Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
Results for Tfap2c
Z-value: 0.67
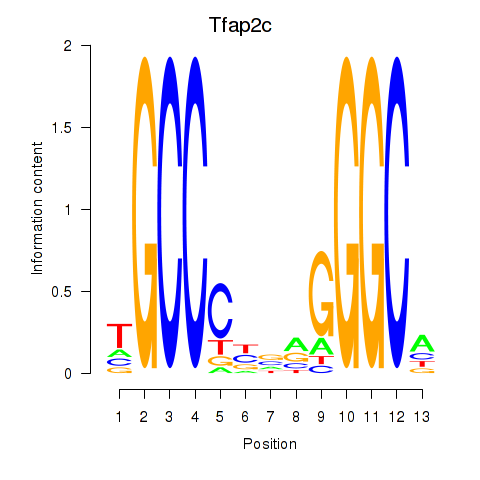
Transcription factors associated with Tfap2c
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tfap2c
|
ENSRNOG00000005246 | transcription factor AP-2 gamma |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tfap2c | rn6_v1_chr3_+_170550314_170550314 | 0.08 | 9.0e-01 | Click! |
Activity profile of Tfap2c motif
Sorted Z-values of Tfap2c motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_9483594 | 0.68 |
ENSRNOT00000089272
|
AABR07062799.2
|
|
| chr1_+_226435979 | 0.67 |
ENSRNOT00000048704
ENSRNOT00000036232 ENSRNOT00000035576 ENSRNOT00000036180 ENSRNOT00000036168 ENSRNOT00000047964 ENSRNOT00000036283 ENSRNOT00000007429 |
Syt7
|
synaptotagmin 7 |
| chr7_+_130532435 | 0.50 |
ENSRNOT00000092672
ENSRNOT00000092535 |
Shank3
|
SH3 and multiple ankyrin repeat domains 3 |
| chr12_-_30304036 | 0.39 |
ENSRNOT00000001218
|
Nupr2
|
nuclear protein 2, transcriptional regulator |
| chr6_-_99783047 | 0.38 |
ENSRNOT00000009028
|
Sptb
|
spectrin, beta, erythrocytic |
| chr4_+_100209951 | 0.31 |
ENSRNOT00000015807
|
LOC691113
|
hypothetical protein LOC691113 |
| chr16_-_71319449 | 0.27 |
ENSRNOT00000029284
|
Fgfr1
|
Fibroblast growth factor receptor 1 |
| chr10_+_61685645 | 0.26 |
ENSRNOT00000003933
|
Mnt
|
MAX network transcriptional repressor |
| chr12_-_41671437 | 0.23 |
ENSRNOT00000001883
|
Lhx5
|
LIM homeobox 5 |
| chr15_+_61692116 | 0.23 |
ENSRNOT00000071455
|
Kbtbd6
|
kelch repeat and BTB domain containing 6 |
| chr14_+_83724933 | 0.22 |
ENSRNOT00000029848
|
Pla2g3
|
phospholipase A2, group III |
| chr6_-_132183434 | 0.22 |
ENSRNOT00000079807
|
AABR07065498.1
|
|
| chr19_-_53688597 | 0.21 |
ENSRNOT00000074448
|
Zcchc14
|
zinc finger CCHC-type containing 14 |
| chr5_+_150080072 | 0.21 |
ENSRNOT00000047917
|
Tmem200b
|
transmembrane protein 200B |
| chr14_-_78707861 | 0.19 |
ENSRNOT00000091927
|
Ppp2r2c
|
protein phosphatase 2, regulatory subunit B, gamma |
| chr6_+_106052212 | 0.19 |
ENSRNOT00000010626
|
Sipa1l1
|
signal-induced proliferation-associated 1 like 1 |
| chr19_-_19832812 | 0.18 |
ENSRNOT00000084657
|
Papd5
|
poly(A) RNA polymerase D5, non-canonical |
| chr8_-_61919874 | 0.18 |
ENSRNOT00000064830
|
RGD1305464
|
similar to human chromosome 15 open reading frame 39 |
| chr19_-_15840990 | 0.17 |
ENSRNOT00000015583
|
Irx3
|
iroquois homeobox 3 |
| chr18_-_57245666 | 0.17 |
ENSRNOT00000080365
|
Ablim3
|
actin binding LIM protein family, member 3 |
| chr10_-_15311189 | 0.16 |
ENSRNOT00000027371
|
Prr35
|
proline rich 35 |
| chr1_+_100299626 | 0.15 |
ENSRNOT00000092327
ENSRNOT00000044257 |
Shank1
|
SH3 and multiple ankyrin repeat domains 1 |
| chr5_+_162031722 | 0.15 |
ENSRNOT00000020483
|
Lrrc38
|
leucine rich repeat containing 38 |
| chrX_+_15114892 | 0.15 |
ENSRNOT00000078168
|
Wdr13
|
WD repeat domain 13 |
| chr12_-_9331195 | 0.15 |
ENSRNOT00000044134
|
Pan3
|
PAN3 poly(A) specific ribonuclease subunit |
| chr3_+_171832500 | 0.15 |
ENSRNOT00000007554
|
Vapb
|
VAMP associated protein B and C |
| chr1_+_35280701 | 0.15 |
ENSRNOT00000061873
|
Ice1
|
interactor of little elongation complex ELL subunit 1 |
| chr1_+_171592797 | 0.14 |
ENSRNOT00000026607
|
Syt9
|
synaptotagmin 9 |
| chr4_-_82133605 | 0.14 |
ENSRNOT00000008023
|
NEWGENE_2813
|
homeobox A2 |
| chr5_-_160619650 | 0.14 |
ENSRNOT00000089981
|
Tmem51
|
transmembrane protein 51 |
| chr3_-_11382004 | 0.14 |
ENSRNOT00000047921
ENSRNOT00000064039 |
Dnm1
|
dynamin 1 |
| chr5_+_79055521 | 0.14 |
ENSRNOT00000010333
|
Col27a1
|
collagen type XXVII alpha 1 chain |
| chr10_-_51669297 | 0.13 |
ENSRNOT00000071595
|
Arhgap44
|
Rho GTPase activating protein 44 |
| chr8_-_68966108 | 0.13 |
ENSRNOT00000012155
|
Smad6
|
SMAD family member 6 |
| chr4_-_117268178 | 0.13 |
ENSRNOT00000043201
ENSRNOT00000084049 |
Fbxo41
|
F-box protein 41 |
| chr17_-_9721542 | 0.13 |
ENSRNOT00000047958
ENSRNOT00000079063 |
Grk6
|
G protein-coupled receptor kinase 6 |
| chr8_-_46750898 | 0.13 |
ENSRNOT00000059951
|
Tbcel
|
tubulin folding cofactor E-like |
| chr3_-_1924827 | 0.12 |
ENSRNOT00000006162
|
Cacna1b
|
calcium voltage-gated channel subunit alpha1 B |
| chr6_-_110904288 | 0.12 |
ENSRNOT00000014645
|
Irf2bpl
|
interferon regulatory factor 2 binding protein-like |
| chr5_-_65073012 | 0.12 |
ENSRNOT00000007957
|
Grin3a
|
glutamate ionotropic receptor NMDA type subunit 3A |
| chr4_-_82160240 | 0.12 |
ENSRNOT00000038775
ENSRNOT00000058985 |
Hoxa4
|
homeo box A4 |
| chr15_+_23665202 | 0.12 |
ENSRNOT00000089226
|
Samd4a
|
sterile alpha motif domain containing 4A |
| chr16_-_71319052 | 0.12 |
ENSRNOT00000050980
|
Fgfr1
|
Fibroblast growth factor receptor 1 |
| chr15_+_61731735 | 0.11 |
ENSRNOT00000015517
|
Kbtbd6
|
kelch repeat and BTB domain containing 6 |
| chr7_-_101140308 | 0.11 |
ENSRNOT00000006279
|
Fam84b
|
family with sequence similarity 84, member B |
| chr18_+_56887354 | 0.11 |
ENSRNOT00000044346
ENSRNOT00000066133 |
Csnk1a1
|
casein kinase 1, alpha 1 |
| chr5_-_86696388 | 0.11 |
ENSRNOT00000007812
|
Megf9
|
multiple EGF-like-domains 9 |
| chr9_+_82120059 | 0.10 |
ENSRNOT00000057368
|
Cdk5r2
|
cyclin-dependent kinase 5 regulatory subunit 2 |
| chr10_-_82197520 | 0.10 |
ENSRNOT00000092024
|
Cacna1g
|
calcium voltage-gated channel subunit alpha1 G |
| chr3_+_138504214 | 0.10 |
ENSRNOT00000091529
|
Kat14
|
lysine acetyltransferase 14 |
| chr10_-_82197848 | 0.10 |
ENSRNOT00000081307
|
Cacna1g
|
calcium voltage-gated channel subunit alpha1 G |
| chr4_+_7377563 | 0.10 |
ENSRNOT00000084826
|
Kcnh2
|
potassium voltage-gated channel subfamily H member 2 |
| chr3_-_11410732 | 0.10 |
ENSRNOT00000034930
|
RGD1561113
|
similar to Hypothetical UPF0184 protein C9orf16 homolog |
| chr9_-_10757954 | 0.09 |
ENSRNOT00000075265
|
Uhrf1
|
ubiquitin-like with PHD and ring finger domains 1 |
| chr3_-_147865393 | 0.09 |
ENSRNOT00000009852
|
Sox12
|
SRY box 12 |
| chr1_-_80515694 | 0.09 |
ENSRNOT00000075144
|
Clasrp
|
CLK4-associating serine/arginine rich protein |
| chr12_+_2180150 | 0.08 |
ENSRNOT00000001322
|
Stxbp2
|
syntaxin binding protein 2 |
| chr10_-_109224370 | 0.08 |
ENSRNOT00000005839
|
Aatk
|
apoptosis-associated tyrosine kinase |
| chr19_-_37938857 | 0.08 |
ENSRNOT00000026730
|
Slc12a4
|
solute carrier family 12 member 4 |
| chr18_+_56887722 | 0.08 |
ENSRNOT00000078783
|
Csnk1a1
|
casein kinase 1, alpha 1 |
| chr7_-_130405347 | 0.08 |
ENSRNOT00000013985
|
Cpt1b
|
carnitine palmitoyltransferase 1B |
| chr20_-_31597830 | 0.08 |
ENSRNOT00000085877
|
Col13a1
|
collagen type XIII alpha 1 chain |
| chr1_+_226252226 | 0.08 |
ENSRNOT00000039369
|
AABR07006258.1
|
|
| chr9_+_17163354 | 0.08 |
ENSRNOT00000026049
|
Polh
|
DNA polymerase eta |
| chr8_+_81766041 | 0.08 |
ENSRNOT00000032280
|
Onecut1
|
one cut homeobox 1 |
| chr8_+_116324040 | 0.07 |
ENSRNOT00000081353
|
Hyal2
|
hyaluronoglucosaminidase 2 |
| chr10_-_68517564 | 0.07 |
ENSRNOT00000086961
|
Asic2
|
acid sensing ion channel subunit 2 |
| chr1_+_220400855 | 0.07 |
ENSRNOT00000027178
|
Slc29a2
|
solute carrier family 29 member 2 |
| chr1_+_220353356 | 0.07 |
ENSRNOT00000080189
|
Cd248
|
CD248 molecule |
| chr5_+_143500441 | 0.07 |
ENSRNOT00000045513
|
Grik3
|
glutamate ionotropic receptor kainate type subunit 3 |
| chr18_+_40853988 | 0.07 |
ENSRNOT00000091759
|
AABR07031963.1
|
uncharacterized protein LOC317165 |
| chr6_+_126170720 | 0.06 |
ENSRNOT00000065246
|
Rin3
|
Ras and Rab interactor 3 |
| chr10_+_13145099 | 0.06 |
ENSRNOT00000091287
|
Srrm2
|
serine/arginine repetitive matrix 2 |
| chr9_-_10757720 | 0.06 |
ENSRNOT00000083848
|
Uhrf1
|
ubiquitin-like with PHD and ring finger domains 1 |
| chr1_+_220362064 | 0.06 |
ENSRNOT00000074361
|
LOC108348067
|
endosialin |
| chr6_+_134958854 | 0.05 |
ENSRNOT00000073950
ENSRNOT00000009377 |
Dync1h1
|
dynein cytoplasmic 1 heavy chain 1 |
| chr2_-_187863503 | 0.05 |
ENSRNOT00000093036
ENSRNOT00000026705 ENSRNOT00000082174 |
Lmna
|
lamin A/C |
| chr1_+_220307394 | 0.05 |
ENSRNOT00000071891
|
LOC108348052
|
equilibrative nucleoside transporter 2 |
| chr8_+_117068582 | 0.05 |
ENSRNOT00000073559
|
Amt
|
aminomethyltransferase |
| chr11_-_45510961 | 0.05 |
ENSRNOT00000002238
|
Tomm70
|
translocase of outer mitochondrial membrane 70 |
| chr6_-_105097054 | 0.05 |
ENSRNOT00000048606
|
Slc8a3
|
solute carrier family 8 member A3 |
| chr20_+_5815837 | 0.05 |
ENSRNOT00000036999
|
Lhfpl5
|
lipoma HMGIC fusion partner-like 5 |
| chr1_-_266934303 | 0.05 |
ENSRNOT00000036202
|
Calhm3
|
calcium homeostasis modulator 3 |
| chr6_-_3182996 | 0.05 |
ENSRNOT00000009359
|
Sos1
|
SOS Ras/Rac guanine nucleotide exchange factor 1 |
| chr10_+_57322331 | 0.05 |
ENSRNOT00000041493
|
Kif1c
|
kinesin family member 1C |
| chr3_+_121632043 | 0.05 |
ENSRNOT00000024882
|
Polr1b
|
RNA polymerase I subunit B |
| chr6_+_126170911 | 0.05 |
ENSRNOT00000077477
|
Rin3
|
Ras and Rab interactor 3 |
| chr1_-_265757389 | 0.05 |
ENSRNOT00000054703
|
C1H10orf76
|
chromosome 1 open reading frame, human C10orf76 |
| chr8_+_48094673 | 0.04 |
ENSRNOT00000008614
|
Nectin1
|
nectin cell adhesion molecule 1 |
| chr10_-_46145548 | 0.04 |
ENSRNOT00000033483
|
Pld6
|
phospholipase D family, member 6 |
| chr3_-_176744377 | 0.04 |
ENSRNOT00000017787
|
Helz2
|
helicase with zinc finger 2, transcriptional coactivator |
| chr8_-_56696968 | 0.04 |
ENSRNOT00000016637
|
Zc3h12c
|
zinc finger CCCH type containing 12C |
| chr16_+_60925093 | 0.04 |
ENSRNOT00000015813
|
Tnks
|
tankyrase |
| chr7_+_142397371 | 0.04 |
ENSRNOT00000040890
ENSRNOT00000065379 |
Slc4a8
|
solute carrier family 4 member 8 |
| chr1_+_154377447 | 0.04 |
ENSRNOT00000084268
ENSRNOT00000092086 ENSRNOT00000091470 ENSRNOT00000025415 |
Picalm
|
phosphatidylinositol binding clathrin assembly protein |
| chr14_+_80248140 | 0.04 |
ENSRNOT00000010852
|
Htra3
|
HtrA serine peptidase 3 |
| chr10_+_106812739 | 0.04 |
ENSRNOT00000074225
|
Syngr2
|
synaptogyrin 2 |
| chr11_-_71387992 | 0.03 |
ENSRNOT00000002411
|
Tnk2
|
tyrosine kinase, non-receptor, 2 |
| chr4_+_49369296 | 0.03 |
ENSRNOT00000007822
|
Wnt16
|
wingless-type MMTV integration site family, member 16 |
| chr16_+_7435634 | 0.03 |
ENSRNOT00000026323
|
Capn7
|
calpain 7 |
| chr4_-_157829241 | 0.03 |
ENSRNOT00000026184
|
Ltbr
|
lymphotoxin beta receptor |
| chr20_-_27308069 | 0.03 |
ENSRNOT00000056047
|
Slc25a16
|
solute carrier family 25 member 16 |
| chr6_+_137243185 | 0.03 |
ENSRNOT00000030879
|
Zbtb42
|
zinc finger and BTB domain containing 42 |
| chr1_-_222242644 | 0.03 |
ENSRNOT00000050891
|
Vegfb
|
vascular endothelial growth factor B |
| chr9_+_121802673 | 0.03 |
ENSRNOT00000086534
|
Yes1
|
YES proto-oncogene 1, Src family tyrosine kinase |
| chr10_+_86860685 | 0.03 |
ENSRNOT00000084644
|
Rara
|
retinoic acid receptor, alpha |
| chr1_-_101853055 | 0.03 |
ENSRNOT00000084869
|
Grin2d
|
glutamate ionotropic receptor NMDA type subunit 2D |
| chr14_-_84978255 | 0.03 |
ENSRNOT00000078179
|
Cabp7
|
calcium binding protein 7 |
| chr12_-_46789726 | 0.03 |
ENSRNOT00000001517
|
Gcn1l1
|
GCN1 eIF2 alpha kinase activator-like 1 |
| chr9_+_16862248 | 0.03 |
ENSRNOT00000080104
ENSRNOT00000024824 |
Ttbk1
|
tau tubulin kinase 1 |
| chr9_-_15700235 | 0.03 |
ENSRNOT00000088713
ENSRNOT00000035907 |
Trerf1
|
transcriptional regulating factor 1 |
| chr5_-_139227196 | 0.03 |
ENSRNOT00000050941
|
Foxo6
|
forkhead box O6 |
| chr4_-_56114254 | 0.03 |
ENSRNOT00000010673
|
Lrrc4
|
leucine rich repeat containing 4 |
| chr17_+_36334147 | 0.03 |
ENSRNOT00000050261
|
E2f3
|
E2F transcription factor 3 |
| chr19_-_57699113 | 0.03 |
ENSRNOT00000026767
|
Egln1
|
egl-9 family hypoxia-inducible factor 1 |
| chr19_-_19832459 | 0.03 |
ENSRNOT00000067582
|
Papd5
|
poly(A) RNA polymerase D5, non-canonical |
| chr1_+_226091774 | 0.02 |
ENSRNOT00000027693
|
Fads3
|
fatty acid desaturase 3 |
| chr9_+_17340341 | 0.02 |
ENSRNOT00000026637
ENSRNOT00000026559 ENSRNOT00000042790 ENSRNOT00000044163 ENSRNOT00000083811 |
Vegfa
|
vascular endothelial growth factor A |
| chr15_-_38165736 | 0.02 |
ENSRNOT00000063962
ENSRNOT00000014820 |
Zdhhc20
|
zinc finger, DHHC-type containing 20 |
| chr1_+_199287710 | 0.02 |
ENSRNOT00000082121
|
Stx4
|
syntaxin 4 |
| chr8_-_107681512 | 0.02 |
ENSRNOT00000077963
|
Mras
|
muscle RAS oncogene homolog |
| chr7_-_29070928 | 0.02 |
ENSRNOT00000080939
|
Chpt1
|
choline phosphotransferase 1 |
| chr11_+_28692708 | 0.02 |
ENSRNOT00000002135
|
Krtap13-1
|
keratin associated protein 13-1 |
| chr7_+_116745061 | 0.02 |
ENSRNOT00000076174
|
Rhpn1
|
rhophilin, Rho GTPase binding protein 1 |
| chr3_+_172385672 | 0.02 |
ENSRNOT00000090989
|
Gnas
|
GNAS complex locus |
| chr1_-_146029840 | 0.02 |
ENSRNOT00000016455
|
Mesdc1
|
mesoderm development candidate 1 |
| chr11_-_83546674 | 0.02 |
ENSRNOT00000044896
|
Ephb3
|
Eph receptor B3 |
| chr10_+_56824505 | 0.02 |
ENSRNOT00000067128
|
Slc16a11
|
solute carrier family 16, member 11 |
| chr20_+_4852671 | 0.02 |
ENSRNOT00000001111
|
Lta
|
lymphotoxin alpha |
| chr4_+_55772377 | 0.02 |
ENSRNOT00000047698
|
Snd1
|
staphylococcal nuclease and tudor domain containing 1 |
| chr12_-_9607168 | 0.01 |
ENSRNOT00000001260
|
Gsx1
|
GS homeobox 1 |
| chr4_+_162937774 | 0.01 |
ENSRNOT00000083188
|
Clec2g
|
C-type lectin domain family 2, member G |
| chr2_-_260884449 | 0.01 |
ENSRNOT00000032874
|
Tyw3
|
tRNA-yW synthesizing protein 3 homolog |
| chr1_-_18058055 | 0.01 |
ENSRNOT00000020988
|
Ptprk
|
protein tyrosine phosphatase, receptor type, K |
| chr7_-_117259791 | 0.01 |
ENSRNOT00000086550
|
Plec
|
plectin |
| chrX_-_32355296 | 0.01 |
ENSRNOT00000081652
ENSRNOT00000065075 |
Ap1s2
|
adaptor-related protein complex 1, sigma 2 subunit |
| chr10_-_83888823 | 0.01 |
ENSRNOT00000055500
|
Ube2z
|
ubiquitin-conjugating enzyme E2Z |
| chr4_+_140886545 | 0.01 |
ENSRNOT00000088273
|
Edem1
|
ER degradation enhancing alpha-mannosidase like protein 1 |
| chr16_-_20686317 | 0.01 |
ENSRNOT00000060097
|
Crlf1
|
cytokine receptor-like factor 1 |
| chr10_+_4578469 | 0.01 |
ENSRNOT00000003332
|
Txndc11
|
thioredoxin domain containing 11 |
| chr7_+_114724610 | 0.01 |
ENSRNOT00000014541
|
Dennd3
|
DENN domain containing 3 |
| chr1_+_221043119 | 0.01 |
ENSRNOT00000028185
|
Map3k11
|
mitogen-activated protein kinase kinase kinase 11 |
| chr9_+_88607508 | 0.01 |
ENSRNOT00000021309
|
Agfg1
|
ArfGAP with FG repeats 1 |
| chr10_+_14598014 | 0.01 |
ENSRNOT00000068336
|
Tsr3
|
TSR3, 20S rRNA accumulation |
| chr7_-_117288018 | 0.01 |
ENSRNOT00000091285
|
Plec
|
plectin |
| chr5_+_151741817 | 0.01 |
ENSRNOT00000081318
|
Kdf1
|
keratinocyte differentiation factor 1 |
| chr5_-_124642569 | 0.01 |
ENSRNOT00000010680
|
Prkaa2
|
protein kinase AMP-activated catalytic subunit alpha 2 |
| chr3_+_123434409 | 0.01 |
ENSRNOT00000028847
|
Atrn
|
attractin |
| chr1_-_84491466 | 0.01 |
ENSRNOT00000034609
|
Map3k10
|
mitogen activated protein kinase kinase kinase 10 |
| chr20_-_31598118 | 0.01 |
ENSRNOT00000046537
|
Col13a1
|
collagen type XIII alpha 1 chain |
| chr1_+_165170645 | 0.01 |
ENSRNOT00000022879
|
Lipt2
|
lipoyl(octanoyl) transferase 2 (putative) |
| chr5_+_151692108 | 0.01 |
ENSRNOT00000086144
|
Fam46b
|
family with sequence similarity 46, member B |
| chr7_+_28956512 | 0.01 |
ENSRNOT00000006946
|
Gnptab
|
N-acetylglucosamine-1-phosphate transferase, alpha and beta subunits |
| chr4_+_157659147 | 0.01 |
ENSRNOT00000048379
|
Iffo1
|
intermediate filament family orphan 1 |
| chr4_+_57050214 | 0.01 |
ENSRNOT00000025165
|
Ahcyl2
|
adenosylhomocysteinase-like 2 |
| chr15_-_51834030 | 0.01 |
ENSRNOT00000024895
|
Ccar2
|
cell cycle and apoptosis regulator 2 |
| chr8_-_63350269 | 0.00 |
ENSRNOT00000042681
|
Cd276
|
Cd276 molecule |
| chr17_+_5382034 | 0.00 |
ENSRNOT00000037597
|
Golm1
|
golgi membrane protein 1 |
| chr10_+_61685241 | 0.00 |
ENSRNOT00000092606
|
Mnt
|
MAX network transcriptional repressor |
| chr5_-_135285311 | 0.00 |
ENSRNOT00000088261
ENSRNOT00000085235 |
Mast2
|
microtubule associated serine/threonine kinase 2 |
| chr6_-_107080524 | 0.00 |
ENSRNOT00000011662
|
Zfyve1
|
zinc finger FYVE-type containing 1 |
| chr5_+_169519212 | 0.00 |
ENSRNOT00000024732
|
Chd5
|
chromodomain helicase DNA binding protein 5 |
| chr1_+_220446425 | 0.00 |
ENSRNOT00000027339
|
LOC100911932
|
endosialin-like |
| chr1_+_81779380 | 0.00 |
ENSRNOT00000065865
ENSRNOT00000080143 ENSRNOT00000089592 ENSRNOT00000080840 |
Arhgef1
|
Rho guanine nucleotide exchange factor 1 |
| chr10_+_65780494 | 0.00 |
ENSRNOT00000013100
|
Poldip2
|
DNA polymerase delta interacting protein 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Tfap2c
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:1990926 | short-term synaptic potentiation(GO:1990926) |
| 0.1 | 0.4 | GO:0048372 | ventricular zone neuroblast division(GO:0021847) fibroblast growth factor receptor signaling pathway involved in orbitofrontal cortex development(GO:0035607) lateral mesodermal cell fate commitment(GO:0048372) lateral mesodermal cell fate specification(GO:0048377) regulation of lateral mesodermal cell fate specification(GO:0048378) |
| 0.1 | 0.5 | GO:1904717 | regulation of AMPA glutamate receptor clustering(GO:1904717) |
| 0.0 | 0.2 | GO:0072086 | specification of loop of Henle identity(GO:0072086) |
| 0.0 | 0.2 | GO:0050893 | sensory processing(GO:0050893) |
| 0.0 | 0.1 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 0.0 | 0.1 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.0 | 0.2 | GO:0010045 | response to nickel cation(GO:0010045) |
| 0.0 | 0.2 | GO:0021937 | cerebellar Purkinje cell-granule cell precursor cell signaling involved in regulation of granule cell precursor cell proliferation(GO:0021937) |
| 0.0 | 0.1 | GO:1902303 | negative regulation of potassium ion export(GO:1902303) |
| 0.0 | 0.2 | GO:0046725 | negative regulation by virus of viral protein levels in host cell(GO:0046725) |
| 0.0 | 0.1 | GO:0003431 | growth plate cartilage chondrocyte development(GO:0003431) |
| 0.0 | 0.1 | GO:0021586 | pons maturation(GO:0021586) |
| 0.0 | 0.1 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.0 | 0.1 | GO:0015864 | pyrimidine nucleoside transport(GO:0015864) |
| 0.0 | 0.2 | GO:0019372 | lipoxygenase pathway(GO:0019372) |
| 0.0 | 0.1 | GO:0010360 | negative regulation of anion channel activity(GO:0010360) |
| 0.0 | 0.2 | GO:0010216 | maintenance of DNA methylation(GO:0010216) |
| 0.0 | 0.1 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 0.0 | 0.2 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 | 0.1 | GO:1904178 | sterol regulatory element binding protein import into nucleus(GO:0035105) negative regulation of adipose tissue development(GO:1904178) |
| 0.0 | 0.2 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.0 | 0.1 | GO:0039663 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 0.0 | 0.0 | GO:0008358 | oocyte construction(GO:0007308) oocyte axis specification(GO:0007309) oocyte anterior/posterior axis specification(GO:0007314) pole plasm assembly(GO:0007315) maternal determination of anterior/posterior axis, embryo(GO:0008358) P granule organization(GO:0030719) |
| 0.0 | 0.0 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) regulation of telomeric DNA binding(GO:1904742) |
| 0.0 | 0.0 | GO:1902962 | regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902959) positive regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902961) regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) regulation of aspartic-type peptidase activity(GO:1905245) positive regulation of aspartic-type peptidase activity(GO:1905247) |
| 0.0 | 0.0 | GO:0033122 | negative regulation of purine nucleotide catabolic process(GO:0033122) |
| 0.0 | 0.3 | GO:0071157 | negative regulation of cell cycle arrest(GO:0071157) |
| 0.0 | 0.4 | GO:0006779 | porphyrin-containing compound biosynthetic process(GO:0006779) |
| 0.0 | 0.0 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.0 | 0.1 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.0 | 0.1 | GO:0071494 | cellular response to UV-C(GO:0071494) |
| 0.0 | 0.2 | GO:0071044 | histone mRNA catabolic process(GO:0071044) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0032009 | early phagosome(GO:0032009) |
| 0.0 | 0.4 | GO:0008091 | spectrin(GO:0008091) spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.1 | GO:0035363 | histone locus body(GO:0035363) |
| 0.0 | 0.1 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.0 | 0.2 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.3 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.1 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.1 | GO:0044194 | cytolytic granule(GO:0044194) tertiary granule(GO:0070820) |
| 0.0 | 0.1 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.0 | GO:0005760 | gamma DNA polymerase complex(GO:0005760) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 0.4 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.2 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.0 | 0.2 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 0.0 | 0.2 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.0 | 0.1 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 0.0 | 0.1 | GO:0031749 | D2 dopamine receptor binding(GO:0031749) |
| 0.0 | 0.1 | GO:0070697 | activin receptor binding(GO:0070697) |
| 0.0 | 0.1 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.0 | 0.1 | GO:0016416 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.0 | 0.1 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.0 | 0.1 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 0.8 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.1 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.0 | 0.1 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 0.4 | GO:0030506 | ankyrin binding(GO:0030506) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.0 | 0.1 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.2 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 0.2 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.4 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |