Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
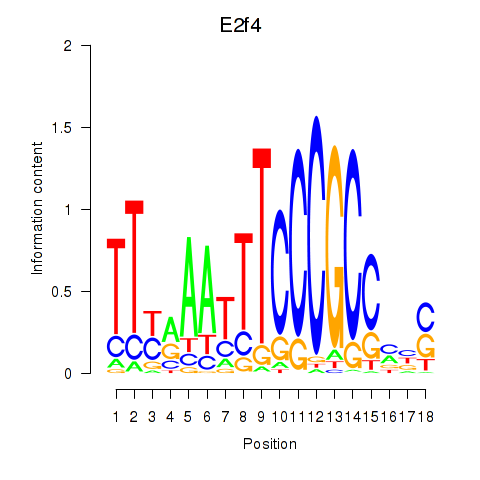
Results for E2f4
Z-value: 1.55
Transcription factors associated with E2f4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
E2f4
|
ENSRNOG00000015708 | E2F transcription factor 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| E2f4 | rn6_v1_chr19_+_37252843_37252843 | 0.79 | 2.2e-69 | Click! |
Activity profile of E2f4 motif
Sorted Z-values of E2f4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_-_108178609 | 75.60 |
ENSRNOT00000004525
|
Cenpf
|
centromere protein F |
| chr13_-_87847263 | 68.88 |
ENSRNOT00000003650
|
Nuf2
|
NUF2, NDC80 kinetochore complex component |
| chr1_+_221448661 | 66.48 |
ENSRNOT00000072493
|
LOC100910252
|
sororin-like |
| chr2_+_264704769 | 61.39 |
ENSRNOT00000012667
|
Depdc1
|
DEP domain containing 1 |
| chr8_+_79054237 | 60.73 |
ENSRNOT00000077613
|
Mns1
|
meiosis-specific nuclear structural 1 |
| chr5_+_152721940 | 58.38 |
ENSRNOT00000039322
|
Aunip
|
aurora kinase A and ninein interacting protein |
| chr1_+_31967978 | 57.21 |
ENSRNOT00000081471
ENSRNOT00000021532 |
Trip13
|
thyroid hormone receptor interactor 13 |
| chr3_+_159421671 | 56.24 |
ENSRNOT00000010343
|
Mybl2
|
myeloblastosis oncogene-like 2 |
| chr15_-_70399924 | 53.31 |
ENSRNOT00000087940
|
Diaph3
|
diaphanous-related formin 3 |
| chr8_+_48665652 | 52.85 |
ENSRNOT00000059715
|
H2afx
|
H2A histone family, member X |
| chr16_+_19800463 | 52.57 |
ENSRNOT00000023065
|
Ankle1
|
ankyrin repeat and LEM domain containing 1 |
| chr5_-_134978125 | 50.57 |
ENSRNOT00000018004
|
Rad54l
|
RAD54 like (S. cerevisiae) |
| chr16_+_41079444 | 48.46 |
ENSRNOT00000015623
|
Neil3
|
nei-like DNA glycosylase 3 |
| chr10_-_89454681 | 46.42 |
ENSRNOT00000028109
|
Brca1
|
BRCA1, DNA repair associated |
| chr15_-_24199341 | 45.10 |
ENSRNOT00000015553
|
Dlgap5
|
DLG associated protein 5 |
| chr15_-_61564695 | 44.53 |
ENSRNOT00000068216
|
Rgcc
|
regulator of cell cycle |
| chr1_-_157700064 | 42.49 |
ENSRNOT00000031974
|
Ddias
|
DNA damage-induced apoptosis suppressor |
| chr8_+_116730641 | 41.33 |
ENSRNOT00000052289
|
Traip
|
TRAF-interacting protein |
| chr9_-_92963697 | 41.22 |
ENSRNOT00000023546
|
Gpr55
|
G protein-coupled receptor 55 |
| chr5_-_150520884 | 41.01 |
ENSRNOT00000085482
|
Rcc1
|
regulator of chromosome condensation 1 |
| chr3_+_1478525 | 35.90 |
ENSRNOT00000008161
|
Psd4
|
pleckstrin and Sec7 domain containing 4 |
| chr10_+_55626741 | 35.86 |
ENSRNOT00000008492
|
Aurkb
|
aurora kinase B |
| chrX_-_31140711 | 32.19 |
ENSRNOT00000082358
|
Fancb
|
Fanconi anemia, complementation group B |
| chr7_-_28715224 | 31.23 |
ENSRNOT00000065899
|
Parpbp
|
PARP1 binding protein |
| chr1_-_94494980 | 30.80 |
ENSRNOT00000020014
|
Ccne1
|
cyclin E1 |
| chr2_+_187447501 | 30.79 |
ENSRNOT00000038589
|
Iqgap3
|
IQ motif containing GTPase activating protein 3 |
| chr3_+_149131785 | 30.52 |
ENSRNOT00000090321
|
Dnmt3b
|
DNA methyltransferase 3 beta |
| chr6_+_129835919 | 30.16 |
ENSRNOT00000036035
|
Vrk1
|
vaccinia related kinase 1 |
| chr10_-_15098791 | 28.47 |
ENSRNOT00000026139
|
Chtf18
|
chromosome transmission fidelity factor 18 |
| chr15_-_60512704 | 27.84 |
ENSRNOT00000049056
|
Tnfsf11
|
tumor necrosis factor superfamily member 11 |
| chr16_-_76110553 | 26.98 |
ENSRNOT00000043384
|
Mcph1
|
microcephalin 1 |
| chr20_+_4967194 | 25.24 |
ENSRNOT00000070846
|
Lsm2
|
LSM2 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr4_+_5841998 | 23.72 |
ENSRNOT00000010025
|
Xrcc2
|
X-ray repair cross complementing 2 |
| chr20_-_7930929 | 22.03 |
ENSRNOT00000000607
|
Tead3
|
TEA domain transcription factor 3 |
| chr12_-_19314016 | 21.93 |
ENSRNOT00000001825
|
Mcm7
|
minichromosome maintenance complex component 7 |
| chr5_-_152198813 | 21.22 |
ENSRNOT00000082953
|
Hmgn2
|
high mobility group nucleosomal binding domain 2 |
| chr13_-_68858781 | 21.03 |
ENSRNOT00000065944
ENSRNOT00000092053 |
Rnf2
|
ring finger protein 2 |
| chr14_-_110883784 | 20.63 |
ENSRNOT00000086299
ENSRNOT00000068190 |
Vrk2
|
vaccinia related kinase 2 |
| chr6_-_26855658 | 20.51 |
ENSRNOT00000011635
|
Agbl5
|
ATP/GTP binding protein-like 5 |
| chr1_-_1392329 | 20.19 |
ENSRNOT00000075346
|
AABR07000137.1
|
|
| chr13_-_90839411 | 19.88 |
ENSRNOT00000010594
|
Slamf9
|
SLAM family member 9 |
| chr20_-_5037022 | 19.72 |
ENSRNOT00000091022
ENSRNOT00000060762 |
Msh5
|
mutS homolog 5 |
| chrX_-_62903530 | 18.94 |
ENSRNOT00000076652
ENSRNOT00000017370 |
Pdk3
|
pyruvate dehydrogenase kinase 3 |
| chr5_-_77031120 | 18.72 |
ENSRNOT00000022987
|
Inip
|
INTS3 and NABP interacting protein |
| chrX_+_15171499 | 18.57 |
ENSRNOT00000008399
|
Suv39h1l1
|
suppressor of variegation 3-9 homolog 1 (Drosophila)-like 1 |
| chr7_-_2941122 | 17.79 |
ENSRNOT00000082107
|
Esyt1
|
extended synaptotagmin 1 |
| chr7_-_117703094 | 17.44 |
ENSRNOT00000019754
|
Tonsl
|
tonsoku-like, DNA repair protein |
| chr1_+_48433079 | 17.34 |
ENSRNOT00000037369
|
Slc22a3
|
solute carrier family 22 member 3 |
| chr20_+_3156170 | 16.73 |
ENSRNOT00000082880
|
RT1-S2
|
RT1 class Ib, locus S2 |
| chr14_+_82356916 | 16.12 |
ENSRNOT00000040229
|
Slbp
|
stem-loop binding protein |
| chr1_+_198214797 | 14.01 |
ENSRNOT00000068543
|
Tbx6
|
T-box 6 |
| chr1_-_1116926 | 13.77 |
ENSRNOT00000062027
|
Raet1l
|
retinoic acid early transcript 1L |
| chr7_-_140951071 | 13.54 |
ENSRNOT00000082521
|
Fam186b
|
family with sequence similarity 186, member B |
| chr20_+_4966817 | 13.16 |
ENSRNOT00000081527
ENSRNOT00000081265 |
Lsm2
|
LSM2 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr10_-_95212111 | 13.05 |
ENSRNOT00000020795
|
Kpna2
|
karyopherin subunit alpha 2 |
| chr13_-_98078555 | 12.23 |
ENSRNOT00000078034
|
Ahctf1
|
AT hook containing transcription factor 1 |
| chr6_+_11644578 | 12.15 |
ENSRNOT00000021923
ENSRNOT00000093689 |
Msh6
|
mutS homolog 6 |
| chr14_+_17210733 | 11.94 |
ENSRNOT00000003075
|
Cxcl10
|
C-X-C motif chemokine ligand 10 |
| chr8_+_62482140 | 11.66 |
ENSRNOT00000026492
|
Edc3
|
enhancer of mRNA decapping 3 |
| chr18_-_27550224 | 11.49 |
ENSRNOT00000037368
|
Cdc25c
|
cell division cycle 25C |
| chr18_-_56115593 | 11.35 |
ENSRNOT00000045041
|
Tcof1
|
treacle ribosome biogenesis factor 1 |
| chr5_-_105212173 | 10.87 |
ENSRNOT00000010383
|
Rps6
|
ribosomal protein S6 |
| chr3_-_163477715 | 10.61 |
ENSRNOT00000009517
|
Prex1
|
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 1 |
| chr1_-_82546937 | 10.51 |
ENSRNOT00000028100
|
Hnrnpul1
|
heterogeneous nuclear ribonucleoprotein U-like 1 |
| chr13_+_70852023 | 10.48 |
ENSRNOT00000003661
|
Shcbp1l
|
SHC binding and spindle associated 1 like |
| chr6_-_55647665 | 10.14 |
ENSRNOT00000007414
|
Bzw2
|
basic leucine zipper and W2 domains 2 |
| chr17_+_78735598 | 9.85 |
ENSRNOT00000020854
|
Hspa14
|
heat shock protein family A, member 14 |
| chr1_+_162342051 | 9.72 |
ENSRNOT00000016478
|
Alg8
|
ALG8, alpha-1,3-glucosyltransferase |
| chr19_+_10024947 | 9.47 |
ENSRNOT00000061392
|
Cfap20
|
cilia and flagella associated protein 20 |
| chr2_+_244521699 | 9.36 |
ENSRNOT00000029382
|
Stpg2
|
sperm-tail PG-rich repeat containing 2 |
| chr5_+_103024376 | 9.25 |
ENSRNOT00000058536
|
AABR07049064.1
|
|
| chr13_-_98078946 | 9.25 |
ENSRNOT00000035875
|
Ahctf1
|
AT hook containing transcription factor 1 |
| chr3_+_91252829 | 9.12 |
ENSRNOT00000006148
|
Traf6
|
TNF receptor associated factor 6 |
| chr6_-_7421456 | 9.03 |
ENSRNOT00000006725
|
Zfp36l2
|
zinc finger protein 36, C3H type-like 2 |
| chrX_-_123731294 | 8.57 |
ENSRNOT00000092574
ENSRNOT00000032618 |
Upf3b
|
UPF3 regulator of nonsense transcripts homolog B (yeast) |
| chr14_+_81819799 | 8.51 |
ENSRNOT00000076840
|
Mxd4
|
Max dimerization protein 4 |
| chr4_-_56438142 | 8.20 |
ENSRNOT00000077079
|
Rbm28
|
RNA binding motif protein 28 |
| chr10_+_82745801 | 8.10 |
ENSRNOT00000005311
|
Col1a1
|
collagen type I alpha 1 chain |
| chr2_+_187915751 | 7.39 |
ENSRNOT00000026994
|
Ubqln4
|
ubiquilin 4 |
| chr8_-_12993651 | 6.41 |
ENSRNOT00000033932
|
Kdm4d
|
lysine demethylase 4D |
| chr10_+_10811937 | 6.28 |
ENSRNOT00000077347
|
Anks3
|
ankyrin repeat and sterile alpha motif domain containing 3 |
| chr11_+_87522971 | 6.12 |
ENSRNOT00000043545
|
Smpd4
|
sphingomyelin phosphodiesterase 4 |
| chr9_-_66845403 | 5.77 |
ENSRNOT00000041546
|
Ica1l
|
islet cell autoantigen 1-like |
| chr13_+_101790865 | 5.75 |
ENSRNOT00000087784
|
Taf1a
|
TATA-box binding protein associated factor, RNA polymerase I subunit A |
| chr6_+_55648021 | 5.60 |
ENSRNOT00000064822
ENSRNOT00000091488 |
Ankmy2
|
ankyrin repeat and MYND domain containing 2 |
| chr5_-_151029233 | 5.51 |
ENSRNOT00000089155
|
Ppp1r8
|
protein phosphatase 1, regulatory subunit 8 |
| chr4_-_10329241 | 5.48 |
ENSRNOT00000017232
|
Fgl2
|
fibrinogen-like 2 |
| chr13_+_110743098 | 5.46 |
ENSRNOT00000075086
|
Rd3
|
retinal degeneration 3 |
| chr1_+_36400443 | 5.46 |
ENSRNOT00000023712
|
Papd7
|
poly(A) RNA polymerase D7, non-canonical |
| chr3_+_122114754 | 5.29 |
ENSRNOT00000006408
|
Sirpa
|
signal-regulatory protein alpha |
| chr14_+_86813082 | 5.20 |
ENSRNOT00000090360
|
Ccm2
|
CCM2 scaffolding protein |
| chr1_+_162549420 | 4.92 |
ENSRNOT00000037050
|
Rsf1
|
remodeling and spacing factor 1 |
| chr9_+_16139101 | 4.61 |
ENSRNOT00000070803
|
LOC100910668
|
uncharacterized LOC100910668 |
| chr8_-_111850393 | 4.50 |
ENSRNOT00000044956
|
Cdv3
|
CDV3 homolog |
| chr1_-_31967915 | 4.30 |
ENSRNOT00000021000
|
Brd9
|
bromodomain containing 9 |
| chr5_+_76860515 | 4.28 |
ENSRNOT00000022866
|
RGD1310951
|
similar to RIKEN cDNA E130308A19 |
| chr7_+_28715300 | 4.12 |
ENSRNOT00000006760
ENSRNOT00000089161 |
Nup37
|
nucleoporin 37 |
| chr4_+_117679342 | 3.94 |
ENSRNOT00000021272
|
Figla
|
folliculogenesis specific bHLH transcription factor |
| chr8_-_62332115 | 3.53 |
ENSRNOT00000025783
|
Mpi
|
mannose phosphate isomerase (mapped) |
| chr9_-_81565416 | 3.49 |
ENSRNOT00000083582
|
Aamp
|
angio-associated, migratory cell protein |
| chr4_-_56438465 | 3.28 |
ENSRNOT00000064007
|
Rbm28
|
RNA binding motif protein 28 |
| chr10_-_45851984 | 3.23 |
ENSRNOT00000058308
|
Zfp496
|
zinc finger protein 496 |
| chr4_-_7281223 | 3.20 |
ENSRNOT00000019666
|
Slc4a2
|
solute carrier family 4 member 2 |
| chr1_-_220067123 | 3.19 |
ENSRNOT00000071020
ENSRNOT00000072373 |
Rbm14
|
RNA binding motif protein 14 |
| chr10_+_39726507 | 3.05 |
ENSRNOT00000036822
|
Meikin
|
meiotic kinetochore factor |
| chr7_-_123088819 | 3.02 |
ENSRNOT00000056077
|
AC096601.1
|
|
| chr10_+_55627025 | 2.99 |
ENSRNOT00000091016
|
Aurkb
|
aurora kinase B |
| chr7_-_123088279 | 2.91 |
ENSRNOT00000071998
|
Tob2
|
transducer of ERBB2, 2 |
| chr13_-_55886679 | 2.89 |
ENSRNOT00000066601
|
RGD1566099
|
similar to novel protein |
| chr5_+_47186558 | 2.44 |
ENSRNOT00000007654
|
LOC100910771
|
mitogen-activated protein kinase kinase kinase 7-like |
| chr20_+_27169267 | 2.36 |
ENSRNOT00000077525
|
Hnrnph3
|
heterogeneous nuclear ribonucleoprotein H3 |
| chr10_+_15099009 | 2.00 |
ENSRNOT00000015206
|
Rpusd1
|
RNA pseudouridylate synthase domain containing 1 |
| chr10_+_4312863 | 1.62 |
ENSRNOT00000091610
|
LOC100911685
|
eukaryotic peptide chain release factor GTP-binding subunit ERF3B-like |
| chr10_+_96663740 | 1.60 |
ENSRNOT00000037067
|
Cep112
|
centrosomal protein 112 |
| chr19_-_55967836 | 1.54 |
ENSRNOT00000051452
|
Sult5a1
|
sulfotransferase family 5A, member 1 |
| chr5_+_9230984 | 1.45 |
ENSRNOT00000009136
|
Vcpip1
|
valosin containing protein interacting protein 1 |
| chr7_+_97778260 | 1.24 |
ENSRNOT00000064264
|
Tbc1d31
|
TBC1 domain family, member 31 |
| chr5_+_162351001 | 1.18 |
ENSRNOT00000065471
|
LOC691162
|
hypothetical protein LOC691162 |
| chr1_-_221448570 | 1.17 |
ENSRNOT00000075165
|
Zfpl1
|
zinc finger protein-like 1 |
| chr20_-_31072469 | 0.92 |
ENSRNOT00000082448
|
Eif4ebp2
|
eukaryotic translation initiation factor 4E binding protein 2 |
| chr4_-_160803558 | 0.91 |
ENSRNOT00000049542
|
Senp18
|
Sumo1/sentrin/SMT3 specific peptidase 18 |
| chr10_+_4313100 | 0.85 |
ENSRNOT00000074487
|
LOC100911685
|
eukaryotic peptide chain release factor GTP-binding subunit ERF3B-like |
| chr4_-_145329878 | 0.78 |
ENSRNOT00000011827
|
Tada3
|
transcriptional adaptor 3 |
| chr9_+_9842585 | 0.78 |
ENSRNOT00000070806
|
Cd70
|
Cd70 molecule |
| chr11_-_36479868 | 0.60 |
ENSRNOT00000075762
|
LOC100911295
|
non-histone chromosomal protein HMG-14-like |
| chr11_+_73738433 | 0.59 |
ENSRNOT00000002353
|
Tmem44
|
transmembrane protein 44 |
| chr8_+_70860671 | 0.55 |
ENSRNOT00000019258
|
Pdcd7
|
programmed cell death 7 |
| chr9_+_93326283 | 0.44 |
ENSRNOT00000024578
|
B3gnt7
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 7 |
| chr5_-_57168610 | 0.21 |
ENSRNOT00000090499
|
B4galt1
|
beta-1,4-galactosyltransferase 1 |
| chr8_-_52814188 | 0.07 |
ENSRNOT00000087265
ENSRNOT00000047633 |
Rexo2
|
RNA exonuclease 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of E2f4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 15.5 | 46.4 | GO:2000620 | positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 15.1 | 30.2 | GO:0043987 | histone H3-S10 phosphorylation(GO:0043987) |
| 14.8 | 44.5 | GO:0003331 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 10.7 | 32.2 | GO:1905168 | positive regulation of double-strand break repair via homologous recombination(GO:1905168) |
| 10.5 | 52.6 | GO:0045950 | negative regulation of mitotic recombination(GO:0045950) |
| 10.2 | 30.5 | GO:0060821 | inactivation of X chromosome by DNA methylation(GO:0060821) |
| 9.7 | 38.8 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 8.2 | 57.2 | GO:0007144 | female meiosis I(GO:0007144) |
| 7.0 | 27.8 | GO:0043397 | corticotropin-releasing hormone secretion(GO:0043396) regulation of corticotropin-releasing hormone secretion(GO:0043397) TNFSF11-mediated signaling pathway(GO:0071847) positive regulation of osteoclast development(GO:2001206) |
| 6.7 | 27.0 | GO:0060623 | regulation of chromosome condensation(GO:0060623) |
| 5.3 | 21.0 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 4.7 | 18.9 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 4.6 | 50.6 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 4.6 | 41.2 | GO:0038171 | cannabinoid signaling pathway(GO:0038171) |
| 4.2 | 42.5 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 4.0 | 11.9 | GO:1901740 | negative regulation of myoblast fusion(GO:1901740) |
| 4.0 | 23.7 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 3.9 | 31.2 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 3.4 | 60.7 | GO:0070986 | left/right axis specification(GO:0070986) |
| 3.2 | 28.5 | GO:1900264 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 3.1 | 21.9 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 2.9 | 20.6 | GO:2000659 | regulation of interleukin-1-mediated signaling pathway(GO:2000659) |
| 2.8 | 14.0 | GO:0014043 | negative regulation of neuron maturation(GO:0014043) |
| 2.7 | 10.9 | GO:0022605 | oogenesis stage(GO:0022605) |
| 2.7 | 16.1 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 2.6 | 30.8 | GO:0033601 | positive regulation of mammary gland epithelial cell proliferation(GO:0033601) |
| 2.5 | 7.4 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 2.4 | 41.3 | GO:0010804 | negative regulation of tumor necrosis factor-mediated signaling pathway(GO:0010804) |
| 2.4 | 12.1 | GO:0043570 | maintenance of DNA repeat elements(GO:0043570) |
| 2.4 | 9.5 | GO:0060295 | regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 2.3 | 20.5 | GO:0035608 | protein deglutamylation(GO:0035608) |
| 2.2 | 22.0 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 2.2 | 30.8 | GO:0001547 | antral ovarian follicle growth(GO:0001547) |
| 2.1 | 6.4 | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 2.1 | 75.6 | GO:0021591 | ventricular system development(GO:0021591) |
| 2.0 | 8.1 | GO:1902618 | cellular response to fluoride(GO:1902618) |
| 1.9 | 18.6 | GO:0000183 | chromatin silencing at rDNA(GO:0000183) |
| 1.8 | 21.5 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 1.7 | 17.3 | GO:0015697 | quaternary ammonium group transport(GO:0015697) |
| 1.7 | 11.7 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 1.6 | 19.7 | GO:0006298 | mismatch repair(GO:0006298) |
| 1.6 | 13.0 | GO:0046015 | regulation of transcription by glucose(GO:0046015) |
| 1.5 | 9.1 | GO:0031666 | positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 1.5 | 35.9 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 1.5 | 52.9 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 1.3 | 21.2 | GO:0040034 | regulation of development, heterochronic(GO:0040034) |
| 1.0 | 5.2 | GO:0060837 | blood vessel endothelial cell differentiation(GO:0060837) |
| 1.0 | 48.5 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.9 | 6.1 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.8 | 9.0 | GO:1904627 | response to phorbol 13-acetate 12-myristate(GO:1904627) cellular response to phorbol 13-acetate 12-myristate(GO:1904628) |
| 0.8 | 11.3 | GO:0042790 | transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:0042790) |
| 0.8 | 135.9 | GO:0007051 | spindle organization(GO:0007051) |
| 0.8 | 3.2 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.8 | 3.0 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.8 | 17.4 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.7 | 9.7 | GO:0006490 | oligosaccharide-lipid intermediate biosynthetic process(GO:0006490) |
| 0.7 | 3.5 | GO:0071554 | GDP-mannose biosynthetic process(GO:0009298) cell wall macromolecule metabolic process(GO:0044036) cell wall organization or biogenesis(GO:0071554) |
| 0.7 | 4.9 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.6 | 2.4 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.4 | 13.8 | GO:0045954 | positive regulation of natural killer cell mediated cytotoxicity(GO:0045954) |
| 0.3 | 1.5 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.3 | 10.6 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.2 | 8.6 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.2 | 5.3 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.2 | 5.5 | GO:0007567 | parturition(GO:0007567) |
| 0.2 | 5.8 | GO:0006360 | transcription from RNA polymerase I promoter(GO:0006360) |
| 0.1 | 16.7 | GO:0019882 | antigen processing and presentation(GO:0019882) |
| 0.1 | 60.6 | GO:0051301 | cell division(GO:0051301) |
| 0.1 | 2.9 | GO:0045671 | negative regulation of osteoclast differentiation(GO:0045671) |
| 0.1 | 15.7 | GO:0010212 | response to ionizing radiation(GO:0010212) |
| 0.1 | 40.4 | GO:0006470 | protein dephosphorylation(GO:0006470) |
| 0.1 | 0.2 | GO:0060058 | apoptotic process involved in mammary gland involution(GO:0060057) positive regulation of apoptotic process involved in mammary gland involution(GO:0060058) positive regulation of apoptotic process involved in morphogenesis(GO:1902339) regulation of mammary gland involution(GO:1903519) positive regulation of mammary gland involution(GO:1903521) positive regulation of apoptotic process involved in development(GO:1904747) |
| 0.1 | 3.2 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.1 | 2.0 | GO:0001522 | pseudouridine synthesis(GO:0001522) |
| 0.1 | 1.6 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.1 | 0.9 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.1 | 0.8 | GO:0090043 | tubulin deacetylation(GO:0090042) regulation of tubulin deacetylation(GO:0090043) |
| 0.1 | 0.8 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.1 | 28.3 | GO:0007283 | spermatogenesis(GO:0007283) |
| 0.1 | 17.8 | GO:0006869 | lipid transport(GO:0006869) |
| 0.1 | 0.6 | GO:0070234 | positive regulation of T cell apoptotic process(GO:0070234) |
| 0.0 | 10.5 | GO:0009615 | response to virus(GO:0009615) |
| 0.0 | 11.2 | GO:0000398 | RNA splicing, via transesterification reactions with bulged adenosine as nucleophile(GO:0000377) mRNA splicing, via spliceosome(GO:0000398) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 13.8 | 68.9 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 11.6 | 46.4 | GO:0031436 | BRCA1-BARD1 complex(GO:0031436) |
| 7.7 | 38.4 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 6.5 | 38.8 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 6.3 | 75.6 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 5.8 | 17.4 | GO:0035101 | FACT complex(GO:0035101) |
| 4.7 | 23.7 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 4.7 | 18.7 | GO:0070876 | SOSS complex(GO:0070876) |
| 4.6 | 110.1 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 4.5 | 45.1 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 4.0 | 12.1 | GO:0032301 | MutSalpha complex(GO:0032301) |
| 3.7 | 18.6 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 3.2 | 28.5 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 3.0 | 21.0 | GO:0001739 | sex chromatin(GO:0001739) |
| 2.6 | 25.6 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 2.4 | 21.9 | GO:0042555 | MCM complex(GO:0042555) |
| 2.0 | 38.5 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 2.0 | 32.2 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 1.8 | 16.1 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 1.3 | 17.8 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 1.2 | 5.8 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.9 | 7.4 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.9 | 10.5 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.8 | 62.2 | GO:0000794 | condensed nuclear chromosome(GO:0000794) |
| 0.8 | 9.1 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.7 | 8.1 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.7 | 30.8 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.7 | 61.4 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.6 | 2.4 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.6 | 12.9 | GO:0019013 | viral nucleocapsid(GO:0019013) |
| 0.5 | 6.4 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.5 | 58.4 | GO:0000922 | spindle pole(GO:0000922) |
| 0.5 | 60.7 | GO:0036126 | sperm flagellum(GO:0036126) |
| 0.5 | 20.4 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.5 | 4.9 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 0.5 | 30.8 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.4 | 21.3 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.3 | 8.6 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.2 | 31.8 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.1 | 7.2 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.1 | 93.1 | GO:0044427 | chromosomal part(GO:0044427) |
| 0.1 | 6.7 | GO:0005844 | polysome(GO:0005844) |
| 0.1 | 11.7 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.1 | 9.5 | GO:0005814 | centriole(GO:0005814) |
| 0.1 | 7.9 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.1 | 11.3 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.1 | 23.3 | GO:0005813 | centrosome(GO:0005813) |
| 0.1 | 0.3 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 5.8 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 11.9 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 32.4 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 7.9 | GO:0005840 | ribosome(GO:0005840) |
| 0.0 | 0.6 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 4.5 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 3.3 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 64.9 | GO:0005634 | nucleus(GO:0005634) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 10.3 | 41.2 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 10.3 | 41.0 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 6.9 | 48.5 | GO:0000405 | bubble DNA binding(GO:0000405) |
| 6.3 | 50.6 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 6.1 | 30.5 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 6.0 | 30.2 | GO:0031493 | nucleosomal histone binding(GO:0031493) |
| 5.6 | 56.2 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 5.3 | 21.0 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 5.1 | 30.8 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 4.7 | 18.9 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 4.0 | 12.1 | GO:0032142 | single guanine insertion binding(GO:0032142) |
| 4.0 | 23.7 | GO:0000150 | recombinase activity(GO:0000150) |
| 3.2 | 9.7 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 2.8 | 28.5 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 2.6 | 38.8 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 2.5 | 71.4 | GO:0070840 | dynein complex binding(GO:0070840) |
| 2.4 | 11.9 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 2.2 | 17.3 | GO:0005329 | dopamine transmembrane transporter activity(GO:0005329) quaternary ammonium group transmembrane transporter activity(GO:0015651) |
| 2.1 | 38.4 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 2.1 | 18.6 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 2.0 | 19.7 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 1.7 | 105.7 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 1.4 | 35.9 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 1.3 | 41.8 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 1.0 | 21.9 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.9 | 30.8 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.9 | 37.7 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.8 | 20.5 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.7 | 5.5 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.7 | 3.3 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.7 | 10.6 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.7 | 21.8 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.6 | 64.2 | GO:0004519 | endonuclease activity(GO:0004519) |
| 0.6 | 8.1 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.5 | 53.3 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.5 | 16.1 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.4 | 11.5 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.4 | 3.5 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.4 | 13.0 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.3 | 14.0 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.3 | 3.4 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.2 | 22.0 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.2 | 41.0 | GO:0005057 | receptor signaling protein activity(GO:0005057) |
| 0.2 | 2.4 | GO:0008079 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.2 | 4.3 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.2 | 4.1 | GO:0015288 | porin activity(GO:0015288) |
| 0.2 | 32.3 | GO:0004721 | phosphoprotein phosphatase activity(GO:0004721) |
| 0.2 | 7.4 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.1 | 2.0 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.1 | 3.2 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.1 | 18.6 | GO:0042393 | histone binding(GO:0042393) |
| 0.1 | 5.3 | GO:0045309 | protein phosphorylated amino acid binding(GO:0045309) |
| 0.1 | 3.2 | GO:0043621 | protein self-association(GO:0043621) |
| 0.1 | 0.6 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.1 | 0.9 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.1 | 17.3 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.1 | 4.0 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.1 | 0.2 | GO:0003945 | N-acetyllactosamine synthase activity(GO:0003945) |
| 0.1 | 6.1 | GO:0004620 | phospholipase activity(GO:0004620) |
| 0.1 | 20.6 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |
| 0.1 | 80.5 | GO:0042802 | identical protein binding(GO:0042802) |
| 0.1 | 5.5 | GO:0016779 | nucleotidyltransferase activity(GO:0016779) |
| 0.0 | 17.8 | GO:0008289 | lipid binding(GO:0008289) |
| 0.0 | 1.5 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.0 | 1.5 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 0.4 | GO:0008378 | galactosyltransferase activity(GO:0008378) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.2 | 130.4 | PID AURORA A PATHWAY | Aurora A signaling |
| 2.4 | 106.4 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 1.6 | 83.5 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 1.6 | 33.4 | PID ATR PATHWAY | ATR signaling pathway |
| 0.9 | 82.9 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.9 | 26.6 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.8 | 52.5 | PID E2F PATHWAY | E2F transcription factor network |
| 0.7 | 13.0 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.6 | 27.8 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.5 | 8.1 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.5 | 41.0 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.5 | 10.9 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.4 | 11.9 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.3 | 10.6 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.3 | 5.2 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.1 | 5.9 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 5.5 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.3 | 99.3 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 3.3 | 50.1 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 3.1 | 87.0 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 2.0 | 21.9 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 1.9 | 17.3 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 1.6 | 11.5 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 1.5 | 32.2 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 1.5 | 16.1 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 1.3 | 133.3 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 1.3 | 18.9 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 1.0 | 9.1 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.8 | 22.0 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.8 | 10.9 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.6 | 5.8 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.5 | 8.1 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.5 | 19.7 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.5 | 41.0 | REACTOME LATE PHASE OF HIV LIFE CYCLE | Genes involved in Late Phase of HIV Life Cycle |
| 0.4 | 4.9 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.4 | 13.3 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.3 | 5.3 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.3 | 8.6 | REACTOME TRANSPORT OF MATURE TRANSCRIPT TO CYTOPLASM | Genes involved in Transport of Mature Transcript to Cytoplasm |
| 0.3 | 53.2 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.2 | 9.4 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.2 | 6.1 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.1 | 10.5 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 3.2 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.6 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |