Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
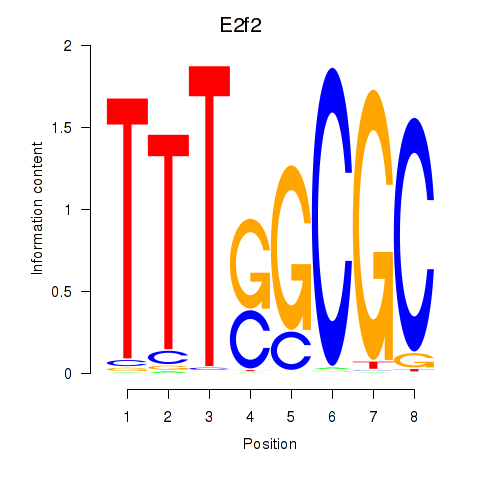
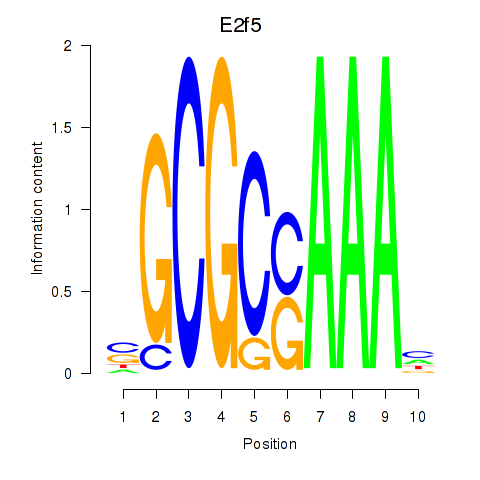
Results for E2f2_E2f5
Z-value: 2.98
Transcription factors associated with E2f2_E2f5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
E2f2
|
ENSRNOG00000047741 | E2F transcription factor 2 |
|
E2f5
|
ENSRNOG00000010760 | E2F transcription factor 5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| E2f2 | rn6_v1_chr5_+_154522119_154522119 | 0.83 | 5.4e-84 | Click! |
| E2f5 | rn6_v1_chr2_-_88366115_88366115 | 0.21 | 1.3e-04 | Click! |
Activity profile of E2f2_E2f5 motif
Sorted Z-values of E2f2_E2f5 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_59153280 | 206.46 |
ENSRNOT00000002066
|
Cdca7
|
cell division cycle associated 7 |
| chr6_+_43884678 | 205.10 |
ENSRNOT00000091551
|
Rrm2
|
ribonucleotide reductase regulatory subunit M2 |
| chr20_+_20576377 | 185.05 |
ENSRNOT00000000783
ENSRNOT00000086806 |
Cdk1
|
cyclin-dependent kinase 1 |
| chr14_-_20953095 | 183.40 |
ENSRNOT00000004440
|
Dck
|
deoxycytidine kinase |
| chr19_+_14523554 | 170.60 |
ENSRNOT00000084271
ENSRNOT00000064731 |
Mcm5
|
minichromosome maintenance complex component 5 |
| chr13_-_45068077 | 157.53 |
ENSRNOT00000004969
|
Mcm6
|
minichromosome maintenance complex component 6 |
| chr9_-_26932201 | 156.83 |
ENSRNOT00000017081
|
Mcm3
|
minichromosome maintenance complex component 3 |
| chr17_+_44528125 | 145.26 |
ENSRNOT00000084538
|
LOC680322
|
similar to Histone H2A type 1 |
| chr8_+_132032944 | 142.64 |
ENSRNOT00000089278
|
Kif15
|
kinesin family member 15 |
| chr1_-_42467586 | 141.33 |
ENSRNOT00000029416
|
Fbxo5
|
F-box protein 5 |
| chr5_+_144581427 | 135.88 |
ENSRNOT00000015227
|
Clspn
|
claspin |
| chr12_-_52452040 | 133.49 |
ENSRNOT00000067453
|
Pole
|
DNA polymerase epsilon, catalytic subunit |
| chr12_-_19314016 | 132.92 |
ENSRNOT00000001825
|
Mcm7
|
minichromosome maintenance complex component 7 |
| chr3_-_105298108 | 131.04 |
ENSRNOT00000010994
|
Arhgap11a
|
Rho GTPase activating protein 11A |
| chr17_-_44738330 | 130.23 |
ENSRNOT00000072195
|
LOC100364835
|
histone cluster 1, H2bd-like |
| chr17_+_42302540 | 123.85 |
ENSRNOT00000025389
|
Gmnn
|
geminin, DNA replication inhibitor |
| chr17_+_44520537 | 122.11 |
ENSRNOT00000077985
|
hist1h2ail2
|
histone cluster 1, H2ai-like2 |
| chr3_+_146695344 | 118.22 |
ENSRNOT00000010955
|
Gins1
|
GINS complex subunit 1 |
| chr17_-_44527801 | 116.84 |
ENSRNOT00000089643
|
Hist1h2bk
|
histone cluster 1 H2B family member k |
| chr3_-_150073721 | 105.97 |
ENSRNOT00000022428
|
E2f1
|
E2F transcription factor 1 |
| chr17_-_44520240 | 105.90 |
ENSRNOT00000086538
|
Hist1h2bh
|
histone cluster 1 H2B family member h |
| chr13_-_110257367 | 104.98 |
ENSRNOT00000005576
|
Dtl
|
denticleless E3 ubiquitin protein ligase homolog |
| chr8_+_48665652 | 104.13 |
ENSRNOT00000059715
|
H2afx
|
H2A histone family, member X |
| chr3_-_124884570 | 103.85 |
ENSRNOT00000028887
|
Pcna
|
proliferating cell nuclear antigen |
| chr4_-_120840111 | 103.53 |
ENSRNOT00000022231
|
Mcm2
|
minichromosome maintenance complex component 2 |
| chr3_+_110918243 | 102.20 |
ENSRNOT00000056432
|
Rad51
|
RAD51 recombinase |
| chr6_-_91518996 | 101.15 |
ENSRNOT00000005835
|
Pole2
|
DNA polymerase epsilon 2, accessory subunit |
| chr7_+_97984862 | 97.06 |
ENSRNOT00000008508
|
Atad2
|
ATPase family, AAA domain containing 2 |
| chr10_+_67427066 | 96.38 |
ENSRNOT00000035642
|
Atad5
|
ATPase family, AAA domain containing 5 |
| chr17_+_44738643 | 92.72 |
ENSRNOT00000087643
|
LOC100910554
|
histone H2A type 1-like |
| chr2_+_243105674 | 91.53 |
ENSRNOT00000013919
|
H2afz
|
H2A histone family, member Z |
| chr6_+_144291974 | 81.32 |
ENSRNOT00000035255
|
Ncapg2
|
non-SMC condensin II complex, subunit G2 |
| chr4_-_77347011 | 80.03 |
ENSRNOT00000008149
|
Ezh2
|
enhancer of zeste 2 polycomb repressive complex 2 subunit |
| chr17_+_44763598 | 79.81 |
ENSRNOT00000079880
|
Hist1h3b
|
histone cluster 1, H3b |
| chr17_+_44748482 | 72.66 |
ENSRNOT00000083765
|
Hist1h2bl
|
histone cluster 1 H2B family member l |
| chr5_+_152681101 | 72.20 |
ENSRNOT00000076052
ENSRNOT00000022574 |
Stmn1
|
stathmin 1 |
| chr6_-_36940868 | 71.96 |
ENSRNOT00000006470
|
Gen1
|
GEN1 Holliday junction 5' flap endonuclease |
| chr17_-_44748188 | 68.03 |
ENSRNOT00000081970
|
LOC103690190
|
histone H2A type 1-E |
| chr8_+_71514281 | 67.68 |
ENSRNOT00000022256
|
Ns5atp9
|
NS5A (hepatitis C virus) transactivated protein 9 |
| chr17_-_1672021 | 65.25 |
ENSRNOT00000036287
|
Zfp367
|
zinc finger protein 367 |
| chr8_-_21968415 | 61.59 |
ENSRNOT00000067325
ENSRNOT00000064932 |
Dnmt1
|
DNA methyltransferase 1 |
| chr2_+_200187179 | 56.37 |
ENSRNOT00000093564
ENSRNOT00000025718 |
Notch2
|
notch 2 |
| chr2_+_198390166 | 54.54 |
ENSRNOT00000081042
|
Hist2h3c2
|
histone cluster 2, H3c2 |
| chr12_+_23151180 | 51.77 |
ENSRNOT00000059486
|
Cux1
|
cut-like homeobox 1 |
| chr5_-_58455819 | 49.46 |
ENSRNOT00000078082
|
Fancg
|
Fanconi anemia, complementation group G |
| chr1_+_221448661 | 45.19 |
ENSRNOT00000072493
|
LOC100910252
|
sororin-like |
| chr5_+_152721940 | 45.13 |
ENSRNOT00000039322
|
Aunip
|
aurora kinase A and ninein interacting protein |
| chr9_+_93545396 | 45.07 |
ENSRNOT00000025093
|
LOC100359583
|
hypothetical protein LOC100359583 |
| chr15_+_83442144 | 43.47 |
ENSRNOT00000039344
|
Bora
|
bora, aurora kinase A activator |
| chr9_-_78369031 | 43.23 |
ENSRNOT00000087870
|
Bard1
|
BRCA1 associated RING domain 1 |
| chr10_+_45289741 | 40.92 |
ENSRNOT00000066190
|
Hist3h2ba
|
histone cluster 3, H2ba |
| chr17_+_77224112 | 38.71 |
ENSRNOT00000024178
|
Mcm10
|
minichromosome maintenance 10 replication initiation factor |
| chr4_+_81244469 | 38.64 |
ENSRNOT00000044407
ENSRNOT00000015828 |
Cbx3
|
chromobox 3 |
| chr8_+_117953444 | 37.99 |
ENSRNOT00000028141
|
Cdc25a
|
cell division cycle 25A |
| chr8_+_111790678 | 37.08 |
ENSRNOT00000013265
|
Topbp1
|
topoisomerase (DNA) II binding protein 1 |
| chr2_+_219628695 | 37.04 |
ENSRNOT00000067324
|
Sass6
|
SAS-6 centriolar assembly protein |
| chr1_+_198214797 | 36.62 |
ENSRNOT00000068543
|
Tbx6
|
T-box 6 |
| chr10_+_13061170 | 36.53 |
ENSRNOT00000004936
|
Pkmyt1
|
protein kinase, membrane associated tyrosine/threonine 1 |
| chrX_-_62698830 | 36.52 |
ENSRNOT00000076359
|
Pola1
|
DNA polymerase alpha 1, catalytic subunit |
| chr4_-_183426439 | 36.26 |
ENSRNOT00000083310
|
Fam60a
|
family with sequence similarity 60, member A |
| chr1_+_274310153 | 35.45 |
ENSRNOT00000054686
|
Smc3
|
structural maintenance of chromosomes 3 |
| chr4_-_170145262 | 34.39 |
ENSRNOT00000074526
|
Hist1h4b
|
histone cluster 1, H4b |
| chr6_-_2886465 | 33.41 |
ENSRNOT00000038448
|
Srsf7
|
serine and arginine rich splicing factor 7 |
| chr7_-_94774569 | 31.78 |
ENSRNOT00000036399
|
Dscc1
|
DNA replication and sister chromatid cohesion 1 |
| chr17_+_43734461 | 31.55 |
ENSRNOT00000072564
|
Hist1h1d
|
histone cluster 1, H1d |
| chr8_-_33661049 | 31.45 |
ENSRNOT00000068037
|
Fli1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr9_-_119818310 | 31.42 |
ENSRNOT00000019359
|
Smchd1
|
structural maintenance of chromosomes flexible hinge domain containing 1 |
| chr15_+_4356261 | 30.99 |
ENSRNOT00000009009
|
Dnajc9
|
DnaJ heat shock protein family (Hsp40) member C9 |
| chr6_+_11644578 | 28.60 |
ENSRNOT00000021923
ENSRNOT00000093689 |
Msh6
|
mutS homolog 6 |
| chr16_+_41079444 | 27.89 |
ENSRNOT00000015623
|
Neil3
|
nei-like DNA glycosylase 3 |
| chr4_+_49017311 | 27.41 |
ENSRNOT00000007476
|
Ing3
|
inhibitor of growth family, member 3 |
| chr18_+_1866080 | 26.47 |
ENSRNOT00000018646
|
Snrpd1
|
small nuclear ribonucleoprotein D1 |
| chr1_+_274309758 | 25.10 |
ENSRNOT00000019560
|
Smc3
|
structural maintenance of chromosomes 3 |
| chr4_+_34282625 | 24.18 |
ENSRNOT00000011138
ENSRNOT00000086735 |
Glcci1
|
glucocorticoid induced 1 |
| chr19_-_9843673 | 23.91 |
ENSRNOT00000015795
|
Gins3
|
GINS complex subunit 3 |
| chr6_+_93462852 | 23.73 |
ENSRNOT00000089473
|
Arid4a
|
AT-rich interaction domain 4A |
| chr4_-_34282351 | 23.09 |
ENSRNOT00000011123
|
Rpa3
|
replication protein A3 |
| chr10_-_65424802 | 23.06 |
ENSRNOT00000018468
|
Traf4
|
Tnf receptor associated factor 4 |
| chr8_+_117347029 | 22.14 |
ENSRNOT00000047717
|
Impdh2
|
inosine monophosphate dehydrogenase 2 |
| chr15_+_45422010 | 21.16 |
ENSRNOT00000012231
|
Rnaseh2b
|
ribonuclease H2, subunit B |
| chr1_-_198706852 | 19.45 |
ENSRNOT00000024074
|
Dctpp1
|
dCTP pyrophosphatase 1 |
| chr2_+_211176556 | 19.43 |
ENSRNOT00000055880
|
Psrc1
|
proline and serine rich coiled-coil 1 |
| chr1_+_142034423 | 19.38 |
ENSRNOT00000090006
|
Ttll13
|
tubulin tyrosine ligase-like family, member 13 |
| chrX_+_15171499 | 19.23 |
ENSRNOT00000008399
|
Suv39h1l1
|
suppressor of variegation 3-9 homolog 1 (Drosophila)-like 1 |
| chr15_-_58068892 | 18.44 |
ENSRNOT00000044254
|
Gtf2f2
|
general transcription factor IIF subunit 2 |
| chr1_+_273563441 | 18.07 |
ENSRNOT00000048691
|
RGD1561333
|
similar to 60S ribosomal protein L8 |
| chr1_+_167538263 | 17.90 |
ENSRNOT00000074058
|
Rrm1
|
ribonucleotide reductase catalytic subunit M1 |
| chr3_+_125470551 | 17.74 |
ENSRNOT00000028898
|
Mcm8
|
minichromosome maintenance 8 homologous recombination repair factor |
| chr7_-_34485034 | 17.25 |
ENSRNOT00000007351
|
Snrpf
|
small nuclear ribonucleoprotein polypeptide F |
| chr5_+_47853818 | 17.18 |
ENSRNOT00000009228
ENSRNOT00000079656 |
Casp8ap2
|
caspase 8 associated protein 2 |
| chr11_+_89511191 | 17.04 |
ENSRNOT00000002510
|
Mcm4
|
minichromosome maintenance complex component 4 |
| chr19_+_37600148 | 16.86 |
ENSRNOT00000023853
|
Ctcf
|
CCCTC-binding factor |
| chr8_-_22111863 | 16.60 |
ENSRNOT00000046983
|
Raver1
|
ribonucleoprotein, PTB-binding 1 |
| chr14_+_81488008 | 16.23 |
ENSRNOT00000018477
|
Tnip2
|
TNFAIP3 interacting protein 2 |
| chr9_-_93377643 | 15.27 |
ENSRNOT00000024712
|
Ncl
|
nucleolin |
| chr7_-_117703094 | 14.99 |
ENSRNOT00000019754
|
Tonsl
|
tonsoku-like, DNA repair protein |
| chr17_+_17965881 | 14.88 |
ENSRNOT00000021751
|
Dek
|
DEK proto-oncogene |
| chr7_+_27438351 | 14.85 |
ENSRNOT00000077976
|
AABR07056503.1
|
|
| chr1_-_71399751 | 14.76 |
ENSRNOT00000020787
|
Zfp444
|
zinc finger protein 444 |
| chr6_+_126636662 | 14.55 |
ENSRNOT00000010739
|
Ubr7
|
ubiquitin protein ligase E3 component n-recognin 7 (putative) |
| chr17_-_82060123 | 14.44 |
ENSRNOT00000025046
|
Nsun6
|
NOP2/Sun RNA methyltransferase family member 6 |
| chr20_+_46429222 | 13.98 |
ENSRNOT00000076818
|
Foxo3
|
forkhead box O3 |
| chr8_+_12355767 | 13.88 |
ENSRNOT00000068445
|
Fam76b
|
family with sequence similarity 76, member B |
| chr1_+_167538744 | 13.48 |
ENSRNOT00000093070
|
Rrm1
|
ribonucleotide reductase catalytic subunit M1 |
| chr14_-_60594844 | 13.23 |
ENSRNOT00000005349
|
Pi4k2b
|
phosphatidylinositol 4-kinase type 2 beta |
| chr1_+_174767960 | 13.14 |
ENSRNOT00000013362
|
Wee1
|
WEE1 G2 checkpoint kinase |
| chr6_+_126636491 | 12.58 |
ENSRNOT00000089601
|
Ubr7
|
ubiquitin protein ligase E3 component n-recognin 7 (putative) |
| chr20_+_6049286 | 12.44 |
ENSRNOT00000042143
ENSRNOT00000092566 |
Brpf3
|
bromodomain and PHD finger containing, 3 |
| chr10_+_67325347 | 12.06 |
ENSRNOT00000079002
ENSRNOT00000085914 |
Suz12
|
suppressor of zeste 12 homolog (Drosophila) |
| chr4_+_170820594 | 11.99 |
ENSRNOT00000032568
|
LOC500354
|
similar to C030030A07Rik protein |
| chr16_+_7103998 | 11.18 |
ENSRNOT00000016581
|
Pbrm1
|
polybromo 1 |
| chr6_+_36941596 | 10.96 |
ENSRNOT00000083383
|
Smc6
|
structural maintenance of chromosomes 6 |
| chr16_-_7103604 | 10.94 |
ENSRNOT00000033677
|
Gnl3
|
G protein nucleolar 3 |
| chrX_+_157353577 | 10.74 |
ENSRNOT00000090596
|
Haus7
|
HAUS augmin-like complex, subunit 7 |
| chr14_-_44375804 | 10.69 |
ENSRNOT00000042825
|
LOC100362751
|
ribosomal protein P2-like |
| chr12_+_8085525 | 10.56 |
ENSRNOT00000089804
ENSRNOT00000001234 |
Slc7a1
|
solute carrier family 7 member 1 |
| chr10_+_11206226 | 10.54 |
ENSRNOT00000006979
|
Tfap4
|
transcription factor AP-4 |
| chr12_+_660011 | 10.06 |
ENSRNOT00000040830
|
Pds5b
|
PDS5 cohesin associated factor B |
| chr9_+_81783349 | 10.02 |
ENSRNOT00000021548
|
Cnot9
|
CCR4-NOT transcription complex subunit 9 |
| chr1_+_243477493 | 9.67 |
ENSRNOT00000021779
ENSRNOT00000085356 |
Dmrt1
|
doublesex and mab-3 related transcription factor 1 |
| chr10_-_14324170 | 9.55 |
ENSRNOT00000035513
ENSRNOT00000090587 |
Hn1l
|
hematological and neurological expressed 1-like |
| chr3_-_176791960 | 9.46 |
ENSRNOT00000018237
|
Gmeb2
|
glucocorticoid modulatory element binding protein 2 |
| chr2_+_207108552 | 9.44 |
ENSRNOT00000027234
|
Slc16a1
|
solute carrier family 16 member 1 |
| chr15_-_44627765 | 9.18 |
ENSRNOT00000058887
|
Dock5
|
dedicator of cytokinesis 5 |
| chr10_+_36098051 | 9.07 |
ENSRNOT00000083971
|
Adamts2
|
ADAM metallopeptidase with thrombospondin type 1 motif, 2 |
| chr1_-_128341240 | 9.06 |
ENSRNOT00000070864
ENSRNOT00000072915 |
Mef2a
|
myocyte enhancer factor 2a |
| chr2_-_4195810 | 8.72 |
ENSRNOT00000018457
|
Slf1
|
SMC5-SMC6 complex localization factor 1 |
| chr18_-_48720472 | 8.68 |
ENSRNOT00000023236
|
Cep120
|
centrosomal protein 120 |
| chr6_+_132805794 | 7.74 |
ENSRNOT00000006281
|
Wdr25
|
WD repeat domain 25 |
| chr1_-_265420503 | 7.65 |
ENSRNOT00000072223
|
Fbxw4
|
F-box and WD repeat domain containing 4 |
| chr3_+_38742072 | 7.49 |
ENSRNOT00000006774
|
Arl6ip6
|
ADP ribosylation factor like GTPase 6 interacting protein 6 |
| chr3_-_117766120 | 7.40 |
ENSRNOT00000056022
|
Fbn1
|
fibrillin 1 |
| chr5_+_154668179 | 7.36 |
ENSRNOT00000015971
|
Hnrnpr
|
heterogeneous nuclear ribonucleoprotein R |
| chr10_-_105795958 | 7.24 |
ENSRNOT00000067739
|
Srsf2
|
serine and arginine rich splicing factor 2 |
| chr5_-_16706909 | 7.17 |
ENSRNOT00000011314
|
Rps20
|
ribosomal protein S20 |
| chr7_+_12006710 | 7.15 |
ENSRNOT00000045421
|
Klf16
|
Kruppel-like factor 16 |
| chrX_+_113510621 | 6.58 |
ENSRNOT00000025912
|
Nxt2
|
nuclear transport factor 2-like export factor 2 |
| chr2_+_104854572 | 6.42 |
ENSRNOT00000064828
ENSRNOT00000090912 |
Hltf
|
helicase-like transcription factor |
| chr3_-_59688692 | 6.21 |
ENSRNOT00000078752
|
Sp3
|
Sp3 transcription factor |
| chr19_-_52206310 | 5.88 |
ENSRNOT00000087886
|
Mbtps1
|
membrane-bound transcription factor peptidase, site 1 |
| chr5_-_128186532 | 5.86 |
ENSRNOT00000012635
|
Prpf38a
|
pre-mRNA processing factor 38A |
| chr4_+_62299044 | 5.83 |
ENSRNOT00000032077
|
Agbl3
|
ATP/GTP binding protein-like 3 |
| chr11_+_86329012 | 5.81 |
ENSRNOT00000072027
|
Cdc45
|
cell division cycle 45 |
| chr10_-_109757550 | 5.76 |
ENSRNOT00000054957
|
Arhgdia
|
Rho GDP dissociation inhibitor alpha |
| chr5_-_173659675 | 5.72 |
ENSRNOT00000027547
|
Klhl17
|
kelch-like family member 17 |
| chr1_-_82546937 | 5.52 |
ENSRNOT00000028100
|
Hnrnpul1
|
heterogeneous nuclear ribonucleoprotein U-like 1 |
| chr14_-_86706626 | 5.52 |
ENSRNOT00000082893
|
H2afv
|
H2A histone family, member V |
| chr13_+_48679774 | 5.38 |
ENSRNOT00000075674
|
Nucks1
|
nuclear casein kinase and cyclin-dependent kinase substrate 1 |
| chr11_-_86276430 | 5.24 |
ENSRNOT00000075164
|
Hira
|
histone cell cycle regulator |
| chr6_-_103935122 | 5.17 |
ENSRNOT00000058354
|
LOC500684
|
hypothetical protein LOC500684 |
| chr1_-_80630038 | 5.10 |
ENSRNOT00000025281
|
Tomm40
|
translocase of outer mitochondrial membrane 40 |
| chr3_-_160608308 | 4.89 |
ENSRNOT00000047560
|
Wfdc15b
|
WAP four-disulfide core domain 15B |
| chr15_-_83442008 | 4.84 |
ENSRNOT00000039400
|
Mzt1
|
mitotic spindle organizing protein 1 |
| chr6_+_135610743 | 4.60 |
ENSRNOT00000010906
|
Traf3
|
Tnf receptor-associated factor 3 |
| chrX_+_71540895 | 4.55 |
ENSRNOT00000004692
ENSRNOT00000082967 |
Ogt
|
O-linked N-acetylglucosamine (GlcNAc) transferase |
| chr3_-_125470413 | 4.39 |
ENSRNOT00000028893
|
Trmt6
|
tRNA methyltransferase 6 |
| chrX_+_156812064 | 4.33 |
ENSRNOT00000077142
|
Hcfc1
|
host cell factor C1 |
| chrY_+_380382 | 4.31 |
ENSRNOT00000092110
|
Rbmy1j
|
RNA binding motif protein, Y-linked, family 1, member J |
| chr8_-_123371257 | 4.29 |
ENSRNOT00000017243
|
Stt3b
|
STT3B, catalytic subunit of the oligosaccharyltransferase complex |
| chr9_+_78862013 | 4.25 |
ENSRNOT00000021105
|
Atic
|
5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase/IMP cyclohydrolase |
| chr3_-_113091770 | 4.11 |
ENSRNOT00000068132
|
Lcmt2
|
leucine carboxyl methyltransferase 2 |
| chr5_-_62001196 | 3.98 |
ENSRNOT00000012602
|
Trmo
|
tRNA methyltransferase O |
| chrX_+_157353730 | 3.87 |
ENSRNOT00000080691
|
Haus7
|
HAUS augmin-like complex, subunit 7 |
| chr3_-_164964702 | 3.84 |
ENSRNOT00000014595
|
Adnp
|
activity-dependent neuroprotector homeobox |
| chrX_-_155862363 | 3.79 |
ENSRNOT00000089847
|
Dkc1
|
dyskerin pseudouridine synthase 1 |
| chr13_+_101698096 | 3.75 |
ENSRNOT00000089309
|
Aida
|
axin interactor, dorsalization associated |
| chr10_+_4312863 | 3.70 |
ENSRNOT00000091610
|
LOC100911685
|
eukaryotic peptide chain release factor GTP-binding subunit ERF3B-like |
| chr6_+_10912383 | 3.39 |
ENSRNOT00000061747
ENSRNOT00000086247 |
Ttc7a
|
tetratricopeptide repeat domain 7A |
| chr10_+_4313100 | 3.33 |
ENSRNOT00000074487
|
LOC100911685
|
eukaryotic peptide chain release factor GTP-binding subunit ERF3B-like |
| chr19_-_25261911 | 3.27 |
ENSRNOT00000045180
ENSRNOT00000009806 |
Cc2d1a
|
coiled-coil and C2 domain containing 1A |
| chr1_+_203509731 | 3.19 |
ENSRNOT00000028007
|
Hmx3
|
H6 family homeobox 3 |
| chr7_+_123308041 | 3.04 |
ENSRNOT00000009428
|
Mei1
|
meiotic double-stranded break formation protein 1 |
| chr18_-_30775401 | 2.93 |
ENSRNOT00000027128
|
Slc25a2
|
solute carrier family 25 member 2 |
| chr10_+_10530365 | 2.93 |
ENSRNOT00000003843
|
Eef2kmt
|
eukaryotic elongation factor 2 lysine methyltransferase |
| chr4_-_85174931 | 2.77 |
ENSRNOT00000014324
ENSRNOT00000088085 |
Nod1
|
nucleotide-binding oligomerization domain containing 1 |
| chr5_+_103024376 | 2.75 |
ENSRNOT00000058536
|
AABR07049064.1
|
|
| chr1_+_278557792 | 2.61 |
ENSRNOT00000023414
|
Atrnl1
|
attractin like 1 |
| chr17_-_30714410 | 2.50 |
ENSRNOT00000022902
ENSRNOT00000081993 |
Prpf4b
|
pre-mRNA processing factor 4B |
| chr13_+_49933155 | 2.48 |
ENSRNOT00000037039
|
Ppp1r15b
|
protein phosphatase 1, regulatory subunit 15B |
| chr3_-_176431423 | 2.28 |
ENSRNOT00000037271
|
Ythdf1
|
YTH N(6)-methyladenosine RNA binding protein 1 |
| chr10_+_4411766 | 2.00 |
ENSRNOT00000003213
|
LOC100911685
|
eukaryotic peptide chain release factor GTP-binding subunit ERF3B-like |
| chr20_-_5037022 | 1.95 |
ENSRNOT00000091022
ENSRNOT00000060762 |
Msh5
|
mutS homolog 5 |
| chr7_-_36499784 | 1.92 |
ENSRNOT00000011948
|
AC124896.1
|
suppressor of cytokine signaling 2 |
| chr1_+_1771710 | 1.55 |
ENSRNOT00000080138
ENSRNOT00000073528 |
Nup43
|
nucleoporin 43 |
| chr4_+_10631073 | 1.39 |
ENSRNOT00000081939
ENSRNOT00000017907 |
Ptpn12
|
protein tyrosine phosphatase, non-receptor type 12 |
| chrX_-_128268285 | 1.38 |
ENSRNOT00000009755
ENSRNOT00000081880 |
Thoc2
|
THO complex 2 |
| chr10_+_59743544 | 1.35 |
ENSRNOT00000093497
ENSRNOT00000056460 |
Tax1bp3
|
Tax1 binding protein 3 |
| chr7_+_13039781 | 1.25 |
ENSRNOT00000067771
ENSRNOT00000090048 |
Mier2
|
mesoderm induction early response 1, family member 2 |
| chr14_+_4125380 | 1.15 |
ENSRNOT00000002882
|
Zfp644
|
zinc finger protein 644 |
| chr2_+_128461224 | 1.04 |
ENSRNOT00000018872
|
Jade1
|
jade family PHD finger 1 |
| chr7_-_123101851 | 0.98 |
ENSRNOT00000090984
ENSRNOT00000005837 |
Phf5a
|
PHD finger protein 5A |
| chr7_-_11783849 | 0.82 |
ENSRNOT00000026202
|
Sf3a2
|
splicing factor 3a, subunit 2 |
| chr10_+_75335146 | 0.73 |
ENSRNOT00000073005
|
Srsf1
|
serine and arginine rich splicing factor 1 |
| chr1_-_201110928 | 0.69 |
ENSRNOT00000027721
|
Nsmce4a
|
NSE4 homolog A, SMC5-SMC6 complex component |
| chrX_+_54734385 | 0.68 |
ENSRNOT00000005023
|
Nr0b1
|
nuclear receptor subfamily 0, group B, member 1 |
| chr10_-_108150511 | 0.55 |
ENSRNOT00000073337
|
Cbx8
|
chromobox 8 |
| chr6_+_36941233 | 0.51 |
ENSRNOT00000007073
|
Smc6
|
structural maintenance of chromosomes 6 |
| chr1_-_200163106 | 0.12 |
ENSRNOT00000027644
|
Mcmbp
|
minichromosome maintenance complex binding protein |
Network of associatons between targets according to the STRING database.
First level regulatory network of E2f2_E2f5
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 72.2 | 505.6 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 47.1 | 141.3 | GO:0007056 | spindle assembly involved in female meiosis(GO:0007056) |
| 43.2 | 129.7 | GO:0071163 | DNA replication preinitiation complex assembly(GO:0071163) |
| 42.5 | 170.0 | GO:0006272 | leading strand elongation(GO:0006272) |
| 30.6 | 183.4 | GO:0046125 | pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 26.4 | 185.1 | GO:0090166 | Golgi disassembly(GO:0090166) |
| 22.1 | 310.0 | GO:0019985 | translesion synthesis(GO:0019985) |
| 21.2 | 106.0 | GO:0071930 | negative regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071930) |
| 19.5 | 19.5 | GO:0046075 | pyrimidine deoxyribonucleoside triphosphate metabolic process(GO:0009211) dTTP metabolic process(GO:0046075) |
| 19.2 | 173.0 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 18.8 | 56.4 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 18.1 | 72.2 | GO:0070495 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 17.4 | 366.1 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 15.4 | 61.6 | GO:0010424 | DNA methylation on cytosine within a CG sequence(GO:0010424) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 14.8 | 236.5 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 14.8 | 118.2 | GO:1902969 | mitotic DNA replication(GO:1902969) |
| 13.3 | 80.0 | GO:0014834 | skeletal muscle satellite cell maintenance involved in skeletal muscle regeneration(GO:0014834) response to trichostatin A(GO:0035983) cellular response to trichostatin A(GO:0035984) |
| 10.5 | 31.4 | GO:0060821 | inactivation of X chromosome by DNA methylation(GO:0060821) |
| 9.0 | 72.0 | GO:0090267 | positive regulation of spindle checkpoint(GO:0090232) positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) |
| 8.8 | 97.1 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 8.4 | 16.9 | GO:0010216 | maintenance of DNA methylation(GO:0010216) |
| 7.9 | 31.8 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 7.4 | 96.4 | GO:0048304 | positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 7.3 | 36.6 | GO:0014043 | negative regulation of neuron maturation(GO:0014043) |
| 6.9 | 116.8 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 6.7 | 60.6 | GO:0044789 | modulation by host of viral release from host cell(GO:0044789) positive regulation by host of viral release from host cell(GO:0044791) |
| 6.5 | 508.4 | GO:0006342 | chromatin silencing(GO:0006342) |
| 5.7 | 22.9 | GO:0043570 | maintenance of DNA repeat elements(GO:0043570) |
| 5.5 | 43.6 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 5.1 | 81.3 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 4.4 | 22.1 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 4.0 | 23.7 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) |
| 3.9 | 43.2 | GO:0085020 | protein K6-linked ubiquitination(GO:0085020) |
| 3.7 | 335.6 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 3.4 | 17.2 | GO:0036337 | Fas signaling pathway(GO:0036337) |
| 3.1 | 9.2 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 2.7 | 62.9 | GO:0060236 | regulation of mitotic spindle organization(GO:0060236) |
| 2.7 | 8.2 | GO:1990166 | protein localization to site of double-strand break(GO:1990166) |
| 2.5 | 7.4 | GO:2001205 | negative regulation of osteoclast development(GO:2001205) |
| 2.4 | 9.7 | GO:0019042 | viral latency(GO:0019042) release from viral latency(GO:0019046) |
| 2.4 | 51.8 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 2.3 | 14.0 | GO:0071386 | cellular response to corticosterone stimulus(GO:0071386) |
| 2.3 | 23.1 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 2.3 | 9.1 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) |
| 2.3 | 9.0 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 2.2 | 15.3 | GO:1901838 | positive regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901838) |
| 2.2 | 8.7 | GO:0010825 | positive regulation of centrosome duplication(GO:0010825) |
| 2.1 | 14.9 | GO:2001032 | regulation of double-strand break repair via nonhomologous end joining(GO:2001032) |
| 2.0 | 16.2 | GO:0023035 | CD40 signaling pathway(GO:0023035) |
| 2.0 | 10.0 | GO:2000325 | regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000325) positive regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000327) |
| 1.9 | 9.7 | GO:0060903 | positive regulation of meiosis I(GO:0060903) |
| 1.9 | 5.8 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 1.8 | 25.5 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 1.6 | 4.8 | GO:0033566 | gamma-tubulin complex localization(GO:0033566) |
| 1.5 | 10.6 | GO:1990822 | arginine transmembrane transport(GO:1903826) basic amino acid transmembrane transport(GO:1990822) |
| 1.3 | 14.7 | GO:1904816 | positive regulation of protein localization to chromosome, telomeric region(GO:1904816) |
| 1.3 | 21.2 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 1.3 | 10.5 | GO:2001269 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001269) |
| 1.3 | 25.0 | GO:0006298 | mismatch repair(GO:0006298) |
| 1.3 | 43.7 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 1.3 | 37.0 | GO:0007099 | centriole replication(GO:0007099) |
| 1.3 | 11.5 | GO:0000722 | telomere maintenance via recombination(GO:0000722) |
| 1.2 | 27.5 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 1.1 | 4.6 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 1.0 | 9.4 | GO:0035879 | plasma membrane lactate transport(GO:0035879) |
| 1.0 | 2.9 | GO:1903352 | L-ornithine transmembrane transport(GO:1903352) |
| 0.9 | 18.4 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.9 | 6.2 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.8 | 5.8 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.7 | 15.0 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.7 | 6.4 | GO:0006301 | postreplication repair(GO:0006301) |
| 0.7 | 4.3 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.7 | 25.9 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.6 | 4.3 | GO:0043686 | co-translational protein modification(GO:0043686) |
| 0.6 | 31.0 | GO:0032781 | positive regulation of ATPase activity(GO:0032781) |
| 0.5 | 3.8 | GO:0010835 | regulation of protein ADP-ribosylation(GO:0010835) |
| 0.5 | 38.6 | GO:0071549 | cellular response to dexamethasone stimulus(GO:0071549) |
| 0.5 | 4.6 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.5 | 44.5 | GO:0001541 | ovarian follicle development(GO:0001541) |
| 0.5 | 12.7 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.5 | 56.1 | GO:0007051 | spindle organization(GO:0007051) |
| 0.5 | 2.8 | GO:1904415 | regulation of xenophagy(GO:1904415) positive regulation of xenophagy(GO:1904417) |
| 0.4 | 3.7 | GO:0043496 | regulation of protein homodimerization activity(GO:0043496) |
| 0.4 | 45.1 | GO:0051092 | positive regulation of NF-kappaB transcription factor activity(GO:0051092) |
| 0.4 | 2.5 | GO:1903912 | negative regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:1903912) |
| 0.4 | 91.5 | GO:0007018 | microtubule-based movement(GO:0007018) |
| 0.3 | 7.4 | GO:0061157 | mRNA destabilization(GO:0061157) |
| 0.3 | 7.2 | GO:0033197 | response to vitamin E(GO:0033197) |
| 0.3 | 25.1 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.3 | 13.2 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.3 | 1.4 | GO:0010793 | regulation of mRNA export from nucleus(GO:0010793) |
| 0.3 | 3.0 | GO:0090220 | meiotic telomere clustering(GO:0045141) chromosome localization to nuclear envelope involved in homologous chromosome segregation(GO:0090220) |
| 0.3 | 7.7 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.3 | 17.7 | GO:0000724 | double-strand break repair via homologous recombination(GO:0000724) |
| 0.2 | 9.1 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.2 | 12.3 | GO:0034644 | cellular response to UV(GO:0034644) |
| 0.2 | 2.3 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.2 | 7.1 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.1 | 6.7 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.1 | 2.9 | GO:0018023 | peptidyl-lysine trimethylation(GO:0018023) |
| 0.1 | 1.0 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.1 | 29.7 | GO:0030336 | negative regulation of cell migration(GO:0030336) |
| 0.1 | 3.2 | GO:0060135 | maternal process involved in female pregnancy(GO:0060135) |
| 0.1 | 1.4 | GO:2000009 | negative regulation of protein localization to cell surface(GO:2000009) |
| 0.1 | 0.7 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.1 | 5.9 | GO:0042990 | regulation of transcription factor import into nucleus(GO:0042990) |
| 0.1 | 3.4 | GO:0006879 | cellular iron ion homeostasis(GO:0006879) |
| 0.1 | 3.6 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.1 | 11.2 | GO:0001890 | placenta development(GO:0001890) |
| 0.1 | 3.8 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 0.7 | GO:0033144 | negative regulation of intracellular steroid hormone receptor signaling pathway(GO:0033144) |
| 0.0 | 3.3 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
| 0.0 | 1.4 | GO:0042058 | regulation of epidermal growth factor receptor signaling pathway(GO:0042058) |
| 0.0 | 0.6 | GO:0016574 | histone ubiquitination(GO:0016574) |
| 0.0 | 0.7 | GO:2001022 | positive regulation of response to DNA damage stimulus(GO:2001022) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 70.1 | 771.3 | GO:0042555 | MCM complex(GO:0042555) |
| 61.7 | 185.1 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 59.1 | 236.5 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 45.2 | 225.8 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 39.4 | 118.2 | GO:0000811 | GINS complex(GO:0000811) |
| 34.6 | 103.8 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 20.1 | 100.6 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 17.7 | 106.0 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 15.4 | 122.9 | GO:0001740 | Barr body(GO:0001740) |
| 11.7 | 1068.1 | GO:0000786 | nucleosome(GO:0000786) |
| 10.8 | 43.2 | GO:0031436 | BRCA1-BARD1 complex(GO:0031436) |
| 10.2 | 81.3 | GO:0000796 | condensin complex(GO:0000796) |
| 9.5 | 28.6 | GO:0032301 | MutSalpha complex(GO:0032301) |
| 8.7 | 60.6 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 7.9 | 102.2 | GO:0000800 | lateral element(GO:0000800) |
| 7.7 | 38.6 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 7.4 | 37.0 | GO:0098536 | deuterosome(GO:0098536) |
| 7.3 | 43.7 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 6.9 | 27.4 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 5.6 | 44.5 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 5.4 | 92.1 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 5.3 | 21.2 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 4.8 | 19.2 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 4.6 | 36.5 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 3.7 | 18.7 | GO:0035061 | interchromatin granule(GO:0035061) |
| 3.5 | 31.8 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 3.1 | 43.5 | GO:0072687 | meiotic spindle(GO:0072687) |
| 3.1 | 49.5 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 3.1 | 37.1 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 3.1 | 15.3 | GO:0001651 | dense fibrillar component(GO:0001651) |
| 2.6 | 36.1 | GO:0016580 | Sin3 complex(GO:0016580) |
| 2.5 | 61.6 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 2.3 | 9.0 | GO:0018444 | translation release factor complex(GO:0018444) |
| 2.1 | 142.6 | GO:0005871 | kinesin complex(GO:0005871) |
| 1.8 | 18.4 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 1.6 | 4.8 | GO:0008275 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) gamma-tubulin small complex(GO:0008275) |
| 1.5 | 4.4 | GO:0031515 | tRNA (m1A) methyltransferase complex(GO:0031515) |
| 1.5 | 14.6 | GO:0070652 | HAUS complex(GO:0070652) |
| 1.3 | 10.5 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.8 | 9.6 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.8 | 3.8 | GO:0090661 | box H/ACA telomerase RNP complex(GO:0090661) |
| 0.7 | 35.7 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.7 | 7.4 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.6 | 160.2 | GO:0005819 | spindle(GO:0005819) |
| 0.6 | 5.1 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.5 | 68.0 | GO:0005874 | microtubule(GO:0005874) |
| 0.5 | 2.4 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.5 | 11.2 | GO:0090544 | BAF-type complex(GO:0090544) |
| 0.4 | 4.6 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.4 | 6.8 | GO:0071011 | precatalytic spliceosome(GO:0071011) |
| 0.3 | 24.5 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.3 | 4.3 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.3 | 4.3 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.2 | 4.8 | GO:0019013 | viral nucleocapsid(GO:0019013) |
| 0.2 | 5.7 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.2 | 2.5 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.2 | 0.7 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.2 | 7.7 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.2 | 1.6 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.1 | 4.6 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.1 | 12.1 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 5.8 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.1 | 1.4 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.1 | 44.5 | GO:0043235 | receptor complex(GO:0043235) |
| 0.1 | 51.7 | GO:0005813 | centrosome(GO:0005813) |
| 0.1 | 10.7 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 7.2 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 234.5 | GO:0005654 | nucleoplasm(GO:0005654) |
| 0.1 | 1.9 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.1 | 14.9 | GO:0043292 | contractile fiber(GO:0043292) |
| 0.1 | 5.6 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.1 | 5.9 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.1 | 4.6 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 1.4 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 16.5 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 45.7 | GO:0005634 | nucleus(GO:0005634) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 45.8 | 183.4 | GO:0001884 | pyrimidine nucleoside binding(GO:0001884) deoxynucleoside kinase activity(GO:0019136) |
| 39.4 | 236.5 | GO:0061731 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 34.6 | 103.8 | GO:0030337 | DNA polymerase processivity factor activity(GO:0030337) |
| 27.2 | 135.9 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 26.7 | 133.5 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 22.8 | 318.6 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 17.0 | 102.2 | GO:0000150 | recombinase activity(GO:0000150) |
| 12.4 | 582.5 | GO:0003678 | DNA helicase activity(GO:0003678) |
| 12.3 | 61.6 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 10.9 | 185.1 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 10.3 | 20.6 | GO:0032564 | dATP binding(GO:0032564) |
| 10.3 | 72.0 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 8.0 | 80.0 | GO:0070878 | primary miRNA binding(GO:0070878) |
| 7.6 | 22.9 | GO:0032142 | single guanine insertion binding(GO:0032142) |
| 7.0 | 56.0 | GO:0019103 | pyrimidine nucleotide binding(GO:0019103) |
| 7.0 | 27.9 | GO:0000405 | bubble DNA binding(GO:0000405) |
| 6.4 | 38.6 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 6.1 | 18.4 | GO:0000991 | transcription factor activity, core RNA polymerase II binding(GO:0000991) |
| 4.9 | 24.3 | GO:0036033 | mediator complex binding(GO:0036033) |
| 4.2 | 21.2 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 3.9 | 101.1 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 3.8 | 15.3 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 3.6 | 14.4 | GO:0016428 | tRNA (cytosine-5-)-methyltransferase activity(GO:0016428) |
| 3.5 | 31.8 | GO:0043142 | single-stranded DNA-dependent ATPase activity(GO:0043142) |
| 2.9 | 26.5 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 2.6 | 13.2 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 2.6 | 91.5 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 2.3 | 122.8 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 2.3 | 81.3 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 2.1 | 17.2 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 2.1 | 19.2 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 2.0 | 42.3 | GO:0031996 | thioesterase binding(GO:0031996) |
| 1.9 | 5.7 | GO:0031208 | POZ domain binding(GO:0031208) |
| 1.9 | 123.8 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 1.6 | 142.6 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 1.6 | 56.4 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 1.5 | 4.6 | GO:0097363 | protein O-GlcNAc transferase activity(GO:0097363) |
| 1.5 | 36.6 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 1.4 | 878.5 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 1.4 | 51.1 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 1.3 | 10.6 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 1.3 | 31.5 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
| 1.2 | 9.4 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 1.0 | 4.0 | GO:0016426 | tRNA (adenine) methyltransferase activity(GO:0016426) |
| 1.0 | 152.3 | GO:0042393 | histone binding(GO:0042393) |
| 0.8 | 108.4 | GO:0001047 | core promoter binding(GO:0001047) |
| 0.8 | 9.0 | GO:0008079 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.6 | 4.3 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.6 | 2.9 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.6 | 5.1 | GO:0015266 | protein channel activity(GO:0015266) |
| 0.5 | 3.8 | GO:0034061 | DNA polymerase activity(GO:0034061) |
| 0.5 | 31.0 | GO:0031072 | heat shock protein binding(GO:0031072) |
| 0.5 | 16.9 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.5 | 10.9 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.4 | 5.2 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.4 | 2.8 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.4 | 2.3 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.4 | 16.2 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.3 | 4.3 | GO:0004579 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.2 | 19.8 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.2 | 1.9 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.2 | 5.8 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.2 | 13.1 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.2 | 0.7 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.2 | 22.1 | GO:0016616 | oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor(GO:0016616) |
| 0.2 | 0.7 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.1 | 27.1 | GO:0016874 | ligase activity(GO:0016874) |
| 0.1 | 2.1 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.1 | 7.4 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.1 | 2.5 | GO:0097472 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) cyclin-dependent protein kinase activity(GO:0097472) |
| 0.1 | 67.2 | GO:0019901 | protein kinase binding(GO:0019901) |
| 0.1 | 10.4 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.1 | 7.4 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 49.7 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.1 | 7.4 | GO:0004386 | helicase activity(GO:0004386) |
| 0.1 | 9.1 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 10.5 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.1 | 3.8 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.1 | 63.9 | GO:0005524 | ATP binding(GO:0005524) |
| 0.1 | 2.9 | GO:0016279 | lysine N-methyltransferase activity(GO:0016278) protein-lysine N-methyltransferase activity(GO:0016279) |
| 0.1 | 4.1 | GO:0008757 | S-adenosylmethionine-dependent methyltransferase activity(GO:0008757) |
| 0.0 | 4.9 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 2.5 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 1.4 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 5.4 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.0 | 5.9 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 23.6 | 236.2 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 10.9 | 511.6 | PID ATR PATHWAY | ATR signaling pathway |
| 7.6 | 106.0 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 7.5 | 195.9 | PID BARD1 PATHWAY | BARD1 signaling events |
| 6.1 | 405.6 | PID E2F PATHWAY | E2F transcription factor network |
| 5.0 | 104.1 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 4.2 | 184.8 | PID PLK1 PATHWAY | PLK1 signaling events |
| 2.9 | 34.8 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 1.4 | 118.4 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 1.4 | 61.6 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 1.3 | 24.3 | PID ATM PATHWAY | ATM pathway |
| 1.2 | 98.2 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 1.2 | 17.2 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 1.0 | 56.4 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.7 | 14.0 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.5 | 19.6 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.5 | 23.1 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.4 | 7.4 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.3 | 16.2 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.3 | 9.1 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.2 | 10.0 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.2 | 5.8 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.1 | 7.7 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.7 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 9.1 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 80.0 | 880.2 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 37.0 | 813.0 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 23.3 | 605.3 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 16.1 | 257.7 | REACTOME REPAIR SYNTHESIS FOR GAP FILLING BY DNA POL IN TC NER | Genes involved in Repair synthesis for gap-filling by DNA polymerase in TC-NER |
| 11.5 | 183.4 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 8.2 | 106.1 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 6.3 | 56.4 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 5.7 | 102.2 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 4.6 | 142.6 | REACTOME KINESINS | Genes involved in Kinesins |
| 4.3 | 38.7 | REACTOME ACTIVATION OF THE PRE REPLICATIVE COMPLEX | Genes involved in Activation of the pre-replicative complex |
| 4.1 | 49.7 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 2.9 | 31.6 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 2.6 | 49.5 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 2.4 | 103.5 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 2.2 | 26.4 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 1.9 | 51.8 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 1.7 | 31.4 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 1.6 | 38.6 | REACTOME RNA POL I TRANSCRIPTION | Genes involved in RNA Polymerase I Transcription |
| 0.9 | 13.2 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.7 | 114.5 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.7 | 9.4 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.6 | 14.0 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.5 | 3.8 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.4 | 4.6 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.4 | 19.0 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.4 | 5.9 | REACTOME ACTIVATION OF CHAPERONES BY ATF6 ALPHA | Genes involved in Activation of Chaperones by ATF6-alpha |
| 0.4 | 9.1 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.3 | 10.6 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.2 | 9.1 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 7.2 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.1 | 9.2 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.1 | 1.9 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.1 | 7.4 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.1 | 2.9 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.1 | 6.7 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.1 | 1.6 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |