Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
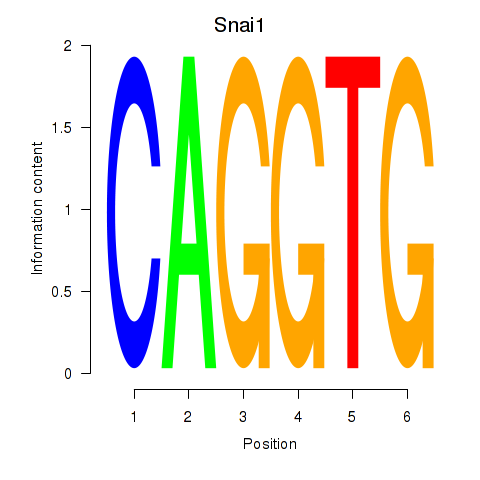
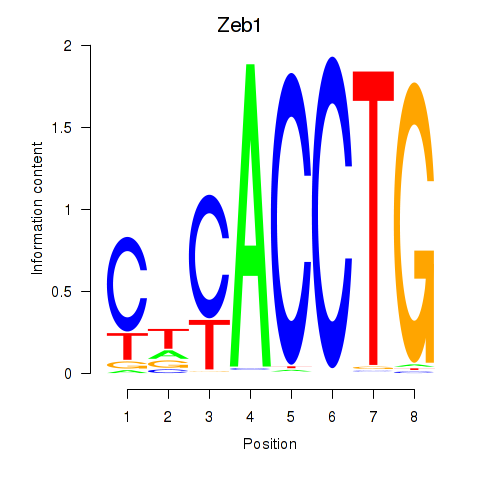
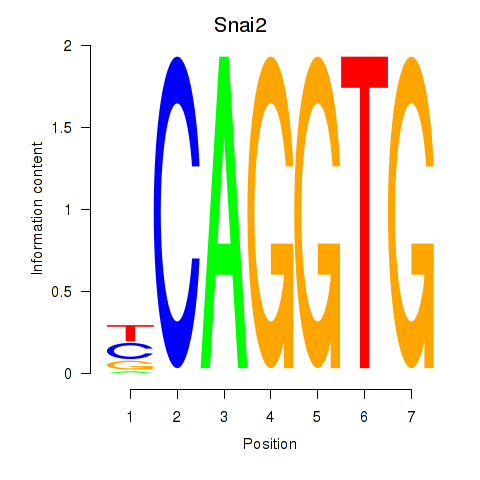
Results for Snai1_Zeb1_Snai2
Z-value: 0.67
Transcription factors associated with Snai1_Zeb1_Snai2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Snai1
|
ENSRNOG00000009594 | snail family transcriptional repressor 1 |
|
Zeb1
|
ENSRNOG00000017863 | zinc finger E-box binding homeobox 1 |
|
Snai2
|
ENSRNOG00000047699 | snail family transcriptional repressor 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Snai1 | rn6_v1_chr3_+_164274710_164274710 | 0.86 | 6.4e-02 | Click! |
| Zeb1 | rn6_v1_chr17_-_54714914_54714914 | 0.74 | 1.5e-01 | Click! |
| Snai2 | rn6_v1_chr11_-_90406797_90406797 | 0.62 | 2.7e-01 | Click! |
Activity profile of Snai1_Zeb1_Snai2 motif
Sorted Z-values of Snai1_Zeb1_Snai2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_9037942 | 1.41 |
ENSRNOT00000036770
|
Ier5l
|
immediate early response 5-like |
| chr8_+_91070073 | 0.56 |
ENSRNOT00000012904
|
Sh3bgrl2
|
SH3 domain binding glutamate-rich protein like 2 |
| chr3_-_123119460 | 0.55 |
ENSRNOT00000028833
|
Avp
|
arginine vasopressin |
| chr4_+_146106386 | 0.50 |
ENSRNOT00000008342
|
Slc6a11
|
solute carrier family 6 member 11 |
| chr18_-_57245666 | 0.40 |
ENSRNOT00000080365
|
Ablim3
|
actin binding LIM protein family, member 3 |
| chr12_+_18074033 | 0.38 |
ENSRNOT00000001727
|
LOC103689975
|
integrator complex subunit 1-like |
| chrX_-_111942749 | 0.38 |
ENSRNOT00000087583
|
AABR07040840.1
|
|
| chr20_-_4879779 | 0.35 |
ENSRNOT00000081924
|
Hspa1b
|
heat shock protein family A (Hsp70) member 1B |
| chr12_-_2174131 | 0.33 |
ENSRNOT00000001313
|
Pcp2
|
Purkinje cell protein 2 |
| chr20_-_2701815 | 0.33 |
ENSRNOT00000061950
|
Hspa1a
|
heat shock protein family A (Hsp70) member 1A |
| chr6_+_137824213 | 0.33 |
ENSRNOT00000056880
|
Pacs2
|
phosphofurin acidic cluster sorting protein 2 |
| chr14_+_16491573 | 0.32 |
ENSRNOT00000002995
|
Sowahb
|
sosondowah ankyrin repeat domain family member B |
| chr18_-_3662654 | 0.32 |
ENSRNOT00000092854
ENSRNOT00000016167 |
Npc1
|
NPC intracellular cholesterol transporter 1 |
| chr20_-_2701637 | 0.31 |
ENSRNOT00000049667
|
Hspa1a
|
heat shock protein family A (Hsp70) member 1A |
| chr1_+_266953139 | 0.30 |
ENSRNOT00000054696
|
Neurl1
|
neuralized E3 ubiquitin protein ligase 1 |
| chr4_-_68597586 | 0.30 |
ENSRNOT00000015921
|
RGD1563986
|
similar to RIKEN cDNA E330009J07 gene |
| chr5_+_139963002 | 0.29 |
ENSRNOT00000048506
|
Col9a2
|
collagen type IX alpha 2 chain |
| chr6_-_99783047 | 0.29 |
ENSRNOT00000009028
|
Sptb
|
spectrin, beta, erythrocytic |
| chr7_-_125465848 | 0.27 |
ENSRNOT00000039588
|
Ldoc1l
|
leucine zipper, down-regulated in cancer 1-like |
| chr5_-_153790157 | 0.27 |
ENSRNOT00000025051
|
Rcan3
|
RCAN family member 3 |
| chr18_-_55891710 | 0.27 |
ENSRNOT00000064686
|
Synpo
|
synaptopodin |
| chr18_-_24735349 | 0.25 |
ENSRNOT00000036537
|
Gpr17
|
G protein-coupled receptor 17 |
| chr16_+_20740826 | 0.25 |
ENSRNOT00000038057
|
Crtc1
|
CREB regulated transcription coactivator 1 |
| chr3_-_152382529 | 0.24 |
ENSRNOT00000027076
|
Scand1
|
SCAN domain-containing 1 |
| chr6_+_104718512 | 0.24 |
ENSRNOT00000007947
|
Smoc1
|
SPARC related modular calcium binding 1 |
| chr2_-_187742747 | 0.23 |
ENSRNOT00000026530
|
Bglap
|
bone gamma-carboxyglutamate protein |
| chr19_+_26106838 | 0.23 |
ENSRNOT00000035987
|
Hook2
|
hook microtubule-tethering protein 2 |
| chrX_-_39956841 | 0.23 |
ENSRNOT00000050708
|
Klhl34
|
kelch-like family member 34 |
| chr17_-_84488480 | 0.22 |
ENSRNOT00000000158
ENSRNOT00000075983 |
Nebl
|
nebulette |
| chr10_+_90731865 | 0.22 |
ENSRNOT00000064429
|
Adam11
|
ADAM metallopeptidase domain 11 |
| chr3_-_148932878 | 0.22 |
ENSRNOT00000013881
|
Nol4l
|
nucleolar protein 4-like |
| chr4_+_87026530 | 0.22 |
ENSRNOT00000018425
|
Avl9
|
AVL9 cell migration associated |
| chr7_+_130532435 | 0.21 |
ENSRNOT00000092672
ENSRNOT00000092535 |
Shank3
|
SH3 and multiple ankyrin repeat domains 3 |
| chr7_+_11737293 | 0.21 |
ENSRNOT00000046059
|
Lingo3
|
leucine rich repeat and Ig domain containing 3 |
| chr2_-_188718704 | 0.21 |
ENSRNOT00000028010
|
Zbtb7b
|
zinc finger and BTB domain containing 7B |
| chr14_-_82055290 | 0.20 |
ENSRNOT00000058062
|
RGD1560394
|
RGD1560394 |
| chr20_-_14620019 | 0.20 |
ENSRNOT00000001779
|
Gnaz
|
G protein subunit alpha z |
| chr4_-_117153907 | 0.20 |
ENSRNOT00000091374
|
Rab11fip5
|
RAB11 family interacting protein 5 |
| chr9_-_80166807 | 0.19 |
ENSRNOT00000079493
|
Igfbp5
|
insulin-like growth factor binding protein 5 |
| chr15_+_39638510 | 0.19 |
ENSRNOT00000037800
|
Rcbtb1
|
RCC1 and BTB domain containing protein 1 |
| chr1_+_220826560 | 0.19 |
ENSRNOT00000027891
|
Fosl1
|
FOS like 1, AP-1 transcription factor subunit |
| chr1_-_222177421 | 0.19 |
ENSRNOT00000078393
|
Esrra
|
estrogen related receptor, alpha |
| chr7_+_11152038 | 0.19 |
ENSRNOT00000006168
|
Nfic
|
nuclear factor I/C |
| chr1_-_31890778 | 0.19 |
ENSRNOT00000039166
|
Tppp
|
tubulin polymerization promoting protein |
| chr12_-_13668515 | 0.19 |
ENSRNOT00000086847
|
Fscn1
|
fascin actin-bundling protein 1 |
| chr1_-_89399039 | 0.18 |
ENSRNOT00000028585
ENSRNOT00000044678 |
Lsr
|
lipolysis stimulated lipoprotein receptor |
| chr1_+_211351350 | 0.18 |
ENSRNOT00000082751
ENSRNOT00000030461 |
Jakmip3
|
janus kinase and microtubule interacting protein 3 |
| chr12_+_22434814 | 0.18 |
ENSRNOT00000076829
|
Slc12a9
|
solute carrier family 12, member 9 |
| chrX_-_75291938 | 0.18 |
ENSRNOT00000003739
ENSRNOT00000083655 |
Abcb7
|
ATP binding cassette subfamily B member 7 |
| chr10_+_86399827 | 0.18 |
ENSRNOT00000009299
|
Grb7
|
growth factor receptor bound protein 7 |
| chr19_+_25123724 | 0.18 |
ENSRNOT00000007407
|
LOC686013
|
hypothetical protein LOC686013 |
| chr3_-_72289310 | 0.18 |
ENSRNOT00000038250
|
Rtn4rl2
|
reticulon 4 receptor-like 2 |
| chr1_-_67390141 | 0.18 |
ENSRNOT00000025808
|
Sbk1
|
SH3 domain binding kinase 1 |
| chr12_-_38691075 | 0.18 |
ENSRNOT00000084192
|
Bcl7a
|
BCL tumor suppressor 7A |
| chr12_-_48857724 | 0.18 |
ENSRNOT00000080783
|
Wscd2
|
WSC domain containing 2 |
| chr5_-_150484207 | 0.17 |
ENSRNOT00000090736
|
Rab42
|
RAB42, member RAS oncogene family |
| chr8_+_127144903 | 0.17 |
ENSRNOT00000013530
|
Eomes
|
eomesodermin |
| chrX_+_116399611 | 0.17 |
ENSRNOT00000050854
|
Zcchc16
|
zinc finger CCHC-type containing 16 |
| chr18_+_56193978 | 0.17 |
ENSRNOT00000041533
ENSRNOT00000080177 |
Camk2a
|
calcium/calmodulin-dependent protein kinase II alpha |
| chr10_+_103874383 | 0.17 |
ENSRNOT00000038935
|
Otop2
|
otopetrin 2 |
| chr12_-_16934706 | 0.17 |
ENSRNOT00000001720
|
Mafk
|
MAF bZIP transcription factor K |
| chr11_+_16826399 | 0.17 |
ENSRNOT00000050701
|
Cxadr
|
coxsackie virus and adenovirus receptor |
| chr1_-_124803363 | 0.17 |
ENSRNOT00000066380
|
Klf13
|
Kruppel-like factor 13 |
| chr9_+_53627208 | 0.17 |
ENSRNOT00000083487
|
Mfsd6
|
major facilitator superfamily domain containing 6 |
| chr10_-_38838272 | 0.17 |
ENSRNOT00000089495
|
Sowaha
|
sosondowah ankyrin repeat domain family member A |
| chr10_-_56008885 | 0.17 |
ENSRNOT00000067677
|
Kdm6b
|
lysine demethylase 6B |
| chr6_+_26399950 | 0.17 |
ENSRNOT00000085424
|
Ift172
|
intraflagellar transport 172 |
| chr3_+_152571121 | 0.17 |
ENSRNOT00000087289
ENSRNOT00000080543 ENSRNOT00000083476 |
Epb41l1
|
erythrocyte membrane protein band 4.1-like 1 |
| chr3_+_95715193 | 0.16 |
ENSRNOT00000089525
|
Pax6
|
paired box 6 |
| chr5_-_172623899 | 0.16 |
ENSRNOT00000080591
|
Ski
|
SKI proto-oncogene |
| chr7_+_12974169 | 0.16 |
ENSRNOT00000010555
|
C2cd4c
|
C2 calcium-dependent domain containing 4C |
| chr8_-_71728654 | 0.16 |
ENSRNOT00000031207
ENSRNOT00000091751 |
Snx22
|
sorting nexin 22 |
| chr14_+_4362717 | 0.16 |
ENSRNOT00000002887
|
Barhl2
|
BarH-like homeobox 2 |
| chr20_+_5067330 | 0.16 |
ENSRNOT00000037482
|
Ly6g6e
|
lymphocyte antigen 6 complex, locus G6E |
| chr18_-_55916220 | 0.16 |
ENSRNOT00000025934
|
Synpo
|
synaptopodin |
| chr17_-_9695292 | 0.16 |
ENSRNOT00000036162
|
Prr7
|
proline rich 7 (synaptic) |
| chr13_-_51930972 | 0.16 |
ENSRNOT00000006969
|
Lgr6
|
leucine-rich repeat-containing G protein-coupled receptor 6 |
| chr9_+_73687743 | 0.16 |
ENSRNOT00000079495
ENSRNOT00000017197 |
Rpe
|
ribulose-5-phosphate-3-epimerase |
| chr1_+_199449973 | 0.16 |
ENSRNOT00000029994
|
Trim72
|
tripartite motif containing 72 |
| chr1_+_220428481 | 0.16 |
ENSRNOT00000027335
|
LOC108348044
|
ras and Rab interactor 1 |
| chr12_+_47551935 | 0.15 |
ENSRNOT00000056932
|
RGD1560398
|
RGD1560398 |
| chr11_-_83926524 | 0.15 |
ENSRNOT00000041777
ENSRNOT00000040029 |
Eif4g1
|
eukaryotic translation initiation factor 4 gamma, 1 |
| chr16_-_81945127 | 0.15 |
ENSRNOT00000023352
|
Mcf2l
|
MCF.2 cell line derived transforming sequence-like |
| chr9_+_82120059 | 0.15 |
ENSRNOT00000057368
|
Cdk5r2
|
cyclin-dependent kinase 5 regulatory subunit 2 |
| chr7_-_71048383 | 0.15 |
ENSRNOT00000005693
|
Gpr182
|
G protein-coupled receptor 182 |
| chr12_+_24978483 | 0.15 |
ENSRNOT00000040069
|
Eln
|
elastin |
| chr10_-_51669297 | 0.15 |
ENSRNOT00000071595
|
Arhgap44
|
Rho GTPase activating protein 44 |
| chr16_+_71058022 | 0.15 |
ENSRNOT00000066901
|
Bag4
|
BCL2-associated athanogene 4 |
| chr8_+_130749838 | 0.15 |
ENSRNOT00000079273
|
Snrk
|
SNF related kinase |
| chr2_-_188559882 | 0.15 |
ENSRNOT00000088199
|
Trim46
|
tripartite motif-containing 46 |
| chr7_-_11018160 | 0.15 |
ENSRNOT00000092061
|
Aes
|
amino-terminal enhancer of split |
| chr8_+_63600663 | 0.15 |
ENSRNOT00000012644
|
Hcn4
|
hyperpolarization activated cyclic nucleotide-gated potassium channel 4 |
| chr4_-_78075398 | 0.15 |
ENSRNOT00000010377
|
Zfp467
|
zinc finger protein 467 |
| chr12_-_47990105 | 0.15 |
ENSRNOT00000080049
|
Ube3b
|
ubiquitin protein ligase E3B |
| chr2_+_195719543 | 0.15 |
ENSRNOT00000028324
|
Celf3
|
CUGBP, Elav-like family member 3 |
| chr4_-_108717309 | 0.15 |
ENSRNOT00000085062
|
AABR07061178.1
|
|
| chr2_+_174542667 | 0.15 |
ENSRNOT00000076793
|
Fstl5
|
follistatin-like 5 |
| chr12_-_51341663 | 0.15 |
ENSRNOT00000066892
ENSRNOT00000052202 |
Pitpnb
|
phosphatidylinositol transfer protein, beta |
| chr14_-_80169431 | 0.15 |
ENSRNOT00000079769
ENSRNOT00000058315 |
Ablim2
|
actin binding LIM protein family, member 2 |
| chr19_+_41482728 | 0.14 |
ENSRNOT00000022943
|
Calb2
|
calbindin 2 |
| chr12_-_2592838 | 0.14 |
ENSRNOT00000079918
|
Evi5l
|
ecotropic viral integration site 5-like |
| chr11_+_61531416 | 0.14 |
ENSRNOT00000093263
|
Atp6v1a
|
ATPase H+ transporting V1 subunit A |
| chr11_+_71151132 | 0.14 |
ENSRNOT00000082594
ENSRNOT00000082435 |
Rubcn
|
RUN and cysteine rich domain containing beclin 1 interacting protein |
| chr1_-_52992213 | 0.14 |
ENSRNOT00000033528
|
Prr18
|
proline rich 18 |
| chr1_-_260992291 | 0.14 |
ENSRNOT00000035415
ENSRNOT00000034758 |
Slit1
|
slit guidance ligand 1 |
| chr4_-_117296082 | 0.14 |
ENSRNOT00000021097
|
Egr4
|
early growth response 4 |
| chr3_-_6626284 | 0.14 |
ENSRNOT00000012494
|
Fcnb
|
ficolin B |
| chr16_-_69132584 | 0.14 |
ENSRNOT00000017776
|
Adgra2
|
adhesion G protein-coupled receptor A2 |
| chr20_+_3830376 | 0.14 |
ENSRNOT00000085547
|
Col11a2
|
collagen type XI alpha 2 chain |
| chr12_+_41385241 | 0.14 |
ENSRNOT00000074974
|
Dtx1
|
deltex E3 ubiquitin ligase 1 |
| chr7_+_23960378 | 0.14 |
ENSRNOT00000006886
|
Prdm4
|
PR/SET domain 4 |
| chr3_+_11198401 | 0.14 |
ENSRNOT00000088576
ENSRNOT00000066368 |
Prrc2b
|
proline-rich coiled-coil 2B |
| chr3_+_11207542 | 0.14 |
ENSRNOT00000013546
|
Prrc2b
|
proline-rich coiled-coil 2B |
| chr17_-_89163113 | 0.13 |
ENSRNOT00000050445
|
LOC100360843
|
ribosomal protein S19-like |
| chr16_-_20888907 | 0.13 |
ENSRNOT00000027428
|
Homer3
|
homer scaffolding protein 3 |
| chr3_+_14889510 | 0.13 |
ENSRNOT00000080760
|
Dab2ip
|
DAB2 interacting protein |
| chr10_+_11912543 | 0.13 |
ENSRNOT00000045192
|
Zfp597
|
zinc finger protein 597 |
| chr14_-_9456990 | 0.13 |
ENSRNOT00000002918
|
Cds1
|
CDP-diacylglycerol synthase 1 |
| chr3_-_164095878 | 0.13 |
ENSRNOT00000079414
|
B4galt5
|
beta-1,4-galactosyltransferase 5 |
| chr7_-_70552897 | 0.13 |
ENSRNOT00000080594
|
Kif5a
|
kinesin family member 5A |
| chr4_-_115516296 | 0.13 |
ENSRNOT00000019399
|
Paip2b
|
poly(A) binding protein interacting protein 2B |
| chr20_-_4823475 | 0.13 |
ENSRNOT00000082536
ENSRNOT00000001114 |
Atp6v1g2
|
ATPase H+ transporting V1 subunit G2 |
| chr5_+_103479767 | 0.13 |
ENSRNOT00000008999
|
Sh3gl2
|
SH3 domain-containing GRB2-like 2 |
| chr10_+_54126486 | 0.13 |
ENSRNOT00000078820
|
Gas7
|
growth arrest specific 7 |
| chr6_-_26445787 | 0.13 |
ENSRNOT00000084242
|
LOC103692570
|
dihydropyrimidinase-related protein 5-like |
| chr12_-_40227297 | 0.13 |
ENSRNOT00000030600
|
Fam109a
|
family with sequence similarity 109, member A |
| chr20_+_3827367 | 0.13 |
ENSRNOT00000079967
|
Rxrb
|
retinoid X receptor beta |
| chr7_+_80713321 | 0.13 |
ENSRNOT00000091355
ENSRNOT00000078354 |
Oxr1
|
oxidation resistance 1 |
| chr1_-_100669684 | 0.13 |
ENSRNOT00000091760
|
Myh14
|
myosin heavy chain 14 |
| chr19_-_25288335 | 0.12 |
ENSRNOT00000050214
|
Nanos3
|
nanos C2HC-type zinc finger 3 |
| chr2_+_157316266 | 0.12 |
ENSRNOT00000015387
|
Tiparp
|
TCDD-inducible poly(ADP-ribose) polymerase |
| chr10_+_56381813 | 0.12 |
ENSRNOT00000019687
|
Zbtb4
|
zinc finger and BTB domain containing 4 |
| chr6_+_26390689 | 0.12 |
ENSRNOT00000079762
ENSRNOT00000091528 |
Ift172
|
intraflagellar transport 172 |
| chr10_-_20818128 | 0.12 |
ENSRNOT00000011061
|
Wwc1
|
WW and C2 domain containing 1 |
| chr12_+_52359310 | 0.12 |
ENSRNOT00000071857
ENSRNOT00000065893 |
Fbrsl1
|
fibrosin-like 1 |
| chr3_+_62648447 | 0.12 |
ENSRNOT00000002111
|
Agps
|
alkylglycerone phosphate synthase |
| chr7_+_11215328 | 0.12 |
ENSRNOT00000061194
|
LOC690617
|
hypothetical protein LOC690617 |
| chr6_+_132219088 | 0.12 |
ENSRNOT00000034883
|
Hhipl1
|
HHIP-like 1 |
| chr15_+_37806836 | 0.12 |
ENSRNOT00000076285
|
Il17d
|
interleukin 17D |
| chr3_-_58181971 | 0.12 |
ENSRNOT00000002076
|
Dlx2
|
distal-less homeobox 2 |
| chr1_+_267607416 | 0.12 |
ENSRNOT00000087894
ENSRNOT00000076367 ENSRNOT00000016851 |
Gsto2
Gsto1
|
glutathione S-transferase omega 2 glutathione S-transferase omega 1 |
| chr6_-_147172022 | 0.12 |
ENSRNOT00000080675
|
Itgb8
|
integrin subunit beta 8 |
| chr1_+_100299626 | 0.12 |
ENSRNOT00000092327
ENSRNOT00000044257 |
Shank1
|
SH3 and multiple ankyrin repeat domains 1 |
| chr3_+_103773459 | 0.12 |
ENSRNOT00000079727
|
Slc12a6
|
solute carrier family 12, member 6 |
| chr20_-_3440769 | 0.12 |
ENSRNOT00000084981
|
Ier3
|
immediate early response 3 |
| chr2_-_186245163 | 0.12 |
ENSRNOT00000089339
|
Dclk2
|
doublecortin-like kinase 2 |
| chr5_-_155772040 | 0.12 |
ENSRNOT00000036788
|
Cela3b
|
chymotrypsin-like elastase family, member 3B |
| chr4_+_157538303 | 0.12 |
ENSRNOT00000086418
|
Zfp384
|
zinc finger protein 384 |
| chr7_-_107634287 | 0.12 |
ENSRNOT00000093672
ENSRNOT00000087116 |
Sla
|
src-like adaptor |
| chr10_-_85435016 | 0.12 |
ENSRNOT00000079921
|
4933428G20Rik
|
RIKEN cDNA 4933428G20 gene |
| chr7_-_119071712 | 0.12 |
ENSRNOT00000037611
|
Myh9l1
|
myosin heavy chain 9-like 1 |
| chr5_+_90800082 | 0.12 |
ENSRNOT00000093696
ENSRNOT00000093206 |
Kdm4c
|
lysine demethylase 4C |
| chr11_-_72378895 | 0.12 |
ENSRNOT00000058885
|
Dlg1
|
discs large MAGUK scaffold protein 1 |
| chr16_-_75530500 | 0.12 |
ENSRNOT00000018580
ENSRNOT00000086035 |
RatNP-3b
|
defensin RatNP-3 precursor |
| chr15_-_4055539 | 0.12 |
ENSRNOT00000090352
|
Sec24c
|
SEC24 homolog C, COPII coat complex component |
| chr20_+_6351458 | 0.12 |
ENSRNOT00000091731
|
Cdkn1a
|
cyclin-dependent kinase inhibitor 1A |
| chr12_+_13323547 | 0.12 |
ENSRNOT00000074138
|
Zfp853
|
zinc finger protein 853 |
| chr2_-_259150479 | 0.12 |
ENSRNOT00000085892
|
AABR07013843.1
|
|
| chr1_-_90956544 | 0.12 |
ENSRNOT00000051422
|
LOC108348128
|
nesprin-4 |
| chr2_+_200572502 | 0.11 |
ENSRNOT00000074666
|
Zfp697
|
zinc finger protein 697 |
| chr12_+_16950728 | 0.11 |
ENSRNOT00000060370
|
Ints1
|
integrator complex subunit 1 |
| chr3_-_175327689 | 0.11 |
ENSRNOT00000082981
|
Taf4
|
TATA-box binding protein associated factor 4 |
| chr1_-_88111293 | 0.11 |
ENSRNOT00000077195
|
Spred3
|
sprouty-related, EVH1 domain containing 3 |
| chr10_-_108473377 | 0.11 |
ENSRNOT00000070934
|
Sgsh
|
N-sulfoglucosamine sulfohydrolase |
| chr10_+_15183803 | 0.11 |
ENSRNOT00000039777
|
Fbxl16
|
F-box and leucine-rich repeat protein 16 |
| chr19_+_59186673 | 0.11 |
ENSRNOT00000071655
|
Slc35f3
|
solute carrier family 35, member F3 |
| chr7_-_101140308 | 0.11 |
ENSRNOT00000006279
|
Fam84b
|
family with sequence similarity 84, member B |
| chr2_-_43393207 | 0.11 |
ENSRNOT00000017968
|
LOC100912399
|
mitogen-activated protein kinase kinase kinase 1-like |
| chr4_+_84597323 | 0.11 |
ENSRNOT00000074054
ENSRNOT00000012755 |
Wipf3
|
WAS/WASL interacting protein family, member 3 |
| chr3_-_123179644 | 0.11 |
ENSRNOT00000028835
|
Lzts3
|
leucine zipper tumor suppressor family member 3 |
| chr9_+_10963723 | 0.11 |
ENSRNOT00000075424
|
Plin4
|
perilipin 4 |
| chr20_+_7136007 | 0.11 |
ENSRNOT00000000580
|
Hmga1
|
high mobility group AT-hook 1 |
| chr2_-_211322719 | 0.11 |
ENSRNOT00000027493
|
RGD1310209
|
similar to KIAA1324 protein |
| chr10_-_13558377 | 0.11 |
ENSRNOT00000084066
ENSRNOT00000081995 |
Tbc1d24
|
TBC1 domain family, member 24 |
| chr5_+_135735825 | 0.11 |
ENSRNOT00000068267
|
Zswim5
|
zinc finger, SWIM-type containing 5 |
| chr6_+_28323647 | 0.11 |
ENSRNOT00000089093
|
Dnmt3a
|
DNA methyltransferase 3 alpha |
| chr9_-_82699551 | 0.11 |
ENSRNOT00000020673
|
Obsl1
|
obscurin-like 1 |
| chr13_-_88642011 | 0.11 |
ENSRNOT00000067037
|
LOC100361087
|
hypothetical LOC100361087 |
| chr6_+_33885495 | 0.11 |
ENSRNOT00000086633
|
Sdc1
|
syndecan 1 |
| chr5_-_169658875 | 0.11 |
ENSRNOT00000015840
|
Kcnab2
|
potassium voltage-gated channel subfamily A regulatory beta subunit 2 |
| chr4_-_183293999 | 0.11 |
ENSRNOT00000079695
|
Ipo8
|
importin 8 |
| chr16_-_19894591 | 0.11 |
ENSRNOT00000085940
|
Ano8
|
anoctamin 8 |
| chr13_+_34483876 | 0.11 |
ENSRNOT00000092998
|
Clasp1
|
cytoplasmic linker associated protein 1 |
| chr6_-_122897997 | 0.11 |
ENSRNOT00000057601
|
Eml5
|
echinoderm microtubule associated protein like 5 |
| chr10_-_13814304 | 0.11 |
ENSRNOT00000012203
|
Dnase1l2
|
deoxyribonuclease 1 like 2 |
| chr11_+_72044096 | 0.11 |
ENSRNOT00000034757
|
Senp5
|
SUMO1/sentrin specific peptidase 5 |
| chr9_+_40972089 | 0.11 |
ENSRNOT00000067928
|
Ptpn18
|
protein tyrosine phosphatase, non-receptor type 18 |
| chr1_-_192057612 | 0.11 |
ENSRNOT00000024471
|
Ndufab1
|
NADH:ubiquinone oxidoreductase subunit AB1 |
| chr7_-_70319346 | 0.11 |
ENSRNOT00000071543
|
Tsfm
|
Ts translation elongation factor, mitochondrial |
| chr6_-_138401621 | 0.11 |
ENSRNOT00000043210
|
LOC103693608
|
integral membrane protein DGCR2/IDD-like |
| chr15_-_28406046 | 0.11 |
ENSRNOT00000015418
|
Zfp219
|
zinc finger protein 219 |
| chr8_+_116324040 | 0.11 |
ENSRNOT00000081353
|
Hyal2
|
hyaluronoglucosaminidase 2 |
| chrX_-_123662350 | 0.10 |
ENSRNOT00000092624
|
Sept6
|
septin 6 |
| chr2_+_189062443 | 0.10 |
ENSRNOT00000028181
|
Adar
|
adenosine deaminase, RNA-specific |
| chr10_+_66942398 | 0.10 |
ENSRNOT00000018986
|
Rab11fip4
|
RAB11 family interacting protein 4 |
| chr5_-_138545404 | 0.10 |
ENSRNOT00000011586
|
Rimkla
|
ribosomal modification protein rimK-like family member A |
| chr8_+_117068582 | 0.10 |
ENSRNOT00000073559
|
Amt
|
aminomethyltransferase |
| chr14_-_114649173 | 0.10 |
ENSRNOT00000083528
|
Sptbn1
|
spectrin, beta, non-erythrocytic 1 |
| chr14_+_36689096 | 0.10 |
ENSRNOT00000083100
|
Usp46
|
ubiquitin specific peptidase 46 |
| chr12_-_44520341 | 0.10 |
ENSRNOT00000066810
|
Nos1
|
nitric oxide synthase 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Snai1_Zeb1_Snai2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0031439 | positive regulation of mRNA cleavage(GO:0031439) positive regulation of endoribonuclease activity(GO:1902380) positive regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904722) |
| 0.1 | 0.6 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 0.1 | 0.5 | GO:0002125 | maternal aggressive behavior(GO:0002125) |
| 0.1 | 0.3 | GO:0032900 | negative regulation of neurotrophin production(GO:0032900) |
| 0.1 | 0.3 | GO:1902630 | regulation of membrane hyperpolarization(GO:1902630) |
| 0.1 | 0.2 | GO:0071691 | cardiac muscle thin filament assembly(GO:0071691) |
| 0.1 | 0.1 | GO:0060086 | circadian temperature homeostasis(GO:0060086) |
| 0.1 | 0.2 | GO:1904204 | regulation of skeletal muscle hypertrophy(GO:1904204) |
| 0.1 | 0.2 | GO:0071298 | cellular response to L-ascorbic acid(GO:0071298) |
| 0.1 | 0.2 | GO:0021905 | pancreatic A cell development(GO:0003322) forebrain-midbrain boundary formation(GO:0021905) somatic motor neuron fate commitment(GO:0021917) regulation of transcription from RNA polymerase II promoter involved in somatic motor neuron fate commitment(GO:0021918) sensory neuron migration(GO:1904937) |
| 0.1 | 0.2 | GO:0070426 | positive regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070426) positive regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070434) |
| 0.1 | 0.4 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.1 | 0.5 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.1 | 0.3 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.0 | 0.1 | GO:1903760 | regulation of voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1903760) |
| 0.0 | 0.3 | GO:0098904 | regulation of AV node cell action potential(GO:0098904) |
| 0.0 | 0.2 | GO:0098989 | NMDA selective glutamate receptor signaling pathway(GO:0098989) |
| 0.0 | 0.0 | GO:0021836 | chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) |
| 0.0 | 0.2 | GO:0021586 | pons maturation(GO:0021586) |
| 0.0 | 0.1 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.0 | 0.2 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.0 | 0.2 | GO:0050893 | sensory processing(GO:0050893) |
| 0.0 | 0.0 | GO:0060764 | cell-cell signaling involved in mammary gland development(GO:0060764) |
| 0.0 | 0.3 | GO:0016056 | rhodopsin mediated signaling pathway(GO:0016056) |
| 0.0 | 0.0 | GO:0001982 | baroreceptor response to decreased systemic arterial blood pressure(GO:0001982) |
| 0.0 | 0.2 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.0 | 0.1 | GO:0098971 | anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) |
| 0.0 | 0.1 | GO:0072714 | response to selenite ion(GO:0072714) |
| 0.0 | 0.2 | GO:0021834 | chemorepulsion involved in embryonic olfactory bulb interneuron precursor migration(GO:0021834) |
| 0.0 | 0.1 | GO:1905218 | cellular response to astaxanthin(GO:1905218) |
| 0.0 | 0.2 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.0 | 0.2 | GO:1904717 | regulation of AMPA glutamate receptor clustering(GO:1904717) |
| 0.0 | 0.1 | GO:0021882 | regulation of transcription from RNA polymerase II promoter involved in forebrain neuron fate commitment(GO:0021882) commitment of multipotent stem cells to neuronal lineage in forebrain(GO:0021898) |
| 0.0 | 0.0 | GO:1904339 | negative regulation of dopaminergic neuron differentiation(GO:1904339) |
| 0.0 | 0.2 | GO:0071879 | positive regulation of adrenergic receptor signaling pathway(GO:0071879) positive regulation of the force of heart contraction(GO:0098735) retrograde trans-synaptic signaling by soluble gas(GO:0098923) trans-synaptic signaling by soluble gas(GO:0099543) regulation of adrenergic receptor signaling pathway involved in heart process(GO:1901204) |
| 0.0 | 0.2 | GO:1990523 | bone regeneration(GO:1990523) |
| 0.0 | 0.2 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.0 | 0.1 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.0 | 0.2 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.0 | 0.1 | GO:0042539 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.0 | 0.1 | GO:0044268 | multicellular organismal protein metabolic process(GO:0044268) |
| 0.0 | 0.1 | GO:0039663 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 0.0 | 0.2 | GO:0010626 | negative regulation of Schwann cell proliferation(GO:0010626) |
| 0.0 | 0.1 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.0 | 0.1 | GO:1902303 | negative regulation of potassium ion export(GO:1902303) |
| 0.0 | 0.1 | GO:0071348 | cellular response to interleukin-11(GO:0071348) |
| 0.0 | 0.1 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.0 | 0.1 | GO:0061744 | motor behavior(GO:0061744) |
| 0.0 | 0.4 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.1 | GO:0019417 | sulfur oxidation(GO:0019417) |
| 0.0 | 0.1 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.0 | 0.1 | GO:0015888 | thiamine transport(GO:0015888) |
| 0.0 | 0.1 | GO:0061181 | regulation of chondrocyte development(GO:0061181) |
| 0.0 | 0.2 | GO:0044027 | hypermethylation of CpG island(GO:0044027) |
| 0.0 | 0.1 | GO:0019853 | L-ascorbic acid biosynthetic process(GO:0019853) |
| 0.0 | 0.2 | GO:1904045 | cellular response to aldosterone(GO:1904045) |
| 0.0 | 0.1 | GO:0070100 | negative regulation of chemokine-mediated signaling pathway(GO:0070100) |
| 0.0 | 0.1 | GO:0001828 | inner cell mass cellular morphogenesis(GO:0001828) |
| 0.0 | 0.2 | GO:0071460 | cellular response to cell-matrix adhesion(GO:0071460) |
| 0.0 | 0.1 | GO:0035986 | senescence-associated heterochromatin focus assembly(GO:0035986) |
| 0.0 | 0.3 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.0 | 0.2 | GO:0032571 | response to vitamin K(GO:0032571) |
| 0.0 | 0.1 | GO:1904685 | positive regulation of metalloendopeptidase activity(GO:1904685) |
| 0.0 | 0.1 | GO:2000474 | regulation of opioid receptor signaling pathway(GO:2000474) |
| 0.0 | 0.2 | GO:0090367 | negative regulation of mRNA modification(GO:0090367) |
| 0.0 | 0.3 | GO:0061525 | hindgut development(GO:0061525) |
| 0.0 | 0.1 | GO:0061642 | chemoattraction of axon(GO:0061642) renal outer medulla development(GO:0072054) |
| 0.0 | 0.1 | GO:2000809 | positive regulation of synaptic vesicle clustering(GO:2000809) |
| 0.0 | 0.1 | GO:2000620 | positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 0.0 | 0.2 | GO:0051365 | cellular response to potassium ion starvation(GO:0051365) |
| 0.0 | 0.3 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.0 | 0.1 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.0 | 0.1 | GO:0036451 | cap mRNA methylation(GO:0036451) |
| 0.0 | 0.1 | GO:0016344 | meiotic chromosome movement towards spindle pole(GO:0016344) |
| 0.0 | 0.4 | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 0.0 | 0.1 | GO:0019464 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.0 | 0.0 | GO:0034241 | positive regulation of macrophage fusion(GO:0034241) |
| 0.0 | 0.1 | GO:0032066 | nucleolus to nucleoplasm transport(GO:0032066) |
| 0.0 | 0.3 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.0 | 0.2 | GO:0098885 | modification of postsynaptic actin cytoskeleton(GO:0098885) |
| 0.0 | 0.0 | GO:0002191 | cap-dependent translational initiation(GO:0002191) |
| 0.0 | 0.2 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.0 | 0.1 | GO:0010728 | regulation of hydrogen peroxide biosynthetic process(GO:0010728) |
| 0.0 | 0.2 | GO:0060574 | intestinal epithelial cell maturation(GO:0060574) |
| 0.0 | 0.1 | GO:0060023 | soft palate development(GO:0060023) |
| 0.0 | 0.0 | GO:0010877 | lipid transport involved in lipid storage(GO:0010877) |
| 0.0 | 0.1 | GO:0035106 | operant conditioning(GO:0035106) |
| 0.0 | 0.1 | GO:0002752 | cell surface pattern recognition receptor signaling pathway(GO:0002752) |
| 0.0 | 0.1 | GO:1903553 | positive regulation of extracellular exosome assembly(GO:1903553) |
| 0.0 | 0.1 | GO:1903373 | positive regulation of endoplasmic reticulum tubular network organization(GO:1903373) |
| 0.0 | 0.2 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.1 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 0.0 | 0.1 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.0 | 0.1 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.0 | 0.0 | GO:1903061 | positive regulation of protein lipidation(GO:1903061) |
| 0.0 | 0.1 | GO:0043324 | eye pigment metabolic process(GO:0042441) pigment metabolic process involved in developmental pigmentation(GO:0043324) pigment metabolic process involved in pigmentation(GO:0043474) rhodopsin metabolic process(GO:0046154) |
| 0.0 | 0.1 | GO:0090271 | positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.0 | 0.3 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.1 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.0 | 0.1 | GO:0098939 | dendritic transport of mitochondrion(GO:0098939) anterograde dendritic transport of mitochondrion(GO:0098972) |
| 0.0 | 0.1 | GO:0006407 | rRNA export from nucleus(GO:0006407) |
| 0.0 | 0.1 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.0 | 0.1 | GO:0048702 | embryonic neurocranium morphogenesis(GO:0048702) |
| 0.0 | 0.1 | GO:0000414 | regulation of histone H3-K36 methylation(GO:0000414) |
| 0.0 | 0.0 | GO:0080033 | cellular response to nitrite(GO:0071250) response to nitrite(GO:0080033) |
| 0.0 | 0.1 | GO:2000019 | negative regulation of male gonad development(GO:2000019) |
| 0.0 | 0.2 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.0 | 0.1 | GO:1903542 | negative regulation of exosomal secretion(GO:1903542) |
| 0.0 | 0.0 | GO:0032701 | negative regulation of interleukin-18 production(GO:0032701) |
| 0.0 | 0.1 | GO:0071681 | cellular response to indole-3-methanol(GO:0071681) |
| 0.0 | 0.2 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.0 | 0.0 | GO:0042998 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) |
| 0.0 | 0.0 | GO:0060060 | post-embryonic retina morphogenesis in camera-type eye(GO:0060060) |
| 0.0 | 0.2 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.1 | GO:0032687 | negative regulation of interferon-alpha production(GO:0032687) |
| 0.0 | 0.3 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 | 0.1 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.0 | 0.1 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 0.0 | 0.3 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.0 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 0.0 | 0.2 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.0 | 0.1 | GO:0032915 | positive regulation of transforming growth factor beta2 production(GO:0032915) |
| 0.0 | 0.1 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.0 | 0.0 | GO:0010182 | carbohydrate mediated signaling(GO:0009756) hexose mediated signaling(GO:0009757) sugar mediated signaling pathway(GO:0010182) glucose mediated signaling pathway(GO:0010255) |
| 0.0 | 0.2 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.0 | 0.1 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.0 | 0.1 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.0 | 0.1 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 0.1 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.0 | 0.6 | GO:0015812 | gamma-aminobutyric acid transport(GO:0015812) |
| 0.0 | 0.1 | GO:0046380 | N-acetylglucosamine biosynthetic process(GO:0006045) N-acetylneuraminate biosynthetic process(GO:0046380) |
| 0.0 | 0.1 | GO:2000328 | regulation of T-helper 17 cell lineage commitment(GO:2000328) |
| 0.0 | 0.1 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) |
| 0.0 | 0.1 | GO:0043311 | regulation of eosinophil degranulation(GO:0043309) positive regulation of eosinophil degranulation(GO:0043311) positive regulation of eosinophil activation(GO:1902568) |
| 0.0 | 0.0 | GO:1904616 | regulation of actin filament binding(GO:1904529) regulation of actin binding(GO:1904616) |
| 0.0 | 0.1 | GO:0061146 | Peyer's patch morphogenesis(GO:0061146) |
| 0.0 | 0.1 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.0 | 0.0 | GO:0071282 | cellular response to iron(II) ion(GO:0071282) |
| 0.0 | 0.1 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.0 | 0.1 | GO:0048341 | paraxial mesoderm formation(GO:0048341) |
| 0.0 | 0.1 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.0 | 0.1 | GO:0071163 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) DNA replication preinitiation complex assembly(GO:0071163) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.0 | 0.2 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.1 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.0 | 0.0 | GO:0034125 | negative regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034125) |
| 0.0 | 0.0 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.0 | 0.2 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.0 | 0.1 | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.0 | 0.1 | GO:0071929 | alpha-tubulin acetylation(GO:0071929) |
| 0.0 | 0.1 | GO:0031989 | bombesin receptor signaling pathway(GO:0031989) |
| 0.0 | 0.1 | GO:0071454 | cellular response to anoxia(GO:0071454) |
| 0.0 | 0.1 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.0 | 0.1 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.0 | 0.1 | GO:0046813 | receptor-mediated virion attachment to host cell(GO:0046813) |
| 0.0 | 0.1 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.0 | 0.1 | GO:0097118 | gephyrin clustering involved in postsynaptic density assembly(GO:0097116) neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.0 | 0.2 | GO:0070561 | vitamin D receptor signaling pathway(GO:0070561) |
| 0.0 | 0.1 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.0 | 0.1 | GO:1990926 | short-term synaptic potentiation(GO:1990926) |
| 0.0 | 0.1 | GO:0046504 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.0 | 0.1 | GO:0002101 | tRNA wobble cytosine modification(GO:0002101) |
| 0.0 | 0.2 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 | 0.1 | GO:0060842 | arterial endothelial cell differentiation(GO:0060842) |
| 0.0 | 0.0 | GO:0009644 | response to high light intensity(GO:0009644) |
| 0.0 | 0.1 | GO:0072021 | ascending thin limb development(GO:0072021) metanephric ascending thin limb development(GO:0072218) |
| 0.0 | 0.1 | GO:0090290 | positive regulation of osteoclast proliferation(GO:0090290) |
| 0.0 | 0.1 | GO:0045900 | negative regulation of translational elongation(GO:0045900) |
| 0.0 | 0.0 | GO:0090063 | positive regulation of microtubule nucleation(GO:0090063) |
| 0.0 | 0.2 | GO:0070862 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.0 | 0.0 | GO:1904350 | regulation of protein catabolic process in the vacuole(GO:1904350) lysosomal protein catabolic process(GO:1905146) |
| 0.0 | 0.1 | GO:0090237 | regulation of arachidonic acid secretion(GO:0090237) |
| 0.0 | 0.0 | GO:0051835 | positive regulation of synapse structural plasticity(GO:0051835) |
| 0.0 | 0.1 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.0 | 0.2 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.0 | 0.0 | GO:0046985 | positive regulation of hemoglobin biosynthetic process(GO:0046985) |
| 0.0 | 0.0 | GO:0042640 | anagen(GO:0042640) regulation of anagen(GO:0051884) |
| 0.0 | 0.0 | GO:1904504 | lipophagy(GO:0061724) regulation of lipophagy(GO:1904502) positive regulation of lipophagy(GO:1904504) |
| 0.0 | 0.0 | GO:0007089 | traversing start control point of mitotic cell cycle(GO:0007089) |
| 0.0 | 0.1 | GO:1903300 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 | 0.1 | GO:0061042 | vascular wound healing(GO:0061042) |
| 0.0 | 0.0 | GO:0031632 | positive regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031632) |
| 0.0 | 0.0 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.0 | 0.1 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.0 | 0.0 | GO:0007468 | regulation of rhodopsin gene expression(GO:0007468) |
| 0.0 | 0.0 | GO:0060605 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.0 | 0.1 | GO:0045105 | intermediate filament polymerization or depolymerization(GO:0045105) |
| 0.0 | 0.0 | GO:0036135 | Schwann cell migration(GO:0036135) regulation of Schwann cell migration(GO:1900147) |
| 0.0 | 0.0 | GO:0071877 | regulation of adrenergic receptor signaling pathway(GO:0071877) |
| 0.0 | 0.0 | GO:0031438 | negative regulation of mRNA cleavage(GO:0031438) negative regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904721) |
| 0.0 | 0.0 | GO:0021816 | lamellipodium assembly involved in ameboidal cell migration(GO:0003363) extension of a leading process involved in cell motility in cerebral cortex radial glia guided migration(GO:0021816) |
| 0.0 | 0.1 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
| 0.0 | 0.1 | GO:0007198 | adenylate cyclase-inhibiting serotonin receptor signaling pathway(GO:0007198) |
| 0.0 | 0.1 | GO:1904426 | positive regulation of GTP binding(GO:1904426) |
| 0.0 | 0.0 | GO:0098758 | response to interleukin-8(GO:0098758) cellular response to interleukin-8(GO:0098759) |
| 0.0 | 0.1 | GO:0045719 | negative regulation of glycogen biosynthetic process(GO:0045719) |
| 0.0 | 0.1 | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.0 | 0.1 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
| 0.0 | 0.1 | GO:2001166 | regulation of histone H2B ubiquitination(GO:2001166) positive regulation of histone H2B ubiquitination(GO:2001168) |
| 0.0 | 0.1 | GO:1902995 | regulation of phospholipid efflux(GO:1902994) positive regulation of phospholipid efflux(GO:1902995) |
| 0.0 | 0.0 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) |
| 0.0 | 0.0 | GO:2000974 | negative regulation of mechanoreceptor differentiation(GO:0045632) negative regulation of pro-B cell differentiation(GO:2000974) negative regulation of inner ear receptor cell differentiation(GO:2000981) |
| 0.0 | 0.1 | GO:0015746 | citrate transport(GO:0015746) |
| 0.0 | 0.1 | GO:0033153 | somatic diversification of T cell receptor genes(GO:0002568) somatic recombination of T cell receptor gene segments(GO:0002681) T cell receptor V(D)J recombination(GO:0033153) |
| 0.0 | 0.1 | GO:0099562 | maintenance of postsynaptic specialization structure(GO:0098880) maintenance of postsynaptic density structure(GO:0099562) |
| 0.0 | 0.1 | GO:0035524 | proline transmembrane transport(GO:0035524) |
| 0.0 | 0.0 | GO:0071680 | response to indole-3-methanol(GO:0071680) |
| 0.0 | 0.1 | GO:1904117 | cellular response to vasopressin(GO:1904117) |
| 0.0 | 0.0 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.0 | 0.0 | GO:0034499 | late endosome to Golgi transport(GO:0034499) |
| 0.0 | 0.2 | GO:0021684 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.0 | 0.0 | GO:0031161 | phosphatidylinositol catabolic process(GO:0031161) |
| 0.0 | 0.2 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.0 | 0.1 | GO:0060596 | mammary placode formation(GO:0060596) |
| 0.0 | 0.1 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 0.0 | 0.0 | GO:0097332 | response to antipsychotic drug(GO:0097332) |
| 0.0 | 0.1 | GO:0010037 | response to carbon dioxide(GO:0010037) |
| 0.0 | 0.1 | GO:0038096 | immune response-regulating cell surface receptor signaling pathway involved in phagocytosis(GO:0002433) Fc-gamma receptor signaling pathway involved in phagocytosis(GO:0038096) |
| 0.0 | 0.0 | GO:0031126 | snoRNA 3'-end processing(GO:0031126) |
| 0.0 | 0.1 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.0 | 0.1 | GO:1903336 | negative regulation of vacuolar transport(GO:1903336) |
| 0.0 | 0.0 | GO:0071579 | regulation of zinc ion transport(GO:0071579) |
| 0.0 | 0.0 | GO:2000669 | negative regulation of dendritic cell apoptotic process(GO:2000669) |
| 0.0 | 0.0 | GO:1903849 | regulation of aorta morphogenesis(GO:1903847) positive regulation of aorta morphogenesis(GO:1903849) |
| 0.0 | 0.0 | GO:0080144 | amino acid homeostasis(GO:0080144) |
| 0.0 | 0.0 | GO:0042723 | thiamine metabolic process(GO:0006772) thiamine diphosphate metabolic process(GO:0042357) thiamine-containing compound metabolic process(GO:0042723) |
| 0.0 | 0.4 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 | 0.1 | GO:1901550 | regulation of endothelial cell development(GO:1901550) regulation of establishment of endothelial barrier(GO:1903140) |
| 0.0 | 0.0 | GO:0010533 | regulation of activation of Janus kinase activity(GO:0010533) |
| 0.0 | 0.1 | GO:0090118 | receptor-mediated endocytosis of low-density lipoprotein particle involved in cholesterol transport(GO:0090118) |
| 0.0 | 0.1 | GO:0045794 | negative regulation of cell volume(GO:0045794) |
| 0.0 | 0.1 | GO:0019471 | 4-hydroxyproline metabolic process(GO:0019471) |
| 0.0 | 0.0 | GO:0003099 | positive regulation of the force of heart contraction by epinephrine-norepinephrine(GO:0001997) positive regulation of the force of heart contraction by chemical signal(GO:0003099) |
| 0.0 | 0.1 | GO:0060856 | establishment of blood-brain barrier(GO:0060856) |
| 0.0 | 0.0 | GO:0035419 | activation of MAPK activity involved in innate immune response(GO:0035419) |
| 0.0 | 0.1 | GO:0002225 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.0 | 0.0 | GO:0021740 | trigeminal sensory nucleus development(GO:0021730) principal sensory nucleus of trigeminal nerve development(GO:0021740) |
| 0.0 | 0.1 | GO:1901552 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.0 | 0.1 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.0 | GO:0033693 | neurofilament bundle assembly(GO:0033693) |
| 0.0 | 0.0 | GO:2001137 | positive regulation of endocytic recycling(GO:2001137) |
| 0.0 | 0.0 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.0 | 0.0 | GO:0048691 | positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) |
| 0.0 | 0.1 | GO:0021960 | anterior commissure morphogenesis(GO:0021960) |
| 0.0 | 0.1 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.0 | 0.0 | GO:0090625 | mRNA cleavage involved in gene silencing by siRNA(GO:0090625) |
| 0.0 | 0.1 | GO:0006930 | substrate-dependent cell migration, cell extension(GO:0006930) |
| 0.0 | 0.1 | GO:2000465 | regulation of glycogen (starch) synthase activity(GO:2000465) positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.0 | 0.0 | GO:0042509 | regulation of tyrosine phosphorylation of STAT protein(GO:0042509) positive regulation of tyrosine phosphorylation of STAT protein(GO:0042531) |
| 0.0 | 0.0 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
| 0.0 | 0.1 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.0 | 0.0 | GO:0090234 | regulation of kinetochore assembly(GO:0090234) |
| 0.0 | 0.0 | GO:0002432 | granuloma formation(GO:0002432) |
| 0.0 | 0.0 | GO:0032364 | oxygen homeostasis(GO:0032364) |
| 0.0 | 0.1 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 0.0 | 0.0 | GO:0003419 | growth plate cartilage chondrocyte proliferation(GO:0003419) |
| 0.0 | 0.0 | GO:2000169 | regulation of peptidyl-cysteine S-nitrosylation(GO:2000169) |
| 0.0 | 0.1 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.0 | 0.2 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.0 | 0.0 | GO:0060478 | acrosomal vesicle exocytosis(GO:0060478) |
| 0.0 | 0.0 | GO:1901423 | response to benzene(GO:1901423) |
| 0.0 | 0.0 | GO:0071895 | odontoblast differentiation(GO:0071895) |
| 0.0 | 0.2 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.0 | 0.1 | GO:0035878 | nail development(GO:0035878) |
| 0.0 | 0.0 | GO:0006362 | transcription elongation from RNA polymerase I promoter(GO:0006362) |
| 0.0 | 0.1 | GO:0035947 | regulation of gluconeogenesis by regulation of transcription from RNA polymerase II promoter(GO:0035947) |
| 0.0 | 0.1 | GO:0099612 | protein localization to axon(GO:0099612) |
| 0.0 | 0.0 | GO:0060447 | bud outgrowth involved in lung branching(GO:0060447) |
| 0.0 | 0.2 | GO:0022038 | corpus callosum development(GO:0022038) |
| 0.0 | 0.4 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 0.0 | GO:1903898 | negative regulation of PERK-mediated unfolded protein response(GO:1903898) |
| 0.0 | 0.0 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.0 | 0.0 | GO:0045575 | basophil activation(GO:0045575) |
| 0.0 | 0.0 | GO:1990743 | protein sialylation(GO:1990743) |
| 0.0 | 0.0 | GO:0015965 | diadenosine polyphosphate biosynthetic process(GO:0015960) diadenosine tetraphosphate metabolic process(GO:0015965) diadenosine tetraphosphate biosynthetic process(GO:0015966) |
| 0.0 | 0.2 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.0 | GO:0044337 | canonical Wnt signaling pathway involved in positive regulation of apoptotic process(GO:0044337) |
| 0.0 | 0.0 | GO:1904640 | response to methionine(GO:1904640) |
| 0.0 | 0.1 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.0 | 0.0 | GO:1901748 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.0 | 0.0 | GO:0071529 | cementum mineralization(GO:0071529) |
| 0.0 | 0.0 | GO:0070142 | synaptic vesicle budding(GO:0070142) |
| 0.0 | 0.5 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.0 | 0.1 | GO:0090166 | Golgi disassembly(GO:0090166) |
| 0.0 | 0.0 | GO:0021849 | neuroblast division in subventricular zone(GO:0021849) |
| 0.0 | 0.0 | GO:0071336 | hair follicle cell proliferation(GO:0071335) regulation of hair follicle cell proliferation(GO:0071336) |
| 0.0 | 0.0 | GO:1900377 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.0 | 0.1 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 0.1 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.0 | 0.0 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) |
| 0.0 | 0.0 | GO:0021913 | regulation of transcription from RNA polymerase II promoter involved in spinal cord motor neuron fate specification(GO:0021912) regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.0 | 0.0 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.0 | 0.0 | GO:1903756 | regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903756) negative regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903758) |
| 0.0 | 0.1 | GO:0030853 | negative regulation of granulocyte differentiation(GO:0030853) |
| 0.0 | 0.1 | GO:0060013 | righting reflex(GO:0060013) |
| 0.0 | 0.1 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.0 | 0.0 | GO:0061738 | late endosomal microautophagy(GO:0061738) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0097444 | spine apparatus(GO:0097444) |
| 0.1 | 0.3 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.1 | 0.2 | GO:0098855 | HCN channel complex(GO:0098855) |
| 0.1 | 0.2 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.1 | 0.2 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.1 | 0.2 | GO:1990032 | parallel fiber(GO:1990032) |
| 0.1 | 0.6 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.2 | GO:0099522 | region of cytosol(GO:0099522) postsynaptic cytosol(GO:0099524) |
| 0.0 | 0.2 | GO:0044393 | microspike(GO:0044393) |
| 0.0 | 0.1 | GO:0031308 | intrinsic component of nuclear outer membrane(GO:0031308) integral component of nuclear outer membrane(GO:0031309) |
| 0.0 | 0.1 | GO:0005592 | collagen type XI trimer(GO:0005592) |
| 0.0 | 0.2 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.2 | GO:0097598 | sperm cytoplasmic droplet(GO:0097598) |
| 0.0 | 0.2 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 0.2 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.0 | 0.1 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.1 | GO:0031592 | centrosomal corona(GO:0031592) |
| 0.0 | 0.2 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.0 | 0.1 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.0 | 0.1 | GO:0034686 | integrin alphav-beta8 complex(GO:0034686) |
| 0.0 | 0.1 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.5 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.1 | GO:1990796 | photoreceptor cell terminal bouton(GO:1990796) |
| 0.0 | 0.1 | GO:0005607 | laminin-2 complex(GO:0005607) |
| 0.0 | 0.1 | GO:0032173 | septin collar(GO:0032173) |
| 0.0 | 0.1 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.0 | 0.3 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.0 | 0.1 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.0 | 0.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.1 | GO:0034667 | integrin alpha3-beta1 complex(GO:0034667) |
| 0.0 | 0.1 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.0 | 0.1 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.1 | GO:0043293 | apoptosome(GO:0043293) |
| 0.0 | 0.0 | GO:0005940 | septin ring(GO:0005940) |
| 0.0 | 0.1 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.0 | 0.1 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 0.1 | GO:1990723 | cytoplasmic periphery of the nuclear pore complex(GO:1990723) |
| 0.0 | 0.2 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.1 | GO:1990393 | 3M complex(GO:1990393) |
| 0.0 | 0.1 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.1 | GO:0036387 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 0.0 | 0.1 | GO:0005745 | m-AAA complex(GO:0005745) |
| 0.0 | 0.1 | GO:0038037 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor complex(GO:0097648) |
| 0.0 | 0.1 | GO:1990415 | Pex17p-Pex14p docking complex(GO:1990415) peroxisomal importomer complex(GO:1990429) |
| 0.0 | 0.1 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.0 | 0.1 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.0 | 0.4 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.5 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 0.1 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.0 | 0.1 | GO:0031094 | platelet dense tubular network(GO:0031094) |
| 0.0 | 0.7 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.1 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 0.3 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.0 | 0.1 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.1 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 0.0 | GO:0070110 | interleukin-6 receptor complex(GO:0005896) ciliary neurotrophic factor receptor complex(GO:0070110) |
| 0.0 | 0.1 | GO:0099571 | postsynaptic actin cytoskeleton(GO:0098871) postsynaptic cytoskeleton(GO:0099571) |
| 0.0 | 0.1 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.0 | 0.1 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.0 | 0.1 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.1 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.1 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 0.1 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.0 | 0.1 | GO:0031074 | nucleocytoplasmic shuttling complex(GO:0031074) |
| 0.0 | 0.0 | GO:0002944 | cyclin K-CDK12 complex(GO:0002944) cyclin K-CDK13 complex(GO:0002945) |
| 0.0 | 0.2 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.0 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.0 | 0.0 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.0 | GO:0033557 | Slx1-Slx4 complex(GO:0033557) |
| 0.0 | 0.0 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 0.0 | 0.0 | GO:0098830 | presynaptic endosome(GO:0098830) presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) |
| 0.0 | 0.1 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 0.0 | GO:0035838 | growing cell tip(GO:0035838) |
| 0.0 | 0.0 | GO:0036454 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
| 0.0 | 0.2 | GO:0001741 | XY body(GO:0001741) |
| 0.0 | 0.1 | GO:0030689 | Noc complex(GO:0030689) |
| 0.0 | 0.0 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.0 | 0.1 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.0 | 0.2 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.1 | GO:0033263 | CORVET complex(GO:0033263) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0031249 | denatured protein binding(GO:0031249) |
| 0.2 | 0.5 | GO:0031893 | vasopressin receptor binding(GO:0031893) |
| 0.1 | 0.5 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.1 | 0.2 | GO:0047783 | steroid 11-beta-monooxygenase activity(GO:0004507) corticosterone 18-monooxygenase activity(GO:0047783) |
| 0.1 | 0.2 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.1 | 0.4 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.1 | 0.2 | GO:0001605 | adrenomedullin receptor activity(GO:0001605) |
| 0.0 | 0.1 | GO:0072590 | N-acetyl-L-aspartate-L-glutamate ligase activity(GO:0072590) |
| 0.0 | 0.2 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.0 | 0.2 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.0 | 0.1 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.0 | 0.1 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.0 | 0.2 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.0 | 0.1 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 0.0 | 0.1 | GO:0031177 | phosphopantetheine binding(GO:0031177) |
| 0.0 | 0.2 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.0 | 0.2 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.0 | 0.1 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.2 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.1 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.3 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.1 | GO:0045174 | oxidoreductase activity, acting on a sulfur group of donors, quinone or similar compound as acceptor(GO:0016672) oxidoreductase activity, acting on phosphorus or arsenic in donors(GO:0030613) oxidoreductase activity, acting on phosphorus or arsenic in donors, disulfide as acceptor(GO:0030614) glutathione dehydrogenase (ascorbate) activity(GO:0045174) methylarsonate reductase activity(GO:0050610) |
| 0.0 | 0.1 | GO:0005302 | L-tyrosine transmembrane transporter activity(GO:0005302) |
| 0.0 | 0.2 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.0 | 0.1 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 0.1 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.3 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 0.1 | GO:1902379 | chemoattractant activity involved in axon guidance(GO:1902379) |
| 0.0 | 0.2 | GO:0071253 | connexin binding(GO:0071253) |
| 0.0 | 0.1 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.1 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.1 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.0 | 0.2 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.1 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.1 | GO:0034485 | phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase activity(GO:0034485) |
| 0.0 | 0.0 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.0 | 0.1 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.1 | GO:0052723 | inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 0.0 | 0.3 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) |
| 0.0 | 0.0 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.1 | GO:0070996 | type 1 melanocortin receptor binding(GO:0070996) |
| 0.0 | 0.2 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.1 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 0.0 | 0.1 | GO:0035827 | rubidium ion transmembrane transporter activity(GO:0035827) |
| 0.0 | 0.2 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.1 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.1 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.0 | 0.3 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.0 | 0.1 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.0 | 0.4 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.1 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.0 | 0.2 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.0 | 0.1 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.2 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.2 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.0 | 0.1 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.0 | 0.1 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.1 | GO:0004920 | interleukin-10 receptor activity(GO:0004920) |
| 0.0 | 0.2 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.1 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.0 | 0.1 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 0.1 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.0 | 0.1 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.0 | 0.1 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.0 | 0.1 | GO:0004445 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) |
| 0.0 | 0.2 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.1 | GO:0004946 | bombesin receptor activity(GO:0004946) |
| 0.0 | 0.1 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.0 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.0 | 0.1 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.0 | 0.1 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.1 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 0.0 | 0.2 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.0 | 0.1 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.0 | 0.0 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.0 | 0.1 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.1 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.0 | 0.3 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.0 | 0.1 | GO:0030250 | cyclase activator activity(GO:0010853) guanylate cyclase activator activity(GO:0030250) |
| 0.0 | 0.2 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.0 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.0 | 0.1 | GO:0015252 | hydrogen ion channel activity(GO:0015252) |
| 0.0 | 0.1 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 0.2 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.1 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.0 | 0.1 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.0 | 0.1 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.0 | 0.2 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.2 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.2 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 0.5 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.0 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.0 | 0.1 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.0 | 0.1 | GO:0015137 | citrate transmembrane transporter activity(GO:0015137) |
| 0.0 | 0.1 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.0 | 0.1 | GO:0098879 | structural constituent of postsynaptic specialization(GO:0098879) structural constituent of postsynaptic density(GO:0098919) |
| 0.0 | 0.3 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.1 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.0 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.0 | 0.1 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.0 | 0.1 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.0 | 0.1 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.0 | 0.1 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.0 | GO:0032810 | sterol response element binding(GO:0032810) |
| 0.0 | 0.1 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.1 | GO:0000403 | Y-form DNA binding(GO:0000403) |
| 0.0 | 0.0 | GO:0004655 | porphobilinogen synthase activity(GO:0004655) |
| 0.0 | 0.0 | GO:0017168 | 5-oxoprolinase (ATP-hydrolyzing) activity(GO:0017168) |
| 0.0 | 0.1 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.2 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.1 | GO:0090554 | phosphatidylcholine-translocating ATPase activity(GO:0090554) |
| 0.0 | 0.1 | GO:0016901 | oxidoreductase activity, acting on the CH-OH group of donors, quinone or similar compound as acceptor(GO:0016901) |
| 0.0 | 0.0 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.0 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.0 | GO:0001225 | RNA polymerase II transcription coactivator binding(GO:0001225) |
| 0.0 | 0.1 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.1 | GO:0051766 | inositol trisphosphate kinase activity(GO:0051766) |
| 0.0 | 0.0 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.0 | 0.0 | GO:0071936 | coreceptor activity involved in Wnt signaling pathway(GO:0071936) |
| 0.0 | 0.1 | GO:0098988 | G-protein coupled glutamate receptor activity(GO:0098988) |
| 0.0 | 0.1 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.0 | 0.2 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.0 | 0.1 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.0 | 0.2 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.1 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.1 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.1 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.1 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.0 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 0.0 | 0.1 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.0 | 0.0 | GO:0004897 | ciliary neurotrophic factor receptor activity(GO:0004897) |
| 0.0 | 0.1 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.0 | 0.1 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.0 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.0 | 0.3 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.1 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.1 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 0.0 | GO:0000829 | inositol heptakisphosphate kinase activity(GO:0000829) |
| 0.0 | 0.1 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 0.1 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.0 | 0.1 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.0 | 0.4 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.2 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.0 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.0 | 0.2 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.2 | GO:0036442 | hydrogen-exporting ATPase activity(GO:0036442) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.0 | 0.2 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.1 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.0 | 0.6 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.2 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 0.6 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.0 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.1 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.4 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 0.0 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 0.0 | ST GA13 PATHWAY | G alpha 13 Pathway |
| 0.0 | 0.1 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.3 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 0.2 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.2 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.0 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.2 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.0 | 0.0 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.5 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.4 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.0 | 0.0 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.7 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.0 | 0.5 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.0 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 0.3 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.3 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.2 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 0.6 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.7 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.0 | REACTOME SIGNALING BY ERBB2 | Genes involved in Signaling by ERBB2 |
| 0.0 | 0.2 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 0.2 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.4 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.0 | 0.2 | REACTOME ACTIVATION OF CHAPERONES BY ATF6 ALPHA | Genes involved in Activation of Chaperones by ATF6-alpha |
| 0.0 | 0.2 | REACTOME TAK1 ACTIVATES NFKB BY PHOSPHORYLATION AND ACTIVATION OF IKKS COMPLEX | Genes involved in TAK1 activates NFkB by phosphorylation and activation of IKKs complex |
| 0.0 | 0.3 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.2 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.4 | REACTOME PI METABOLISM | Genes involved in PI Metabolism |
| 0.0 | 0.0 | REACTOME G ALPHA1213 SIGNALLING EVENTS | Genes involved in G alpha (12/13) signalling events |
| 0.0 | 0.2 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.2 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.0 | 0.1 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.0 | 0.1 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.1 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.0 | 0.3 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.1 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |