Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
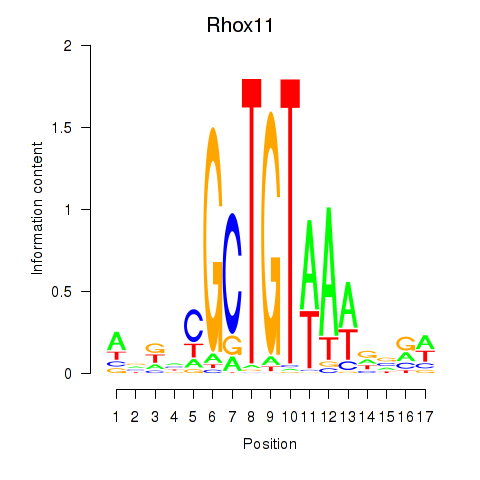
Results for Rhox11
Z-value: 0.40
Transcription factors associated with Rhox11
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Rhox11
|
ENSRNOG00000028044 | reproductive homeobox 11 |
Activity profile of Rhox11 motif
Sorted Z-values of Rhox11 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_252808380 | 0.08 |
ENSRNOT00000025856
|
Ch25h
|
cholesterol 25-hydroxylase |
| chr7_+_15422479 | 0.08 |
ENSRNOT00000066520
|
Zfp563
|
zinc finger protein 563 |
| chr1_-_266914093 | 0.08 |
ENSRNOT00000027526
|
Calhm2
|
calcium homeostasis modulator 2 |
| chr8_-_77599781 | 0.07 |
ENSRNOT00000087980
|
Aqp9
|
aquaporin 9 |
| chr6_-_49324450 | 0.07 |
ENSRNOT00000066904
|
Sntg2
|
syntrophin, gamma 2 |
| chr9_-_52830457 | 0.07 |
ENSRNOT00000073557
|
Slc40a1
|
solute carrier family 40 member 1 |
| chr10_+_72272248 | 0.07 |
ENSRNOT00000003908
|
Car4
|
carbonic anhydrase 4 |
| chr13_+_87986240 | 0.06 |
ENSRNOT00000003705
|
Rgs5
|
regulator of G-protein signaling 5 |
| chr3_-_43117337 | 0.06 |
ENSRNOT00000078001
|
Nr4a2
|
nuclear receptor subfamily 4, group A, member 2 |
| chr4_-_78342863 | 0.06 |
ENSRNOT00000049038
|
Gimap6
|
GTPase, IMAP family member 6 |
| chr11_+_74014983 | 0.05 |
ENSRNOT00000040884
|
NEWGENE_2724
|
glycoprotein V platelet |
| chr3_+_177164359 | 0.05 |
ENSRNOT00000066837
|
RGD1561282
|
RGD1561282 |
| chr11_+_38035450 | 0.05 |
ENSRNOT00000083067
|
Mx2
|
MX dynamin like GTPase 2 |
| chr2_+_55747318 | 0.05 |
ENSRNOT00000050655
|
Dab2
|
DAB2, clathrin adaptor protein |
| chr8_+_113936991 | 0.05 |
ENSRNOT00000017743
|
Aste1
|
asteroid homolog 1 (Drosophila) |
| chr7_-_29233392 | 0.04 |
ENSRNOT00000064241
|
Spic
|
Spi-C transcription factor |
| chr15_-_61648267 | 0.04 |
ENSRNOT00000071805
|
Naa16
|
N(alpha)-acetyltransferase 16, NatA auxiliary subunit |
| chr17_+_89452814 | 0.04 |
ENSRNOT00000058760
|
RGD1561231
|
similar to MAP/microtubule affinity-regulating kinase 4 (MAP/microtubule affinity-regulating kinase like 1) |
| chr1_+_85386470 | 0.04 |
ENSRNOT00000093332
ENSRNOT00000044326 |
Plekhg2
|
pleckstrin homology and RhoGEF domain containing G2 |
| chr7_+_137155481 | 0.04 |
ENSRNOT00000089895
|
Ano6
|
anoctamin 6 |
| chr15_+_3436361 | 0.03 |
ENSRNOT00000016387
|
Ap3m1
|
adaptor-related protein complex 3, mu 1 subunit |
| chr2_+_263212051 | 0.03 |
ENSRNOT00000089396
|
Negr1
|
neuronal growth regulator 1 |
| chr19_+_38422164 | 0.03 |
ENSRNOT00000017174
|
Nqo1
|
NAD(P)H quinone dehydrogenase 1 |
| chr3_+_72080630 | 0.03 |
ENSRNOT00000008911
|
Med19
|
mediator complex subunit 19 |
| chr7_+_24638208 | 0.03 |
ENSRNOT00000009135
|
Mterf2
|
mitochondrial transcription termination factor 2 |
| chr5_+_74727494 | 0.03 |
ENSRNOT00000076683
|
Rn50_5_0791.1
|
|
| chr6_-_127337791 | 0.03 |
ENSRNOT00000032015
|
Ifi27l2b
|
interferon, alpha-inducible protein 27 like 2B |
| chr5_+_141530211 | 0.03 |
ENSRNOT00000056546
|
4933427I04Rik
|
Riken cDNA 4933427I04 gene |
| chr1_-_199655147 | 0.03 |
ENSRNOT00000026979
|
LOC103691238
|
zinc finger protein 239-like |
| chr14_+_82350734 | 0.03 |
ENSRNOT00000023259
|
Tmem129
|
transmembrane protein 129 |
| chrX_+_908044 | 0.03 |
ENSRNOT00000072087
|
Zfp300
|
zinc finger protein 300 |
| chr14_-_77810147 | 0.03 |
ENSRNOT00000035427
|
Cytl1
|
cytokine like 1 |
| chr1_+_87655618 | 0.03 |
ENSRNOT00000035994
ENSRNOT00000073521 |
Zfp84
|
zinc finger protein 84 |
| chr6_-_88917070 | 0.03 |
ENSRNOT00000000767
|
Mdga2
|
MAM domain containing glycosylphosphatidylinositol anchor 2 |
| chr16_-_59721227 | 0.03 |
ENSRNOT00000015166
|
Prag1
|
PEAK1 related kinase activating pseudokinase 1 |
| chr8_-_48672732 | 0.03 |
ENSRNOT00000079275
|
Hmbs
|
hydroxymethylbilane synthase |
| chr4_-_9060554 | 0.03 |
ENSRNOT00000046248
|
AABR07059215.1
|
|
| chr2_-_199354793 | 0.03 |
ENSRNOT00000023548
|
Bcl9
|
B-cell CLL/lymphoma 9 |
| chr1_-_87777668 | 0.02 |
ENSRNOT00000037288
|
AABR07002870.1
|
|
| chr1_-_67284864 | 0.02 |
ENSRNOT00000082908
|
LOC691661
|
similar to zinc finger and SCAN domain containing 4 |
| chr9_-_30321323 | 0.02 |
ENSRNOT00000018234
|
Fam135a
|
family with sequence similarity 135, member A |
| chr19_-_10976396 | 0.02 |
ENSRNOT00000073239
|
Nlrc5
|
NLR family, CARD domain containing 5 |
| chrM_+_3904 | 0.02 |
ENSRNOT00000040993
|
Mt-nd2
|
mitochondrially encoded NADH dehydrogenase 2 |
| chr15_-_19282873 | 0.02 |
ENSRNOT00000075184
|
NEWGENE_1359268
|
Smad nuclear interacting protein 1 |
| chr7_-_138483612 | 0.02 |
ENSRNOT00000085620
|
Slc38a4
|
solute carrier family 38, member 4 |
| chr4_+_152437966 | 0.02 |
ENSRNOT00000013070
|
Rad52
|
RAD52 homolog, DNA repair protein |
| chr8_-_14880644 | 0.02 |
ENSRNOT00000015977
|
Fat3
|
FAT atypical cadherin 3 |
| chr10_-_90657185 | 0.02 |
ENSRNOT00000003685
|
Ccdc43
|
coiled-coil domain containing 43 |
| chr2_-_13696426 | 0.02 |
ENSRNOT00000046251
|
Rasa1
|
RAS p21 protein activator 1 |
| chr1_-_167093560 | 0.02 |
ENSRNOT00000027301
|
Il18bp
|
interleukin 18 binding protein |
| chr1_+_234252757 | 0.02 |
ENSRNOT00000091814
|
Rorb
|
RAR-related orphan receptor B |
| chr14_+_109533792 | 0.02 |
ENSRNOT00000067690
|
LOC108348154
|
ankyrin repeat domain-containing protein 29-like |
| chr2_-_122756842 | 0.02 |
ENSRNOT00000080181
|
Ccdc144b
|
coiled-coil domain containing 144B |
| chr10_-_73690860 | 0.02 |
ENSRNOT00000004876
ENSRNOT00000075843 |
Ints2
|
integrator complex subunit 2 |
| chr3_-_120373500 | 0.02 |
ENSRNOT00000067727
|
Nphp1
|
nephrocystin 1 |
| chrX_-_15347591 | 0.02 |
ENSRNOT00000037066
|
Timm17b
|
translocase of inner mitochondrial membrane 17b |
| chr5_+_119727839 | 0.02 |
ENSRNOT00000014354
|
Cachd1
|
cache domain containing 1 |
| chr4_-_155561324 | 0.02 |
ENSRNOT00000085447
|
Slc2a3
|
solute carrier family 2 member 3 |
| chrX_-_17252877 | 0.02 |
ENSRNOT00000060213
|
LOC102547118
|
retinitis pigmentosa 1-like 1 protein-like |
| chr6_+_99883959 | 0.02 |
ENSRNOT00000010588
|
Fntb
|
farnesyltransferase, CAAX box, beta |
| chr8_-_71337746 | 0.02 |
ENSRNOT00000021554
|
Zfp609
|
zinc finger protein 609 |
| chr20_+_3363737 | 0.02 |
ENSRNOT00000079654
|
Atat1
|
alpha tubulin acetyltransferase 1 |
| chr5_+_118574801 | 0.02 |
ENSRNOT00000035949
|
Ube2u
|
ubiquitin-conjugating enzyme E2U (putative) |
| chr15_-_103340239 | 0.02 |
ENSRNOT00000067309
ENSRNOT00000088909 |
Tgds
|
TDP-glucose 4,6-dehydratase |
| chr9_+_95274707 | 0.02 |
ENSRNOT00000045163
|
Ugt1a5
|
UDP glucuronosyltransferase family 1 member A5 |
| chr2_-_261402880 | 0.01 |
ENSRNOT00000052396
ENSRNOT00000077839 ENSRNOT00000088561 |
Fpgt
|
fucose-1-phosphate guanylyltransferase |
| chr7_+_140608434 | 0.01 |
ENSRNOT00000001163
|
AC114446.1
|
|
| chr20_+_27975549 | 0.01 |
ENSRNOT00000092075
|
Lims1
|
LIM zinc finger domain containing 1 |
| chr10_-_70339578 | 0.01 |
ENSRNOT00000076618
|
Slfn13
|
schlafen family member 13 |
| chr3_+_2262253 | 0.01 |
ENSRNOT00000042100
ENSRNOT00000061303 ENSRNOT00000048137 ENSRNOT00000048353 ENSRNOT00000012129 |
Nsmf
|
NMDA receptor synaptonuclear signaling and neuronal migration factor |
| chr7_+_23012070 | 0.01 |
ENSRNOT00000042460
|
LOC300308
|
similar to hypothetical protein 4930509O22 |
| chr3_+_35175313 | 0.01 |
ENSRNOT00000081744
|
Kif5c
|
kinesin family member 5C |
| chr14_-_70098021 | 0.01 |
ENSRNOT00000004912
|
Med28
|
mediator complex subunit 28 |
| chrX_+_65800059 | 0.01 |
ENSRNOT00000076428
|
Gpr165
|
G protein-coupled receptor 165 |
| chr9_-_113846053 | 0.01 |
ENSRNOT00000017253
|
LOC100910483
|
ankyrin repeat domain-containing protein 12-like |
| chr13_+_47454591 | 0.01 |
ENSRNOT00000005791
|
LOC498222
|
similar to specifically androgen-regulated protein |
| chr1_+_64031859 | 0.01 |
ENSRNOT00000090873
|
Mboat7l1
|
membrane bound O-acyltransferase domain containing 7-like 1 |
| chr11_-_38420032 | 0.01 |
ENSRNOT00000002209
ENSRNOT00000080681 |
C2cd2
|
C2 calcium-dependent domain containing 2 |
| chr4_+_147844658 | 0.01 |
ENSRNOT00000042673
|
H1foo
|
H1 histone family, member O, oocyte-specific |
| chr2_-_84436912 | 0.01 |
ENSRNOT00000059565
|
Ankrd33b
|
ankyrin repeat domain 33B |
| chrX_+_35599258 | 0.01 |
ENSRNOT00000072627
ENSRNOT00000005061 |
Cdkl5
|
cyclin-dependent kinase-like 5 |
| chr3_-_151106557 | 0.01 |
ENSRNOT00000025657
|
Gss
|
glutathione synthetase |
| chr3_+_56355431 | 0.01 |
ENSRNOT00000037188
|
Myo3b
|
myosin IIIB |
| chr5_-_172335892 | 0.01 |
ENSRNOT00000018494
|
Tnfrsf14
|
TNF receptor superfamily member 14 |
| chr2_-_88314254 | 0.01 |
ENSRNOT00000038546
|
Car13
|
carbonic anhydrase 13 |
| chr15_+_37418885 | 0.01 |
ENSRNOT00000043836
|
RGD1565170
|
similar to 60S ribosomal protein L23a |
| chr17_-_78561613 | 0.01 |
ENSRNOT00000020220
|
Fam107b
|
family with sequence similarity 107, member B |
| chr12_-_2661565 | 0.01 |
ENSRNOT00000064859
|
Cd209f
|
CD209f antigen |
| chr5_+_133819726 | 0.01 |
ENSRNOT00000081075
|
Stil
|
Scl/Tal1 interrupting locus |
| chr2_-_178521038 | 0.01 |
ENSRNOT00000033107
|
Rxfp1
|
relaxin/insulin-like family peptide receptor 1 |
| chr2_+_55775274 | 0.01 |
ENSRNOT00000018545
|
C9
|
complement C9 |
| chr12_+_1889331 | 0.01 |
ENSRNOT00000066281
|
Arhgef18
|
Rho/Rac guanine nucleotide exchange factor 18 |
| chr2_+_252090669 | 0.01 |
ENSRNOT00000020656
|
Lpar3
|
lysophosphatidic acid receptor 3 |
| chr1_+_61268248 | 0.01 |
ENSRNOT00000082730
|
LOC102552527
|
zinc finger protein 420-like |
| chr9_+_67699379 | 0.01 |
ENSRNOT00000091237
ENSRNOT00000088183 |
Ctla4
|
cytotoxic T-lymphocyte-associated protein 4 |
| chr1_-_67134827 | 0.01 |
ENSRNOT00000045214
|
Vom1r45
|
vomeronasal 1 receptor 45 |
| chr1_+_84679614 | 0.01 |
ENSRNOT00000025581
|
AABR07002774.1
|
|
| chr2_+_200076054 | 0.01 |
ENSRNOT00000025327
|
Sec22b
|
SEC22 homolog B, vesicle trafficking protein |
| chr4_-_14490446 | 0.01 |
ENSRNOT00000009132
|
Sema3c
|
semaphorin 3C |
| chr3_+_153491151 | 0.01 |
ENSRNOT00000047047
|
Manbal
|
mannosidase beta like |
| chr2_-_183213228 | 0.01 |
ENSRNOT00000067618
|
Trim2
|
tripartite motif-containing 2 |
| chr16_+_19874034 | 0.01 |
ENSRNOT00000023579
|
Mrpl34
|
mitochondrial ribosomal protein L34 |
| chr7_-_123156558 | 0.01 |
ENSRNOT00000005943
|
Polr3h
|
RNA polymerase III subunit H |
| chr9_+_30419001 | 0.01 |
ENSRNOT00000018116
|
Col9a1
|
collagen type IX alpha 1 chain |
| chr5_-_138462864 | 0.01 |
ENSRNOT00000011325
|
Ccdc30
|
coiled-coil domain containing 30 |
| chr4_-_165192647 | 0.00 |
ENSRNOT00000086461
|
Klra5
|
killer cell lectin-like receptor, subfamily A, member 5 |
| chr5_+_159484370 | 0.00 |
ENSRNOT00000010593
|
Sdhb
|
succinate dehydrogenase complex iron sulfur subunit B |
| chr8_-_53816447 | 0.00 |
ENSRNOT00000011454
|
Ttc12
|
tetratricopeptide repeat domain 12 |
| chrX_-_1346181 | 0.00 |
ENSRNOT00000042736
|
LOC102555453
|
60S ribosomal protein L12-like |
| chrX_-_18890090 | 0.00 |
ENSRNOT00000041039
|
Klf8
|
Kruppel-like factor 8 |
| chr11_-_81379871 | 0.00 |
ENSRNOT00000089294
|
Eif4a2
|
eukaryotic translation initiation factor 4A2 |
| chr9_-_121972055 | 0.00 |
ENSRNOT00000089735
|
AABR07068851.1
|
clusterin-like protein 1 |
| chr3_-_5617225 | 0.00 |
ENSRNOT00000008327
|
Tmem8c
|
transmembrane protein 8C |
| chr4_+_117780533 | 0.00 |
ENSRNOT00000021211
|
Tprkb
|
Tp53rk binding protein |
| chr14_+_113867209 | 0.00 |
ENSRNOT00000067591
|
Ccdc88a
|
coiled coil domain containing 88A |
| chr1_+_22332090 | 0.00 |
ENSRNOT00000091252
|
AABR07000663.1
|
trace amine-associated receptor 8c (Taar8c), mRNA |
| chr3_-_161272460 | 0.00 |
ENSRNOT00000020740
|
Acot8
|
acyl-CoA thioesterase 8 |
| chr5_-_138222534 | 0.00 |
ENSRNOT00000009691
|
Zfp691
|
zinc finger protein 691 |
| chr4_+_9882904 | 0.00 |
ENSRNOT00000016909
|
Dnajc2
|
DnaJ heat shock protein family (Hsp40) member C2 |
| chr3_-_52510507 | 0.00 |
ENSRNOT00000091259
|
Scn1a
|
sodium voltage-gated channel alpha subunit 1 |
| chr4_+_45034123 | 0.00 |
ENSRNOT00000078255
|
ST7
|
suppression of tumorigenicity 7 |
| chr18_+_58325319 | 0.00 |
ENSRNOT00000065802
ENSRNOT00000077726 |
Napg
|
NSF attachment protein gamma |
| chr1_-_146289465 | 0.00 |
ENSRNOT00000017362
|
Abhd17c
|
abhydrolase domain containing 17C |
| chrX_-_40086870 | 0.00 |
ENSRNOT00000010027
|
Smpx
|
small muscle protein, X-linked |
| chr11_-_35749464 | 0.00 |
ENSRNOT00000078818
ENSRNOT00000078425 |
Erg
|
ERG, ETS transcription factor |
| chr14_+_77079402 | 0.00 |
ENSRNOT00000042200
|
Slc2a9
|
solute carrier family 2 member 9 |
| chr4_+_174692331 | 0.00 |
ENSRNOT00000011770
|
Plekha5
|
pleckstrin homology domain containing A5 |
| chr7_-_135850542 | 0.00 |
ENSRNOT00000038032
|
Twf1
|
twinfilin actin-binding protein 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Rhox11
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0034395 | regulation of transcription from RNA polymerase II promoter in response to iron(GO:0034395) |
| 0.0 | 0.1 | GO:1904823 | purine nucleobase transmembrane transport(GO:1904823) |
| 0.0 | 0.0 | GO:2000298 | regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 0.0 | 0.1 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 0.0 | 0.0 | GO:1902304 | positive regulation of potassium ion export(GO:1902304) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0015265 | urea channel activity(GO:0015265) |