Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
Results for E2f8
Z-value: 1.21
Transcription factors associated with E2f8
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
E2f8
|
ENSRNOG00000022537 | E2F transcription factor 8 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| E2f8 | rn6_v1_chr1_-_104202591_104202591 | 0.83 | 1.5e-83 | Click! |
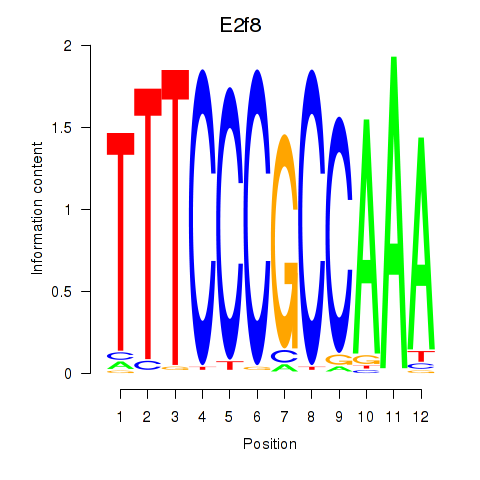
Activity profile of E2f8 motif
Sorted Z-values of E2f8 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_159421671 | 69.78 |
ENSRNOT00000010343
|
Mybl2
|
myeloblastosis oncogene-like 2 |
| chr9_-_10757720 | 62.39 |
ENSRNOT00000083848
|
Uhrf1
|
ubiquitin-like with PHD and ring finger domains 1 |
| chr9_-_10757954 | 61.85 |
ENSRNOT00000075265
|
Uhrf1
|
ubiquitin-like with PHD and ring finger domains 1 |
| chr19_+_14523554 | 56.81 |
ENSRNOT00000084271
ENSRNOT00000064731 |
Mcm5
|
minichromosome maintenance complex component 5 |
| chr12_-_38274036 | 51.21 |
ENSRNOT00000063990
|
Kntc1
|
kinetochore associated 1 |
| chr6_+_43884678 | 49.47 |
ENSRNOT00000091551
|
Rrm2
|
ribonucleotide reductase regulatory subunit M2 |
| chr13_-_45068077 | 40.43 |
ENSRNOT00000004969
|
Mcm6
|
minichromosome maintenance complex component 6 |
| chr1_+_257614731 | 36.85 |
ENSRNOT00000075680
|
LOC100911660
|
lymphocyte-specific helicase-like |
| chr1_+_257901985 | 36.20 |
ENSRNOT00000073015
|
Hells
|
helicase, lymphoid specific |
| chr1_+_257766691 | 35.48 |
ENSRNOT00000088148
|
Hells
|
helicase, lymphoid specific |
| chr1_+_257615088 | 35.19 |
ENSRNOT00000083183
|
LOC100911660
|
lymphocyte-specific helicase-like |
| chr4_-_157798868 | 35.03 |
ENSRNOT00000044425
|
Tuba3b
|
tubulin, alpha 3B |
| chr7_+_102586313 | 34.98 |
ENSRNOT00000006188
|
Myc
|
myelocytomatosis oncogene |
| chr9_-_11027506 | 32.70 |
ENSRNOT00000071107
|
Chaf1a
|
chromatin assembly factor 1 subunit A |
| chr8_-_39201588 | 32.62 |
ENSRNOT00000011226
|
Stt3a
|
STT3A, catalytic subunit of the oligosaccharyltransferase complex |
| chr10_-_4910305 | 31.36 |
ENSRNOT00000033122
|
Rmi2
|
RecQ mediated genome instability 2 |
| chr2_-_190100276 | 28.00 |
ENSRNOT00000015351
|
S100a9
|
S100 calcium binding protein A9 |
| chr5_+_154522119 | 25.25 |
ENSRNOT00000072618
|
E2f2
|
E2F transcription factor 2 |
| chr4_+_179905116 | 24.89 |
ENSRNOT00000052352
|
Tuba3a
|
tubulin, alpha 3A |
| chr12_-_21720094 | 24.68 |
ENSRNOT00000032838
|
LOC100910636
|
zinc finger CW-type PWWP domain protein 1-like |
| chr4_-_120840111 | 22.37 |
ENSRNOT00000022231
|
Mcm2
|
minichromosome maintenance complex component 2 |
| chr4_+_62703779 | 19.24 |
ENSRNOT00000014832
|
Nup205
|
nucleoporin 205 |
| chr3_-_150073721 | 17.79 |
ENSRNOT00000022428
|
E2f1
|
E2F transcription factor 1 |
| chr14_+_48726045 | 16.59 |
ENSRNOT00000086646
|
Arap2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr2_+_197682000 | 15.95 |
ENSRNOT00000066821
|
Hormad1
|
HORMA domain containing 1 |
| chr10_+_67325347 | 15.66 |
ENSRNOT00000079002
ENSRNOT00000085914 |
Suz12
|
suppressor of zeste 12 homolog (Drosophila) |
| chr11_+_89511191 | 15.26 |
ENSRNOT00000002510
|
Mcm4
|
minichromosome maintenance complex component 4 |
| chr5_+_152681101 | 15.13 |
ENSRNOT00000076052
ENSRNOT00000022574 |
Stmn1
|
stathmin 1 |
| chr12_+_660011 | 14.99 |
ENSRNOT00000040830
|
Pds5b
|
PDS5 cohesin associated factor B |
| chr2_-_28423762 | 14.24 |
ENSRNOT00000022864
|
Btf3
|
basic transcription factor 3 |
| chr5_-_147375009 | 14.22 |
ENSRNOT00000009436
|
S100pbp
|
S100P binding protein |
| chr1_+_220009397 | 13.88 |
ENSRNOT00000084410
|
Rbm4b
|
RNA binding motif protein 4B |
| chr19_+_37652969 | 12.68 |
ENSRNOT00000041970
|
Carmil2
|
capping protein regulator and myosin 1 linker 2 |
| chr9_-_61975640 | 12.04 |
ENSRNOT00000085744
|
Boll
|
boule homolog, RNA binding protein |
| chr8_+_111790678 | 11.64 |
ENSRNOT00000013265
|
Topbp1
|
topoisomerase (DNA) II binding protein 1 |
| chr9_-_64573660 | 11.38 |
ENSRNOT00000021299
|
LOC108348134
|
protein boule-like |
| chr12_-_16395029 | 11.27 |
ENSRNOT00000001700
|
Nudt1
|
nudix hydrolase 1 |
| chr14_+_110676090 | 10.96 |
ENSRNOT00000029513
|
Fancl
|
Fanconi anemia, complementation group L |
| chr5_+_147375350 | 10.05 |
ENSRNOT00000010674
|
Yars
|
tyrosyl-tRNA synthetase |
| chr14_+_82356916 | 9.91 |
ENSRNOT00000040229
|
Slbp
|
stem-loop binding protein |
| chrX_-_112159458 | 9.88 |
ENSRNOT00000087403
|
Tex13b
|
testis expressed 13B |
| chr9_-_61974898 | 9.67 |
ENSRNOT00000091519
|
Boll
|
boule homolog, RNA binding protein |
| chr15_+_31469953 | 9.57 |
ENSRNOT00000077849
|
AABR07017825.4
|
|
| chr9_-_64573076 | 8.81 |
ENSRNOT00000084658
|
LOC108348134
|
protein boule-like |
| chr15_+_30621374 | 8.70 |
ENSRNOT00000090632
|
AABR07017748.2
|
|
| chr1_+_221558093 | 8.34 |
ENSRNOT00000077274
|
Majin
|
membrane anchored junction protein |
| chr10_+_86819472 | 8.22 |
ENSRNOT00000081424
|
Cdc6
|
cell division cycle 6 |
| chr10_+_86819929 | 7.09 |
ENSRNOT00000038228
|
Cdc6
|
cell division cycle 6 |
| chr4_+_149970237 | 7.07 |
ENSRNOT00000019529
|
Hnrnpf
|
heterogeneous nuclear ribonucleoprotein F |
| chr20_-_5485837 | 6.32 |
ENSRNOT00000092272
ENSRNOT00000000559 ENSRNOT00000092597 |
Daxx
|
death-domain associated protein |
| chr4_+_149970567 | 6.30 |
ENSRNOT00000091765
|
Hnrnpf
|
heterogeneous nuclear ribonucleoprotein F |
| chr6_+_128973956 | 5.38 |
ENSRNOT00000075399
|
Fam181a
|
family with sequence similarity 181, member A |
| chr5_-_144996431 | 5.23 |
ENSRNOT00000016992
|
Zmym4
|
zinc finger MYM-type containing 4 |
| chr8_-_36314811 | 5.18 |
ENSRNOT00000013243
|
St3gal4
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 4 |
| chr7_+_73273985 | 4.78 |
ENSRNOT00000077730
|
Pop1
|
POP1 homolog, ribonuclease P/MRP subunit |
| chr14_+_106393959 | 4.53 |
ENSRNOT00000092168
|
Wdpcp
|
WD repeat containing planar cell polarity effector |
| chr4_+_157554794 | 4.43 |
ENSRNOT00000024116
|
Ing4
|
inhibitor of growth family, member 4 |
| chr14_+_44580216 | 4.30 |
ENSRNOT00000088674
ENSRNOT00000003907 |
Rfc1
|
replication factor C subunit 1 |
| chr2_+_24546536 | 3.72 |
ENSRNOT00000014036
|
Otp
|
orthopedia homeobox |
| chr10_+_90622992 | 3.23 |
ENSRNOT00000032856
|
Meioc
|
meiosis specific with coiled-coil domain |
| chr7_-_125497691 | 3.01 |
ENSRNOT00000049445
|
AABR07058578.1
|
|
| chrX_-_15504165 | 2.63 |
ENSRNOT00000006233
|
Otud5
|
OTU deubiquitinase 5 |
| chr3_-_151486693 | 1.80 |
ENSRNOT00000073736
ENSRNOT00000071099 |
Gdf5
|
growth differentiation factor 5 |
| chr13_+_96195836 | 1.40 |
ENSRNOT00000042547
|
RGD1563812
|
similar to basic transcription factor 3 |
| chr10_+_93354003 | 1.22 |
ENSRNOT00000008140
|
Mettl2b
|
methyltransferase like 2B |
| chr5_+_147185474 | 1.16 |
ENSRNOT00000000134
|
Ak2
|
adenylate kinase 2 |
| chr6_+_95816749 | 0.77 |
ENSRNOT00000008880
|
Six6
|
SIX homeobox 6 |
| chr1_-_200163106 | 0.70 |
ENSRNOT00000027644
|
Mcmbp
|
minichromosome maintenance complex binding protein |
| chr2_+_186630278 | 0.12 |
ENSRNOT00000021604
|
LOC365839
|
similar to elongation protein 4 homolog |
| chr1_+_174767960 | 0.03 |
ENSRNOT00000013362
|
Wee1
|
WEE1 G2 checkpoint kinase |
Network of associatons between targets according to the STRING database.
First level regulatory network of E2f8
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 24.5 | 195.9 | GO:0010216 | maintenance of DNA methylation(GO:0010216) |
| 11.7 | 35.0 | GO:0044330 | canonical Wnt signaling pathway involved in positive regulation of wound healing(GO:0044330) metanephric cap development(GO:0072185) metanephric cap morphogenesis(GO:0072186) metanephric cap mesenchymal cell proliferation involved in metanephros development(GO:0090094) response to putrescine(GO:1904585) cellular response to putrescine(GO:1904586) |
| 11.2 | 78.1 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 9.3 | 28.0 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 7.2 | 43.0 | GO:1990086 | lens fiber cell apoptotic process(GO:1990086) |
| 4.7 | 32.6 | GO:0043686 | co-translational protein modification(GO:0043686) |
| 3.8 | 15.1 | GO:0070495 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 3.2 | 12.7 | GO:2000813 | negative regulation of barbed-end actin filament capping(GO:2000813) |
| 3.1 | 49.5 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 2.9 | 82.1 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 2.8 | 11.3 | GO:0046061 | dATP catabolic process(GO:0046061) |
| 2.3 | 16.0 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 2.1 | 6.3 | GO:0071454 | cellular response to anoxia(GO:0071454) |
| 1.7 | 9.9 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 1.6 | 15.7 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 1.5 | 4.5 | GO:0016476 | regulation of embryonic cell shape(GO:0016476) |
| 1.4 | 19.2 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 1.2 | 32.7 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 1.2 | 4.8 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 1.1 | 69.8 | GO:0090307 | mitotic spindle assembly(GO:0090307) |
| 1.0 | 21.7 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.9 | 2.6 | GO:1905076 | regulation of interleukin-17 secretion(GO:1905076) negative regulation of interleukin-17 secretion(GO:1905077) |
| 0.8 | 8.3 | GO:0044821 | telomere tethering at nuclear periphery(GO:0034398) meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.6 | 1.8 | GO:0043932 | ossification involved in bone remodeling(GO:0043932) |
| 0.5 | 47.3 | GO:0007093 | mitotic cell cycle checkpoint(GO:0007093) |
| 0.5 | 31.4 | GO:0033045 | regulation of sister chromatid segregation(GO:0033045) |
| 0.5 | 3.7 | GO:0021979 | hypothalamus cell differentiation(GO:0021979) |
| 0.5 | 13.9 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.5 | 3.2 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.4 | 4.4 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.4 | 1.2 | GO:0046060 | dATP metabolic process(GO:0046060) |
| 0.2 | 8.2 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.1 | 7.4 | GO:0006513 | protein monoubiquitination(GO:0006513) |
| 0.1 | 58.9 | GO:0007017 | microtubule-based process(GO:0007017) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 17.1 | 51.2 | GO:1990423 | RZZ complex(GO:1990423) |
| 12.4 | 49.5 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 12.3 | 135.6 | GO:0042555 | MCM complex(GO:0042555) |
| 10.9 | 32.7 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 6.4 | 19.2 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 3.5 | 159.2 | GO:0000791 | euchromatin(GO:0000791) |
| 3.0 | 17.8 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 2.6 | 15.7 | GO:0001739 | sex chromatin(GO:0001739) |
| 2.3 | 71.7 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 1.9 | 32.6 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 1.6 | 4.8 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 1.3 | 12.7 | GO:0008290 | F-actin capping protein complex(GO:0008290) pinosome(GO:0044352) macropinosome(GO:0044354) |
| 1.1 | 9.9 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.7 | 4.3 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.7 | 11.0 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.6 | 8.2 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.5 | 11.6 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.4 | 59.9 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.4 | 8.3 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.4 | 6.3 | GO:0001741 | XY body(GO:0001741) |
| 0.4 | 16.0 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.2 | 4.5 | GO:0044447 | axoneme part(GO:0044447) |
| 0.1 | 15.3 | GO:0000922 | spindle pole(GO:0000922) |
| 0.1 | 5.2 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.1 | 33.8 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.1 | 9.4 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.1 | 11.3 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.1 | 4.4 | GO:0000123 | histone acetyltransferase complex(GO:0000123) |
| 0.1 | 21.4 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 15.1 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 12.1 | GO:0044427 | chromosomal part(GO:0044427) |
| 0.0 | 1.2 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 31.1 | 124.2 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 8.2 | 49.5 | GO:0016728 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 7.0 | 69.8 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 5.5 | 77.5 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 4.7 | 28.0 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 4.1 | 32.7 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 3.8 | 11.3 | GO:0008413 | 8-oxo-7,8-dihydroguanosine triphosphate pyrophosphatase activity(GO:0008413) 8-oxo-7,8-dihydrodeoxyguanosine triphosphate pyrophosphatase activity(GO:0035539) |
| 2.4 | 21.7 | GO:0008494 | translation activator activity(GO:0008494) |
| 2.3 | 32.6 | GO:0004579 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 1.8 | 55.7 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 1.6 | 4.8 | GO:0000171 | ribonuclease MRP activity(GO:0000171) |
| 1.3 | 5.2 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 1.0 | 19.2 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.7 | 35.0 | GO:0070888 | E-box binding(GO:0070888) |
| 0.6 | 6.3 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.5 | 59.9 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.5 | 15.7 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.4 | 2.6 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.4 | 4.3 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.4 | 13.4 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.3 | 16.6 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.3 | 9.9 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.2 | 43.0 | GO:0001047 | core promoter binding(GO:0001047) |
| 0.2 | 14.2 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.2 | 1.8 | GO:0036122 | BMP binding(GO:0036122) |
| 0.2 | 8.2 | GO:0016876 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.1 | 4.4 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.1 | 1.2 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.1 | 58.0 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.1 | 54.7 | GO:0008270 | zinc ion binding(GO:0008270) |
| 0.0 | 15.1 | GO:0015631 | tubulin binding(GO:0015631) |
| 0.0 | 57.9 | GO:0005524 | ATP binding(GO:0005524) |
| 0.0 | 12.7 | GO:0005543 | phospholipid binding(GO:0005543) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 223.2 | PID E2F PATHWAY | E2F transcription factor network |
| 1.2 | 28.0 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 1.2 | 15.1 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 1.1 | 71.7 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.6 | 22.4 | PID ATR PATHWAY | ATR signaling pathway |
| 0.4 | 11.0 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.4 | 16.6 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.2 | 6.3 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.2 | 15.3 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.2 | 4.3 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 12.1 | 133.2 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 3.8 | 82.6 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 3.1 | 50.0 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 2.5 | 25.3 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 1.4 | 35.0 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.9 | 9.9 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.7 | 19.2 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.5 | 11.0 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.3 | 32.6 | REACTOME ASPARAGINE N LINKED GLYCOSYLATION | Genes involved in Asparagine N-linked glycosylation |
| 0.3 | 8.2 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.3 | 24.2 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.1 | 2.6 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.1 | 9.4 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.1 | 1.2 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 1.8 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |