation of anion transmembrane transport(GO:1903959) |
| 0.4 | 1.2 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.4 | 17.9 | GO:0006182 | cGMP biosynthetic process(GO:0006182) |
| 0.3 | 3.3 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.3 | 2.2 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.3 | 2.2 | GO:0019482 | beta-alanine metabolic process(GO:0019482) |
| 0.3 | 5.6 | GO:0030225 | macrophage differentiation(GO:0030225) |
| 0.3 | 28.5 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.3 | 5.0 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.3 | 4.1 | GO:0045954 | positive regulation of natural killer cell mediated cytotoxicity(GO:0045954) |
| 0.3 | 20.2 | GO:0050868 | negative regulation of T cell activation(GO:0050868) |
| 0.3 | 9.2 | GO:0035116 | embryonic hindlimb morphogenesis(GO:0035116) |
| 0.3 | 6.3 | GO:1903427 | negative regulation of reactive oxygen species biosynthetic process(GO:1903427) |
| 0.3 | 22.7 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.2 | 1.0 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.2 | 18.1 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.2 | 2.2 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.2 | 7.5 | GO:1901998 | toxin transport(GO:1901998) |
| 0.2 | 15.7 | GO:0030317 | sperm motility(GO:0030317) |
| 0.2 | 9.3 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.2 | 1.9 | GO:0035067 | negative regulation of histone acetylation(GO:0035067) |
| 0.1 | 3.5 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.1 | 33.1 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.1 | 4.9 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.1 | 9.8 | GO:0042113 | B cell activation(GO:0042113) |
| 0.1 | 0.7 | GO:0032703 | negative regulation of interleukin-2 production(GO:0032703) |
| 0.1 | 1.7 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.1 | 4.3 | GO:0044380 | protein localization to cytoskeleton(GO:0044380) |
| 0.1 | 2.3 | GO:0010592 | positive regulation of lamellipodium assembly(GO:0010592) |
| 0.1 | 0.9 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.1 | 0.8 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.1 | 1.3 | GO:0048820 | hair follicle maturation(GO:0048820) |
| 0.1 | 2.8 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.1 | 0.1 | GO:0003350 | pulmonary myocardium development(GO:0003350) |
| 0.1 | 1.2 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.1 | 1.5 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.1 | 2.1 | GO:0006911 | phagocytosis, engulfment(GO:0006911) |
| 0.1 | 0.2 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.1 | 4.7 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.1 | 8.2 | GO:0002250 | adaptive immune response(GO:0002250) |
| 0.1 | 0.7 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.1 | 0.3 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.1 | 5.5 | GO:0030216 | keratinocyte differentiation(GO:0030216) |
| 0.0 | 0.7 | GO:2000249 | regulation of actin cytoskeleton reorganization(GO:2000249) |
| 0.0 | 5.7 | GO:0045930 | negative regulation of mitotic cell cycle(GO:0045930) |
| 0.0 | 6.0 | GO:0007596 | blood coagulation(GO:0007596) |
| 0.0 | 0.2 | GO:0016321 | female meiosis chromosome segregation(GO:0016321) |
| 0.0 | 14.7 | GO:0007169 | transmembrane receptor protein tyrosine kinase signaling pathway(GO:0007169) |
| 0.0 | 0.5 | GO:0036344 | platelet formation(GO:0030220) platelet morphogenesis(GO:0036344) |
| 0.0 | 0.4 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.0 | 2.3 | GO:0043401 | steroid hormone mediated signaling pathway(GO:0043401) |
| 0.0 | 1.0 | GO:0008593 | regulation of Notch signaling pathway(GO:0008593) |
| 0.0 | 6.1 | GO:0008380 | RNA splicing(GO:0008380) |
| 0.0 | 3.4 | GO:0090305 | nucleic acid phosphodiester bond hydrolysis(GO:0090305) |
| 0.0 | 0.6 | GO:0019933 | cAMP-mediated signaling(GO:0019933) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 10.7 | 32.0 | GO:0030690 | Noc1p-Noc2p complex(GO:0030690) |
| 4.9 | 29.4 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 4.2 | 20.9 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 3.9 | 19.3 | GO:0031904 | endosome lumen(GO:0031904) |
| 3.0 | 12.2 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 2.2 | 38.8 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 1.8 | 51.8 | GO:0001891 | phagocytic cup(GO:0001891) |
| 1.8 | 9.1 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 1.8 | 12.7 | GO:1990357 | terminal web(GO:1990357) |
| 1.5 | 6.1 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 1.2 | 12.5 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 1.2 | 3.5 | GO:0005760 | gamma DNA polymerase complex(GO:0005760) |
| 1.1 | 15.8 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 1.0 | 10.3 | GO:0097227 | sperm annulus(GO:0097227) |
| 0.8 | 27.0 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.8 | 2.4 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.8 | 4.7 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.8 | 7.8 | GO:0051286 | cell tip(GO:0051286) |
| 0.8 | 109.9 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.7 | 21.5 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.7 | 17.9 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.7 | 3.5 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 0.7 | 6.8 | GO:0070187 | telosome(GO:0070187) |
| 0.6 | 5.0 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.5 | 53.2 | GO:0000786 | nucleosome(GO:0000786) |
| 0.5 | 26.8 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.4 | 3.1 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.4 | 5.1 | GO:0045180 | basal cortex(GO:0045180) |
| 0.4 | 15.7 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.4 | 6.0 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.4 | 25.7 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.4 | 15.0 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.3 | 45.5 | GO:0098857 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.3 | 27.2 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.3 | 2.6 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.3 | 28.5 | GO:0005884 | actin filament(GO:0005884) |
| 0.3 | 0.9 | GO:0044609 | DBIRD complex(GO:0044609) |
| 0.3 | 86.1 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.3 | 3.5 | GO:0042581 | specific granule(GO:0042581) |
| 0.2 | 7.2 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.2 | 3.5 | GO:0032039 | integrator complex(GO:0032039) |
| 0.2 | 5.4 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.2 | 0.7 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.2 | 60.2 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.2 | 21.5 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.2 | 14.2 | GO:0005776 | autophagosome(GO:0005776) |
| 0.1 | 2.1 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.1 | 6.9 | GO:0030286 | dynein complex(GO:0030286) |
| 0.1 | 8.2 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 1.3 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.1 | 0.8 | GO:0090543 | Flemming body(GO:0090543) |
| 0.1 | 1.2 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.1 | 10.9 | GO:0000776 | kinetochore(GO:0000776) |
| 0.1 | 1.1 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.1 | 9.5 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 2.3 | GO:0032420 | stereocilium(GO:0032420) |
| 0.1 | 9.0 | GO:0005769 | early endosome(GO:0005769) |
| 0.1 | 25.0 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.1 | 5.9 | GO:0030133 | transport vesicle(GO:0030133) |
| 0.1 | 3.3 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.1 | 13.9 | GO:0009986 | cell surface(GO:0009986) |
| 0.1 | 4.8 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 1.7 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.0 | 4.9 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 0.4 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 2.8 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.8 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 8.8 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 13.5 | GO:0005813 | centrosome(GO:0005813) |
| 0.0 | 3.0 | GO:0005819 | spindle(GO:0005819) |
| 0.0 | 1.4 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 117.3 | GO:0005576 | extracellular region(GO:0005576) |
| 0.0 | 1.4 | GO:0010008 | endosome membrane(GO:0010008) |
| 0.0 | 4.7 | GO:0098793 | presynapse(GO:0098793) |
| 0.0 | 0.9 | GO:0000123 | histone acetyltransferase complex(GO:0000123) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.0 | 32.0 | GO:0035877 | death effector domain binding(GO:0035877) |
| 7.5 | 37.7 | GO:0043120 | tumor necrosis factor binding(GO:0043120) |
| 6.9 | 20.6 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) glycoprotein transporter activity(GO:0034437) |
| 5.8 | 11.5 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 4.9 | 19.6 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 4.7 | 18.6 | GO:0047619 | acylcarnitine hydrolase activity(GO:0047619) |
| 4.0 | 32.1 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 3.9 | 15.8 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 3.3 | 9.9 | GO:0002114 | interleukin-33 receptor activity(GO:0002114) |
| 3.2 | 18.9 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 2.9 | 31.7 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 2.6 | 31.3 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 2.2 | 123.4 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 2.2 | 15.1 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 2.2 | 51.8 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 2.1 | 21.0 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 2.1 | 27.2 | GO:0003796 | lysozyme activity(GO:0003796) |
| 2.0 | 6.0 | GO:0046592 | polyamine oxidase activity(GO:0046592) |
| 1.8 | 5.3 | GO:0004731 | purine nucleobase binding(GO:0002060) purine-nucleoside phosphorylase activity(GO:0004731) |
| 1.5 | 6.1 | GO:0030172 | troponin C binding(GO:0030172) |
| 1.5 | 14.6 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 1.4 | 8.2 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 1.2 | 27.5 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 1.1 | 17.0 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 1.1 | 6.4 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 1.1 | 6.4 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 1.1 | 9.5 | GO:1990948 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 1.0 | 38.6 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 1.0 | 17.6 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 1.0 | 7.8 | GO:0030911 | TPR domain binding(GO:0030911) |
| 1.0 | 4.8 | GO:0004008 | copper-exporting ATPase activity(GO:0004008) copper-dependent protein binding(GO:0032767) copper-transporting ATPase activity(GO:0043682) |
| 0.9 | 3.7 | GO:1990955 | G-rich single-stranded DNA binding(GO:1990955) |
| 0.9 | 16.4 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.9 | 3.5 | GO:0016890 | site-specific endodeoxyribonuclease activity, specific for altered base(GO:0016890) |
| 0.9 | 4.3 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.9 | 6.0 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.8 | 25.7 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.8 | 19.4 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.8 | 8.8 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.8 | 16.9 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.7 | 10.3 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 0.7 | 14.0 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.7 | 8.7 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.7 | 17.9 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.7 | 24.1 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.7 | 5.6 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.7 | 9.0 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.7 | 6.1 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.7 | 19.4 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.5 | 20.4 | GO:0022829 | wide pore channel activity(GO:0022829) |
| 0.5 | 2.1 | GO:0004487 | methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.5 | 9.1 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.5 | 33.1 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.4 | 3.1 | GO:0098505 | ankyrin repeat binding(GO:0071532) G-rich strand telomeric DNA binding(GO:0098505) |
| 0.4 | 2.2 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.4 | 1.7 | GO:0004609 | phosphatidylserine decarboxylase activity(GO:0004609) |
| 0.4 | 1.2 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.3 | 36.6 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.3 | 4.7 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.3 | 7.0 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.3 | 6.3 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.3 | 6.4 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
| 0.3 | 6.1 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.3 | 9.1 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.3 | 3.4 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.2 | 2.0 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.2 | 97.7 | GO:0005525 | GTP binding(GO:0005525) |
| 0.2 | 4.3 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.2 | 1.5 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.2 | 1.2 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.2 | 1.8 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.2 | 9.3 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.2 | 8.2 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.2 | 54.8 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.2 | 4.7 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.2 | 18.5 | GO:0004702 | receptor signaling protein serine/threonine kinase activity(GO:0004702) |
| 0.1 | 10.3 | GO:0004004 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) |
| 0.1 | 9.3 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.1 | 11.5 | GO:0004527 | exonuclease activity(GO:0004527) |
| 0.1 | 11.3 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.1 | 39.3 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.1 | 11.4 | GO:0005178 | integrin binding(GO:0005178) |
| 0.1 | 8.3 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.1 | 10.0 | GO:0004620 | phospholipase activity(GO:0004620) |
| 0.1 | 1.9 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.1 | 1.5 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.1 | 53.0 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.1 | 9.1 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.1 | 43.5 | GO:0003779 | actin binding(GO:0003779) |
| 0.1 | 6.7 | GO:0002039 | p53 binding(GO:0002039) |
| 0.1 | 1.2 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.1 | 2.0 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.1 | 46.9 | GO:0003690 | double-stranded DNA binding(GO:0003690) |
| 0.1 | 2.6 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.1 | 0.7 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.1 | 1.2 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.1 | 0.2 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 0.1 | 7.2 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.1 | 0.8 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.1 | 9.3 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 1.5 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 16.4 | GO:0019904 | protein domain specific binding(GO:0019904) |
| 0.0 | 1.0 | GO:0003678 | DNA helicase activity(GO:0003678) |
| 0.0 | 1.2 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.0 | 0.4 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.9 | 32.0 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 2.2 | 31.3 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 1.3 | 18.8 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 1.2 | 23.0 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 1.1 | 40.0 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 1.1 | 21.7 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 1.0 | 55.0 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 1.0 | 33.8 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.7 | 26.5 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.7 | 22.3 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.7 | 27.0 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.6 | 9.8 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.5 | 5.6 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.5 | 11.5 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.4 | 8.7 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.4 | 7.3 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.4 | 13.9 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.3 | 14.6 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.3 | 12.2 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.2 | 3.9 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.2 | 2.6 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.2 | 11.4 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.2 | 8.8 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.2 | 13.3 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.2 | 6.9 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.2 | 10.6 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.1 | 4.9 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.1 | 4.7 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.1 | 3.3 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.1 | 1.1 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.1 | 1.2 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.1 | 20.4 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 1.7 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 1.8 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 7.1 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.6 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.2 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 0.3 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.9 | 38.8 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 4.7 | 18.9 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 2.9 | 32.0 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 2.4 | 31.3 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 1.9 | 15.1 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 1.8 | 35.1 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 1.8 | 47.8 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 1.5 | 28.5 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 1.4 | 13.7 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 1.2 | 21.9 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 1.2 | 33.8 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 1.1 | 22.3 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 1.0 | 19.4 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 1.0 | 9.0 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.8 | 8.0 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.8 | 54.2 | REACTOME SIGNALING BY SCF KIT | Genes involved in Signaling by SCF-KIT |
| 0.7 | 11.6 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.6 | 9.2 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.6 | 12.0 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.5 | 5.3 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.5 | 7.3 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.5 | 5.6 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.4 | 6.2 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.4 | 10.3 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.3 | 6.0 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.3 | 8.8 | REACTOME NUCLEOTIDE BINDING DOMAIN LEUCINE RICH REPEAT CONTAINING RECEPTOR NLR SIGNALING PATHWAYS | Genes involved in Nucleotide-binding domain, leucine rich repeat containing receptor (NLR) signaling pathways |
| 0.3 | 4.9 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.3 | 0.3 | REACTOME THROMBOXANE SIGNALLING THROUGH TP RECEPTOR | Genes involved in Thromboxane signalling through TP receptor |
| 0.3 | 3.5 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.2 | 4.7 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.2 | 36.3 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.2 | 2.7 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.2 | 18.9 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.2 | 4.0 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.2 | 4.9 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.2 | 18.6 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.2 | 8.7 | REACTOME NGF SIGNALLING VIA TRKA FROM THE PLASMA MEMBRANE | Genes involved in NGF signalling via TRKA from the plasma membrane |
| 0.2 | 2.9 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.2 | 6.1 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.1 | 21.0 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |
| 0.1 | 4.0 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 3.3 | REACTOME SIGNALING BY EGFR IN CANCER | Genes involved in Signaling by EGFR in Cancer |
| 0.1 | 1.2 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.1 | 9.1 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.1 | 1.1 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.1 | 11.5 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.1 | 1.7 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.1 | 2.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 1.1 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 1.2 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.0 | 2.3 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.0 | 1.8 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 1.8 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |