Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
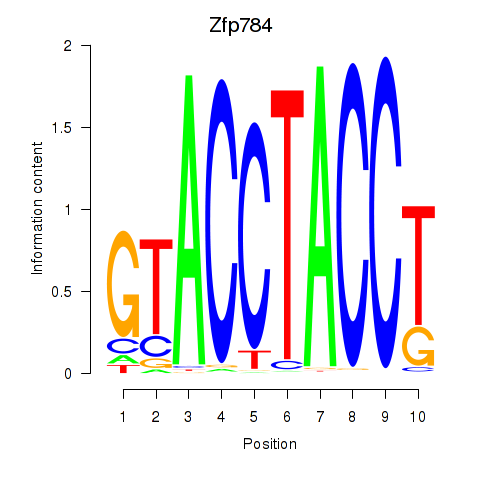
Results for Zfp784
Z-value: 0.42
Transcription factors associated with Zfp784
| Gene Symbol | Gene ID | Gene Info |
|---|
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zfp865 | rn6_v1_chr1_+_72356880_72356880 | 0.08 | 8.9e-01 | Click! |
Activity profile of Zfp784 motif
Sorted Z-values of Zfp784 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_+_11101306 | 0.18 |
ENSRNOT00000030893
|
Drd1
|
dopamine receptor D1 |
| chr1_-_16203838 | 0.16 |
ENSRNOT00000018556
ENSRNOT00000078586 |
Pde7b
|
phosphodiesterase 7B |
| chr1_+_168964202 | 0.14 |
ENSRNOT00000089102
|
LOC103694855
|
hemoglobin subunit beta-2-like |
| chr7_+_11582984 | 0.13 |
ENSRNOT00000026893
|
Gng7
|
G protein subunit gamma 7 |
| chr12_-_39850567 | 0.13 |
ENSRNOT00000001712
|
Hvcn1
|
hydrogen voltage-gated channel 1 |
| chr18_+_36371041 | 0.12 |
ENSRNOT00000025408
|
Sh3rf2
|
SH3 domain containing ring finger 2 |
| chr1_-_168972725 | 0.12 |
ENSRNOT00000090422
|
Hbb
|
hemoglobin subunit beta |
| chr1_+_168945449 | 0.12 |
ENSRNOT00000087661
ENSRNOT00000019913 |
LOC103694855
|
hemoglobin subunit beta-2-like |
| chr2_+_252263386 | 0.11 |
ENSRNOT00000092913
ENSRNOT00000084034 ENSRNOT00000041186 ENSRNOT00000092931 |
Ssx2ip
|
SSX family member 2 interacting protein |
| chr4_-_157252104 | 0.09 |
ENSRNOT00000082739
|
Ptpn6
|
protein tyrosine phosphatase, non-receptor type 6 |
| chr17_+_49322205 | 0.09 |
ENSRNOT00000017713
|
Pou6f2
|
POU domain, class 6, transcription factor 2 |
| chr15_+_51756978 | 0.09 |
ENSRNOT00000024067
|
Egr3
|
early growth response 3 |
| chr16_+_19051965 | 0.09 |
ENSRNOT00000016399
|
Slc35e1
|
solute carrier family 35, member E1 |
| chr1_+_219403970 | 0.08 |
ENSRNOT00000029607
|
Ptprcap
|
protein tyrosine phosphatase, receptor type, C-associated protein |
| chr17_+_57031766 | 0.07 |
ENSRNOT00000092187
ENSRNOT00000068545 |
Crem
|
cAMP responsive element modulator |
| chr5_-_72287669 | 0.07 |
ENSRNOT00000022255
|
Klf4
|
Kruppel like factor 4 |
| chr20_-_3978845 | 0.07 |
ENSRNOT00000000532
|
Psmb9
|
proteasome subunit beta 9 |
| chr3_+_110442637 | 0.07 |
ENSRNOT00000010471
|
Pak6
|
p21 (RAC1) activated kinase 6 |
| chr3_-_10226286 | 0.07 |
ENSRNOT00000093627
|
Fubp3
|
far upstream element binding protein 3 |
| chr7_+_142912316 | 0.06 |
ENSRNOT00000010171
|
Nr4a1
|
nuclear receptor subfamily 4, group A, member 1 |
| chr1_-_205777167 | 0.06 |
ENSRNOT00000024279
|
Uros
|
uroporphyrinogen III synthase |
| chr6_-_91518996 | 0.06 |
ENSRNOT00000005835
|
Pole2
|
DNA polymerase epsilon 2, accessory subunit |
| chr19_+_25043680 | 0.06 |
ENSRNOT00000043971
|
Adgrl1
|
adhesion G protein-coupled receptor L1 |
| chr5_+_118574801 | 0.06 |
ENSRNOT00000035949
|
Ube2u
|
ubiquitin-conjugating enzyme E2U (putative) |
| chr11_-_11585078 | 0.05 |
ENSRNOT00000088878
|
Robo2
|
roundabout guidance receptor 2 |
| chr8_+_118333706 | 0.05 |
ENSRNOT00000028278
|
Cspg5
|
chondroitin sulfate proteoglycan 5 |
| chr6_-_42616548 | 0.05 |
ENSRNOT00000081433
|
Atp6v1c2
|
ATPase H+ transporting V1 subunit C2 |
| chr8_-_49308806 | 0.05 |
ENSRNOT00000047291
|
Cd3e
|
CD3e molecule |
| chr11_-_66759402 | 0.05 |
ENSRNOT00000003326
|
Hcls1
|
hematopoietic cell specific Lyn substrate 1 |
| chr14_+_86828355 | 0.05 |
ENSRNOT00000088611
|
Ccm2
|
CCM2 scaffolding protein |
| chr5_-_148392689 | 0.05 |
ENSRNOT00000018464
ENSRNOT00000080166 |
Tinagl1
|
tubulointerstitial nephritis antigen-like 1 |
| chr11_-_14304603 | 0.05 |
ENSRNOT00000040202
ENSRNOT00000082143 |
Samsn1
|
SAM domain, SH3 domain and nuclear localization signals, 1 |
| chr8_+_127144903 | 0.04 |
ENSRNOT00000013530
|
Eomes
|
eomesodermin |
| chr1_-_164143818 | 0.04 |
ENSRNOT00000022557
|
Dgat2
|
diacylglycerol O-acyltransferase 2 |
| chr16_+_50111306 | 0.04 |
ENSRNOT00000019302
|
Cyp4v3
|
cytochrome P450, family 4, subfamily v, polypeptide 3 |
| chr15_-_52332097 | 0.04 |
ENSRNOT00000017723
|
Fgf17
|
fibroblast growth factor 17 |
| chr7_-_144837583 | 0.04 |
ENSRNOT00000055289
|
Cbx5
|
chromobox 5 |
| chr8_-_130127392 | 0.04 |
ENSRNOT00000026159
|
Cck
|
cholecystokinin |
| chrX_+_48196916 | 0.04 |
ENSRNOT00000045976
|
Da2-19
|
uncharacterized LOC302729 |
| chr9_+_66058047 | 0.04 |
ENSRNOT00000071819
|
Cdk15
|
cyclin-dependent kinase 15 |
| chr1_-_176079125 | 0.03 |
ENSRNOT00000047044
|
RGD1566189
|
similar to ferritin light chain |
| chr1_+_276659542 | 0.03 |
ENSRNOT00000019681
|
Tcf7l2
|
transcription factor 7 like 2 |
| chr14_-_5006594 | 0.03 |
ENSRNOT00000076571
|
Zfp326
|
zinc finger protein 326 |
| chr9_-_100306829 | 0.03 |
ENSRNOT00000038563
|
RGD1563692
|
similar to hypothetical protein FLJ22671 |
| chr10_-_64657089 | 0.03 |
ENSRNOT00000080703
|
Abr
|
active BCR-related |
| chr1_-_101449829 | 0.03 |
ENSRNOT00000028315
|
LOC100360087
|
ferritin light chain 1-like |
| chr12_+_32103198 | 0.03 |
ENSRNOT00000085464
|
Tmem132d
|
transmembrane protein 132D |
| chr15_-_52317219 | 0.03 |
ENSRNOT00000016555
|
Dmtn
|
dematin actin binding protein |
| chr6_+_50528823 | 0.03 |
ENSRNOT00000008321
|
Lamb1
|
laminin subunit beta 1 |
| chr10_+_57185347 | 0.03 |
ENSRNOT00000040488
ENSRNOT00000086374 |
Mink1
|
misshapen-like kinase 1 |
| chr8_-_49038169 | 0.03 |
ENSRNOT00000047303
|
Phldb1
|
pleckstrin homology-like domain, family B, member 1 |
| chr1_-_72339395 | 0.03 |
ENSRNOT00000021772
|
Zfp580
|
zinc finger protein 580 |
| chr1_-_72464492 | 0.03 |
ENSRNOT00000068550
|
Nat14
|
N-acetyltransferase 14 |
| chr9_-_100306194 | 0.03 |
ENSRNOT00000087584
|
RGD1563692
|
similar to hypothetical protein FLJ22671 |
| chr14_-_84189266 | 0.03 |
ENSRNOT00000005934
|
Tcn2
|
transcobalamin 2 |
| chr6_+_9483594 | 0.03 |
ENSRNOT00000089272
|
AABR07062799.2
|
|
| chr7_-_124367630 | 0.03 |
ENSRNOT00000055978
|
Ttll1
|
tubulin tyrosine ligase like 1 |
| chr7_+_15385868 | 0.03 |
ENSRNOT00000042108
|
RGD1560687
|
similar to Ferritin light chain (Ferritin L subunit) |
| chrX_-_139476206 | 0.03 |
ENSRNOT00000049856
|
Ftl1l1
|
ferritin light chain 1-like 1 |
| chr7_-_144837395 | 0.03 |
ENSRNOT00000089024
|
Cbx5
|
chromobox 5 |
| chr10_-_86393141 | 0.03 |
ENSRNOT00000009485
|
Mien1
|
migration and invasion enhancer 1 |
| chr20_-_31956649 | 0.03 |
ENSRNOT00000072429
|
Hk1
|
hexokinase 1 |
| chr4_+_42202838 | 0.03 |
ENSRNOT00000082453
|
Mdfic
|
MyoD family inhibitor domain containing |
| chr16_-_49522341 | 0.03 |
ENSRNOT00000081642
|
LOC100911140
|
coiled-coil domain-containing protein 110-like |
| chr5_+_145079803 | 0.02 |
ENSRNOT00000084202
|
Sfpq
|
splicing factor proline and glutamine rich |
| chr4_-_55398941 | 0.02 |
ENSRNOT00000075324
|
Grm8
|
glutamate metabotropic receptor 8 |
| chr3_-_153893847 | 0.02 |
ENSRNOT00000085938
|
LOC100911769
|
band 4.1-like protein 1-like |
| chr4_+_157513414 | 0.02 |
ENSRNOT00000078769
|
Pianp
|
PILR alpha associated neural protein |
| chr9_+_43889473 | 0.02 |
ENSRNOT00000024330
|
Inpp4a
|
inositol polyphosphate-4-phosphatase type I A |
| chr16_-_20097287 | 0.02 |
ENSRNOT00000025162
|
Unc13a
|
unc-13 homolog A |
| chr13_-_91776397 | 0.02 |
ENSRNOT00000073147
|
Mptx1
|
mucosal pentraxin 1 |
| chr7_-_116063078 | 0.02 |
ENSRNOT00000076932
ENSRNOT00000035496 |
Gml
|
glycosylphosphatidylinositol anchored molecule like |
| chr7_-_124286478 | 0.02 |
ENSRNOT00000088125
|
Pacsin2
|
protein kinase C and casein kinase substrate in neurons 2 |
| chr8_-_115981910 | 0.02 |
ENSRNOT00000019867
|
Dock3
|
dedicator of cyto-kinesis 3 |
| chr7_-_70842405 | 0.02 |
ENSRNOT00000047449
|
Nxph4
|
neurexophilin 4 |
| chr3_-_164055561 | 0.02 |
ENSRNOT00000064849
|
B4galt5
|
beta-1,4-galactosyltransferase 5 |
| chr17_-_77527894 | 0.02 |
ENSRNOT00000032173
|
Bend7
|
BEN domain containing 7 |
| chr2_+_205568935 | 0.02 |
ENSRNOT00000025248
|
Ampd1
|
adenosine monophosphate deaminase 1 |
| chr9_+_100489852 | 0.02 |
ENSRNOT00000022854
|
Ppp1r7
|
protein phosphatase 1, regulatory subunit 7 |
| chr5_-_24255606 | 0.02 |
ENSRNOT00000037483
|
Plekhf2
|
pleckstrin homology and FYVE domain containing 2 |
| chrX_+_105575759 | 0.02 |
ENSRNOT00000088760
|
Armcx3
|
armadillo repeat containing, X-linked 3 |
| chr1_+_82452469 | 0.02 |
ENSRNOT00000028026
|
Exosc5
|
exosome component 5 |
| chr13_-_1946508 | 0.02 |
ENSRNOT00000043890
|
Dsel
|
dermatan sulfate epimerase-like |
| chr5_+_152325871 | 0.02 |
ENSRNOT00000044711
|
Ftl1
|
ferritin light chain 1 |
| chr7_+_145068286 | 0.02 |
ENSRNOT00000088956
ENSRNOT00000065753 |
Nckap1l
|
NCK associated protein 1 like |
| chr8_+_111600532 | 0.02 |
ENSRNOT00000081952
|
Rab6b
|
RAB6B, member RAS oncogene family |
| chr16_-_47163898 | 0.02 |
ENSRNOT00000074490
|
Ccdc110
|
coiled-coil domain containing 110 |
| chr4_+_116933341 | 0.02 |
ENSRNOT00000031768
|
RGD1560795
|
similar to Sepiapterin reductase (SPR) |
| chr17_+_8299131 | 0.02 |
ENSRNOT00000083687
|
Gm45623
|
predicted gene 45623 |
| chr3_-_29996865 | 0.02 |
ENSRNOT00000080382
|
Zeb2
|
zinc finger E-box binding homeobox 2 |
| chrX_-_79909678 | 0.02 |
ENSRNOT00000050336
|
Brwd3
|
bromodomain and WD repeat domain containing 3 |
| chr5_+_147714163 | 0.02 |
ENSRNOT00000012663
|
Marcksl1
|
MARCKS-like 1 |
| chr3_-_9822182 | 0.02 |
ENSRNOT00000076637
ENSRNOT00000010115 |
RGD1305178
|
similar to Hypothetical protein MGC11690 |
| chr5_+_74727494 | 0.02 |
ENSRNOT00000076683
|
Rn50_5_0791.1
|
|
| chr7_-_120026755 | 0.02 |
ENSRNOT00000077677
|
Card10
|
caspase recruitment domain family, member 10 |
| chr19_+_54553419 | 0.02 |
ENSRNOT00000025392
|
Jph3
|
junctophilin 3 |
| chr6_-_39363367 | 0.02 |
ENSRNOT00000088687
ENSRNOT00000065531 |
Fam84a
|
family with sequence similarity 84, member A |
| chr1_+_255040426 | 0.02 |
ENSRNOT00000092151
|
Pcgf5
|
polycomb group ring finger 5 |
| chr1_-_259674425 | 0.02 |
ENSRNOT00000091116
|
LOC108348083
|
delta-1-pyrroline-5-carboxylate synthase |
| chr7_-_120027026 | 0.02 |
ENSRNOT00000011215
|
Card10
|
caspase recruitment domain family, member 10 |
| chr7_+_117723263 | 0.02 |
ENSRNOT00000085152
|
Kifc2
|
kinesin family member C2 |
| chr13_-_26768708 | 0.01 |
ENSRNOT00000092763
|
Bcl2
|
BCL2, apoptosis regulator |
| chr18_+_72550454 | 0.01 |
ENSRNOT00000092173
|
Smad2
|
SMAD family member 2 |
| chr15_+_33885106 | 0.01 |
ENSRNOT00000024483
|
Dhrs2
|
dehydrogenase/reductase 2 |
| chr3_+_145032200 | 0.01 |
ENSRNOT00000068210
|
Syndig1
|
synapse differentiation inducing 1 |
| chr12_-_38274036 | 0.01 |
ENSRNOT00000063990
|
Kntc1
|
kinetochore associated 1 |
| chr20_+_55594676 | 0.01 |
ENSRNOT00000057010
|
Sim1
|
single-minded family bHLH transcription factor 1 |
| chr2_+_54466280 | 0.01 |
ENSRNOT00000033112
|
C6
|
complement C6 |
| chr4_-_119889949 | 0.01 |
ENSRNOT00000033687
|
H1fx
|
H1 histone family, member X |
| chr1_-_104202591 | 0.01 |
ENSRNOT00000035512
|
E2f8
|
E2F transcription factor 8 |
| chr1_-_259089632 | 0.01 |
ENSRNOT00000020940
ENSRNOT00000091297 |
Aldh18a1
|
aldehyde dehydrogenase 18 family, member A1 |
| chr17_-_43584152 | 0.01 |
ENSRNOT00000023241
|
Slc17a2
|
solute carrier family 17, member 2 |
| chr2_-_184263564 | 0.01 |
ENSRNOT00000015279
|
Fbxw7
|
F-box and WD repeat domain containing 7 |
| chr5_-_126856292 | 0.01 |
ENSRNOT00000013575
|
Lrrc42
|
leucine rich repeat containing 42 |
| chr5_-_78267063 | 0.01 |
ENSRNOT00000040894
ENSRNOT00000087648 |
Cdc26
|
cell division cycle 26 |
| chr2_-_241545250 | 0.01 |
ENSRNOT00000073875
|
Bank1
|
B-cell scaffold protein with ankyrin repeats 1 |
| chr8_-_131899023 | 0.01 |
ENSRNOT00000034690
|
Zfp445
|
zinc finger protein 445 |
| chr3_+_152552822 | 0.01 |
ENSRNOT00000089719
|
Epb41l1
|
erythrocyte membrane protein band 4.1-like 1 |
| chr17_+_86066833 | 0.01 |
ENSRNOT00000022661
|
Msrb2
|
methionine sulfoxide reductase B2 |
| chr18_+_72550219 | 0.01 |
ENSRNOT00000046847
|
Smad2
|
SMAD family member 2 |
| chr10_-_76263866 | 0.01 |
ENSRNOT00000003219
|
Scpep1
|
serine carboxypeptidase 1 |
| chr10_-_62032400 | 0.01 |
ENSRNOT00000086378
ENSRNOT00000004161 |
Dph1
|
diphthamide biosynthesis 1 |
| chr11_+_84396033 | 0.01 |
ENSRNOT00000002316
|
Abcc5
|
ATP binding cassette subfamily C member 5 |
| chr3_+_56861396 | 0.01 |
ENSRNOT00000000008
ENSRNOT00000084375 |
Gad1
|
glutamate decarboxylase 1 |
| chr10_+_107455845 | 0.01 |
ENSRNOT00000004373
|
C1qtnf1
|
C1q and tumor necrosis factor related protein 1 |
| chr10_+_45659143 | 0.01 |
ENSRNOT00000058327
|
Wnt9a
|
wingless-type MMTV integration site family, member 9A |
| chr6_+_99625306 | 0.01 |
ENSRNOT00000008573
|
Plekhg3
|
pleckstrin homology and RhoGEF domain containing G3 |
| chr8_+_71167305 | 0.01 |
ENSRNOT00000021337
|
Rbpms2
|
RNA binding protein with multiple splicing 2 |
| chr7_+_24939498 | 0.01 |
ENSRNOT00000010601
|
Ckap4
|
cytoskeleton-associated protein 4 |
| chr18_+_51492196 | 0.01 |
ENSRNOT00000020570
|
Gramd3
|
GRAM domain containing 3 |
| chr7_-_145575981 | 0.01 |
ENSRNOT00000074622
|
LOC102555445
|
16.5 kDa submandibular gland glycoprotein-like |
| chr1_-_48825364 | 0.01 |
ENSRNOT00000024213
|
Agpat4
|
1-acylglycerol-3-phosphate O-acyltransferase 4 |
| chr15_-_48833285 | 0.01 |
ENSRNOT00000019117
|
Pnoc
|
prepronociceptin |
| chr15_-_24199341 | 0.01 |
ENSRNOT00000015553
|
Dlgap5
|
DLG associated protein 5 |
| chr4_+_7377563 | 0.01 |
ENSRNOT00000084826
|
Kcnh2
|
potassium voltage-gated channel subfamily H member 2 |
| chrX_-_43592200 | 0.01 |
ENSRNOT00000005033
|
Acot9
|
acyl-CoA thioesterase 9 |
| chr3_+_38559914 | 0.01 |
ENSRNOT00000085545
|
Fmnl2
|
formin-like 2 |
| chr15_-_12889102 | 0.01 |
ENSRNOT00000061214
ENSRNOT00000084687 |
RGD1306063
|
similar to HT021 |
| chrX_-_13116743 | 0.01 |
ENSRNOT00000004305
|
Mid1ip1
|
MID1 interacting protein 1 |
| chr1_-_88826302 | 0.01 |
ENSRNOT00000028275
|
Lrfn3
|
leucine rich repeat and fibronectin type III domain containing 3 |
| chr7_-_117257402 | 0.01 |
ENSRNOT00000081021
|
Plec
|
plectin |
| chr18_-_37096132 | 0.01 |
ENSRNOT00000041188
|
Ppp2r2b
|
protein phosphatase 2, regulatory subunit B, beta |
| chr10_+_53778662 | 0.01 |
ENSRNOT00000045718
|
Myh2
|
myosin heavy chain 2 |
| chr8_-_128754514 | 0.01 |
ENSRNOT00000025019
|
Cx3cr1
|
C-X3-C motif chemokine receptor 1 |
| chr5_+_119097715 | 0.01 |
ENSRNOT00000045987
|
Ror1
|
receptor tyrosine kinase-like orphan receptor 1 |
| chr18_-_60095354 | 0.01 |
ENSRNOT00000038652
|
Atp8b1
|
ATPase phospholipid transporting 8B1 |
| chr8_-_67708304 | 0.00 |
ENSRNOT00000009368
|
Fem1b
|
fem-1 homolog B |
| chr7_-_140617721 | 0.00 |
ENSRNOT00000081355
|
Tuba1b
|
tubulin, alpha 1B |
| chr17_+_43900450 | 0.00 |
ENSRNOT00000023622
|
Btn1a1
|
butyrophilin, subfamily 1, member A1 |
| chr12_+_22153983 | 0.00 |
ENSRNOT00000080775
ENSRNOT00000001894 |
Pcolce
|
procollagen C-endopeptidase enhancer |
| chrX_-_104726816 | 0.00 |
ENSRNOT00000005165
|
Tspan6
|
tetraspanin 6 |
| chr11_-_68197772 | 0.00 |
ENSRNOT00000003060
ENSRNOT00000081875 |
Hspbap1
|
Hspb associated protein 1 |
| chr4_+_58693384 | 0.00 |
ENSRNOT00000081707
|
Mkln1
|
muskelin 1 |
| chr8_+_132241134 | 0.00 |
ENSRNOT00000006055
|
Clec3b
|
C-type lectin domain family 3, member B |
| chr11_-_33003021 | 0.00 |
ENSRNOT00000084134
|
Runx1
|
runt-related transcription factor 1 |
| chr7_+_40217269 | 0.00 |
ENSRNOT00000082090
|
Cep290
|
centrosomal protein 290 |
| chr1_+_29225707 | 0.00 |
ENSRNOT00000018849
|
Ncoa7
|
nuclear receptor coactivator 7 |
| chrX_+_78196300 | 0.00 |
ENSRNOT00000048695
|
P2ry10
|
purinergic receptor P2Y10 |
| chr9_+_20048121 | 0.00 |
ENSRNOT00000014791
|
Mep1a
|
meprin 1 subunit alpha |
| chr14_+_106153575 | 0.00 |
ENSRNOT00000010264
|
Vps54
|
VPS54 GARP complex subunit |
| chr17_+_76410585 | 0.00 |
ENSRNOT00000024016
|
Cdc123
|
cell division cycle 123 |
| chr17_-_56909992 | 0.00 |
ENSRNOT00000021801
|
Bambi
|
BMP and activin membrane-bound inhibitor |
| chr19_+_33928356 | 0.00 |
ENSRNOT00000058162
|
Ednra
|
endothelin receptor type A |
| chrX_-_139229702 | 0.00 |
ENSRNOT00000042507
|
AABR07041771.1
|
|
| chr2_-_231409988 | 0.00 |
ENSRNOT00000080637
ENSRNOT00000037255 ENSRNOT00000074002 |
Ank2
|
ankyrin 2 |
| chr6_-_105097054 | 0.00 |
ENSRNOT00000048606
|
Slc8a3
|
solute carrier family 8 member A3 |
| chr5_-_164844586 | 0.00 |
ENSRNOT00000011287
|
Clcn6
|
chloride voltage-gated channel 6 |
| chr14_-_21553949 | 0.00 |
ENSRNOT00000031642
|
Smr3b
|
submaxillary gland androgen regulated protein 3B |
| chr7_-_116926555 | 0.00 |
ENSRNOT00000010637
|
Naprt
|
nicotinate phosphoribosyltransferase |
| chrX_-_23144324 | 0.00 |
ENSRNOT00000000178
ENSRNOT00000081239 |
Pfkfb1
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 |
| chrX_+_123404518 | 0.00 |
ENSRNOT00000015085
|
Slc25a5
|
solute carrier family 25 member 5 |
| chr4_-_58250798 | 0.00 |
ENSRNOT00000048436
|
Klf14
|
Kruppel-like factor 14 |
| chr7_+_144605058 | 0.00 |
ENSRNOT00000031404
|
Hoxc8
|
homeobox C8 |
| chr2_+_199199845 | 0.00 |
ENSRNOT00000020932
|
LOC100909441
|
tubulin alpha-1C chain-like |
| chr2_-_200692335 | 0.00 |
ENSRNOT00000026290
|
Hsd3b5
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 5 |
| chr10_-_98544447 | 0.00 |
ENSRNOT00000073149
|
Abca6
|
ATP binding cassette subfamily A member 6 |
| chr9_-_17120677 | 0.00 |
ENSRNOT00000025769
|
Yipf3
|
Yip1 domain family, member 3 |
| chr15_+_15275541 | 0.00 |
ENSRNOT00000012153
|
Cadps
|
calcium dependent secretion activator |
| chr3_-_8751324 | 0.00 |
ENSRNOT00000030623
|
LOC499770
|
similar to LOC495800 protein |
| chr6_-_129509720 | 0.00 |
ENSRNOT00000006131
|
Atg2b
|
autophagy related 2B |
| chr1_-_147077568 | 0.00 |
ENSRNOT00000074040
|
LOC100911002
|
synaptonemal complex protein 3-like |
Network of associatons between targets according to the STRING database.
First level regulatory network of Zfp784
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0031161 | phosphatidylinositol catabolic process(GO:0031161) |
| 0.0 | 0.4 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.1 | GO:2000342 | negative regulation of chemokine (C-X-C motif) ligand 2 production(GO:2000342) |
| 0.0 | 0.1 | GO:0033277 | abortive mitotic cell cycle(GO:0033277) |
| 0.0 | 0.1 | GO:1901423 | response to benzene(GO:1901423) |
| 0.0 | 0.1 | GO:0042531 | regulation of tyrosine phosphorylation of STAT protein(GO:0042509) positive regulation of tyrosine phosphorylation of STAT protein(GO:0042531) |
| 0.0 | 0.1 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.0 | 0.0 | GO:0048619 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) embryonic hindgut morphogenesis(GO:0048619) |
| 0.0 | 0.1 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.0 | 0.0 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) |
| 0.0 | 0.1 | GO:0050925 | negative regulation of negative chemotaxis(GO:0050925) |
| 0.0 | 0.1 | GO:0043435 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.0 | 0.0 | GO:0021812 | neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.0 | 0.2 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.0 | 0.1 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.1 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 0.1 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.0 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.1 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.0 | 0.0 | GO:0044609 | DBIRD complex(GO:0044609) |
| 0.0 | 0.0 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.0 | 0.1 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.0 | 0.1 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.0 | 0.3 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 0.1 | GO:0015252 | hydrogen ion channel activity(GO:0015252) |
| 0.0 | 0.1 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.0 | 0.1 | GO:0001010 | transcription factor activity, sequence-specific DNA binding transcription factor recruiting(GO:0001010) |
| 0.0 | 0.0 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.0 | 0.1 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |