Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
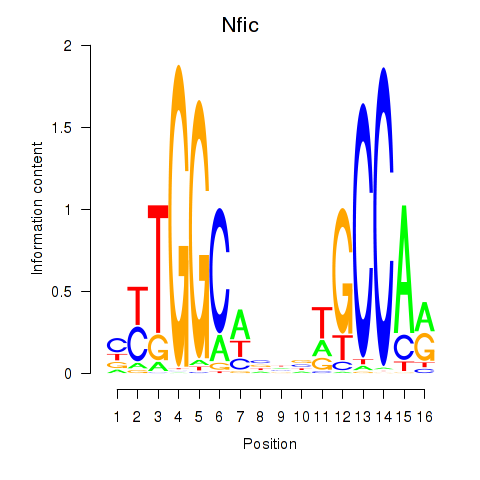
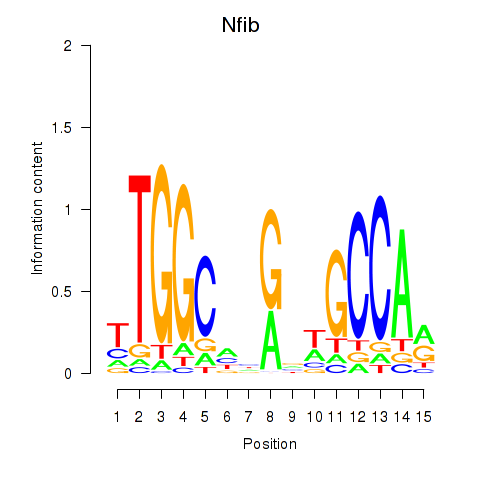
Results for Nfic_Nfib
Z-value: 0.78
Transcription factors associated with Nfic_Nfib
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nfic
|
ENSRNOG00000004505 | nuclear factor I/C |
|
Nfib
|
ENSRNOG00000009795 | nuclear factor I/B |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nfib | rn6_v1_chr5_-_100647298_100647298 | -0.42 | 4.8e-01 | Click! |
| Nfic | rn6_v1_chr7_+_11152038_11152038 | 0.42 | 4.8e-01 | Click! |
Activity profile of Nfic_Nfib motif
Sorted Z-values of Nfic_Nfib motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_80416273 | 1.21 |
ENSRNOT00000023457
|
Bloc1s3
|
biogenesis of lysosomal organelles complex-1, subunit 3 |
| chr4_-_147163467 | 0.68 |
ENSRNOT00000010748
|
Timp4
|
tissue inhibitor of metalloproteinase 4 |
| chr15_-_34550850 | 0.66 |
ENSRNOT00000027794
ENSRNOT00000090228 |
Cbln3
|
cerebellin 3 precursor |
| chr4_+_8256611 | 0.58 |
ENSRNOT00000061894
|
AABR07059198.1
|
|
| chr5_+_154522119 | 0.56 |
ENSRNOT00000072618
|
E2f2
|
E2F transcription factor 2 |
| chr20_+_5008508 | 0.55 |
ENSRNOT00000001153
|
Vwa7
|
von Willebrand factor A domain containing 7 |
| chr14_+_83724933 | 0.47 |
ENSRNOT00000029848
|
Pla2g3
|
phospholipase A2, group III |
| chr20_-_9855443 | 0.47 |
ENSRNOT00000090275
ENSRNOT00000066266 |
Tff3
|
trefoil factor 3 |
| chrX_-_23187341 | 0.46 |
ENSRNOT00000000180
|
Alas2
|
5'-aminolevulinate synthase 2 |
| chr4_+_7377563 | 0.45 |
ENSRNOT00000084826
|
Kcnh2
|
potassium voltage-gated channel subfamily H member 2 |
| chr15_-_27815261 | 0.43 |
ENSRNOT00000032992
|
Klhl33
|
kelch-like family member 33 |
| chr5_-_134526089 | 0.42 |
ENSRNOT00000013321
|
Cyp4b1
|
cytochrome P450, family 4, subfamily b, polypeptide 1 |
| chr7_-_119441487 | 0.37 |
ENSRNOT00000067635
|
Pvalb
|
parvalbumin |
| chr4_+_169161585 | 0.36 |
ENSRNOT00000079785
|
Emp1
|
epithelial membrane protein 1 |
| chr16_+_54291251 | 0.35 |
ENSRNOT00000079006
|
Mtus1
|
microtubule associated tumor suppressor 1 |
| chr19_+_55669626 | 0.34 |
ENSRNOT00000033352
|
Cdh15
|
cadherin 15 |
| chr1_-_124803363 | 0.33 |
ENSRNOT00000066380
|
Klf13
|
Kruppel-like factor 13 |
| chr2_-_195888216 | 0.31 |
ENSRNOT00000056436
|
Tuft1
|
tuftelin 1 |
| chr4_-_85386231 | 0.30 |
ENSRNOT00000015316
|
Inmt
|
indolethylamine N-methyltransferase |
| chr17_+_6909728 | 0.30 |
ENSRNOT00000061231
|
LOC681410
|
hypothetical protein LOC681410 |
| chr10_+_56662242 | 0.30 |
ENSRNOT00000086919
|
Asgr1
|
asialoglycoprotein receptor 1 |
| chr5_-_156734541 | 0.30 |
ENSRNOT00000021036
|
LOC100909857
|
cytidine deaminase-like |
| chr7_-_12609694 | 0.30 |
ENSRNOT00000093287
ENSRNOT00000079485 |
Kiss1r
|
KISS1 receptor |
| chr1_+_147021436 | 0.28 |
ENSRNOT00000042490
|
F8a1
|
coagulation factor VIII-associated 1 |
| chr14_+_84237520 | 0.27 |
ENSRNOT00000083580
|
Gal3st1
|
galactose-3-O-sulfotransferase 1 |
| chr4_+_169147243 | 0.26 |
ENSRNOT00000011580
|
Emp1
|
epithelial membrane protein 1 |
| chr17_-_61332391 | 0.26 |
ENSRNOT00000034599
|
LOC100362965
|
SNRPN upstream reading frame protein-like |
| chr7_+_116632506 | 0.26 |
ENSRNOT00000009811
|
Gpihbp1
|
glycosylphosphatidylinositol anchored high density lipoprotein binding protein 1 |
| chr18_+_79406381 | 0.26 |
ENSRNOT00000022303
ENSRNOT00000058295 ENSRNOT00000058296 ENSRNOT00000022280 |
Mbp
|
myelin basic protein |
| chr2_-_195908037 | 0.24 |
ENSRNOT00000079776
|
Tuft1
|
tuftelin 1 |
| chr5_+_136669674 | 0.23 |
ENSRNOT00000048770
|
Slc6a9
|
solute carrier family 6 member 9 |
| chr4_-_100232559 | 0.22 |
ENSRNOT00000016984
|
Vamp5
|
vesicle-associated membrane protein 5 |
| chr11_+_31002451 | 0.21 |
ENSRNOT00000002838
|
Eva1c
|
eva-1 homolog C |
| chr17_+_80882666 | 0.21 |
ENSRNOT00000024430
|
Vim
|
vimentin |
| chr9_+_117795132 | 0.21 |
ENSRNOT00000086943
|
Akain1
|
A-kinase anchor inhibitor 1 |
| chr10_-_104681377 | 0.21 |
ENSRNOT00000046026
|
Trim65
|
tripartite motif-containing 65 |
| chr2_+_95320283 | 0.20 |
ENSRNOT00000015537
|
Hey1
|
hes-related family bHLH transcription factor with YRPW motif 1 |
| chr10_-_104163634 | 0.20 |
ENSRNOT00000005198
|
Mif4gd
|
MIF4G domain containing |
| chr1_-_89543967 | 0.19 |
ENSRNOT00000079631
|
Hpn
|
hepsin |
| chr8_-_117932518 | 0.19 |
ENSRNOT00000028130
|
Camp
|
cathelicidin antimicrobial peptide |
| chr5_-_138697641 | 0.18 |
ENSRNOT00000012062
|
Guca2b
|
guanylate cyclase activator 2B |
| chr10_-_45534570 | 0.18 |
ENSRNOT00000058362
|
Gjc2
|
gap junction protein, gamma 2 |
| chr4_+_62220736 | 0.18 |
ENSRNOT00000086377
|
Cald1
|
caldesmon 1 |
| chr3_+_14467330 | 0.17 |
ENSRNOT00000078939
|
Gsn
|
gelsolin |
| chr10_+_49231730 | 0.16 |
ENSRNOT00000065335
|
Trim16
|
tripartite motif-containing 16 |
| chr1_+_221792221 | 0.16 |
ENSRNOT00000054828
|
Nrxn2
|
neurexin 2 |
| chr10_-_15166457 | 0.16 |
ENSRNOT00000026676
|
Metrn
|
meteorin, glial cell differentiation regulator |
| chr4_-_123494742 | 0.15 |
ENSRNOT00000073268
|
Slc41a3
|
solute carrier family 41, member 3 |
| chr2_+_266141581 | 0.15 |
ENSRNOT00000078187
ENSRNOT00000051951 |
Rpe65
|
RPE65, retinoid isomerohydrolase |
| chr13_+_47454591 | 0.15 |
ENSRNOT00000005791
|
LOC498222
|
similar to specifically androgen-regulated protein |
| chr5_-_12526962 | 0.15 |
ENSRNOT00000092104
|
St18
|
suppression of tumorigenicity 18 |
| chr9_+_50526811 | 0.14 |
ENSRNOT00000036990
|
RGD1305645
|
similar to RIKEN cDNA 1500015O10 |
| chrX_-_106558366 | 0.14 |
ENSRNOT00000042126
|
Bex2
|
brain expressed X-linked 2 |
| chr3_-_160853650 | 0.14 |
ENSRNOT00000018844
|
Matn4
|
matrilin 4 |
| chr1_-_221281180 | 0.14 |
ENSRNOT00000028379
|
Cdc42ep2
|
CDC42 effector protein 2 |
| chr3_-_2689084 | 0.13 |
ENSRNOT00000020926
|
Ptgds
|
prostaglandin D2 synthase |
| chr12_+_24978483 | 0.13 |
ENSRNOT00000040069
|
Eln
|
elastin |
| chr10_-_62648844 | 0.13 |
ENSRNOT00000034282
|
Abhd15
|
abhydrolase domain containing 15 |
| chr13_+_89524329 | 0.13 |
ENSRNOT00000004279
|
Mpz
|
myelin protein zero |
| chr8_-_45375435 | 0.13 |
ENSRNOT00000010873
|
Ubash3b
|
ubiquitin associated and SH3 domain containing, B |
| chr15_-_60289763 | 0.13 |
ENSRNOT00000038579
|
Fam216b
|
family with sequence similarity 216, member B |
| chr3_-_153188915 | 0.13 |
ENSRNOT00000079893
|
Soga1
|
suppressor of glucose, autophagy associated 1 |
| chr5_-_155772040 | 0.13 |
ENSRNOT00000036788
|
Cela3b
|
chymotrypsin-like elastase family, member 3B |
| chr9_+_73493027 | 0.12 |
ENSRNOT00000074427
ENSRNOT00000089478 |
Unc80
|
unc-80 homolog, NALCN activator |
| chr5_+_162031722 | 0.12 |
ENSRNOT00000020483
|
Lrrc38
|
leucine rich repeat containing 38 |
| chr11_-_39448503 | 0.12 |
ENSRNOT00000047347
|
LOC102553613
|
SH3 domain-binding glutamic acid-rich protein-like |
| chr1_-_188713270 | 0.12 |
ENSRNOT00000082192
ENSRNOT00000065892 |
Gprc5b
|
G protein-coupled receptor, class C, group 5, member B |
| chr3_-_8766433 | 0.12 |
ENSRNOT00000021865
|
Kyat1
|
kynurenine aminotransferase 1 |
| chr10_-_104575890 | 0.12 |
ENSRNOT00000050223
|
H3f3b
|
H3 histone family member 3B |
| chr16_-_74122889 | 0.12 |
ENSRNOT00000025763
|
Plat
|
plasminogen activator, tissue type |
| chr6_+_107245820 | 0.11 |
ENSRNOT00000012757
|
Papln
|
papilin, proteoglycan-like sulfated glycoprotein |
| chr14_-_8510138 | 0.11 |
ENSRNOT00000080758
|
Arhgap24
|
Rho GTPase activating protein 24 |
| chr1_+_219764001 | 0.11 |
ENSRNOT00000082388
|
Pc
|
pyruvate carboxylase |
| chr5_+_135029955 | 0.11 |
ENSRNOT00000074860
|
LOC100911669
|
uncharacterized LOC100911669 |
| chr18_+_70263359 | 0.11 |
ENSRNOT00000019573
|
Cfap53
|
cilia and flagella associated protein 53 |
| chr3_+_63510293 | 0.11 |
ENSRNOT00000058093
|
Dfnb59
|
deafness, autosomal recessive 59 |
| chr5_-_160620334 | 0.10 |
ENSRNOT00000019171
|
Tmem51
|
transmembrane protein 51 |
| chr9_-_80167033 | 0.10 |
ENSRNOT00000023530
|
Igfbp5
|
insulin-like growth factor binding protein 5 |
| chr20_+_13778178 | 0.10 |
ENSRNOT00000058314
|
Gstt4
|
glutathione S-transferase, theta 4 |
| chr7_+_130296897 | 0.10 |
ENSRNOT00000044854
|
Adm2
|
adrenomedullin 2 |
| chr6_-_111222858 | 0.10 |
ENSRNOT00000074707
|
Tmed8
|
transmembrane p24 trafficking protein 8 |
| chr12_+_50407843 | 0.10 |
ENSRNOT00000073763
|
Cryba4
|
crystallin, beta A4 |
| chr18_+_32273770 | 0.10 |
ENSRNOT00000087408
|
Fgf1
|
fibroblast growth factor 1 |
| chr3_+_109862117 | 0.10 |
ENSRNOT00000083351
ENSRNOT00000070912 |
Thbs1
|
thrombospondin 1 |
| chr4_+_145427367 | 0.10 |
ENSRNOT00000037788
|
Il17rc
|
interleukin 17 receptor C |
| chr1_+_89162639 | 0.10 |
ENSRNOT00000028508
|
Atp4a
|
ATPase H+/K+ transporting alpha subunit |
| chr13_-_50497466 | 0.10 |
ENSRNOT00000076072
|
Etnk2
|
ethanolamine kinase 2 |
| chr2_+_225827504 | 0.10 |
ENSRNOT00000018343
|
Gclm
|
glutamate cysteine ligase, modifier subunit |
| chr1_-_91042230 | 0.10 |
ENSRNOT00000073107
|
LOC103690054
|
DNA-directed RNA polymerase II subunit RPB9 |
| chr1_+_221099998 | 0.10 |
ENSRNOT00000028262
|
Ltbp3
|
latent transforming growth factor beta binding protein 3 |
| chr9_+_98668231 | 0.10 |
ENSRNOT00000027528
|
Asb1
|
ankyrin repeat and SOCS box-containing 1 |
| chr4_+_117962319 | 0.10 |
ENSRNOT00000057441
|
Tgfa
|
transforming growth factor alpha |
| chr7_-_144993652 | 0.10 |
ENSRNOT00000087748
|
Itga5
|
integrin subunit alpha 5 |
| chr3_+_172195844 | 0.09 |
ENSRNOT00000034915
|
Npepl1
|
aminopeptidase-like 1 |
| chr10_-_90999506 | 0.09 |
ENSRNOT00000034401
|
Gfap
|
glial fibrillary acidic protein |
| chrX_+_63343546 | 0.09 |
ENSRNOT00000076315
|
Klhl15
|
kelch-like family member 15 |
| chr10_+_13836128 | 0.09 |
ENSRNOT00000012720
|
Pgp
|
phosphoglycolate phosphatase |
| chr1_+_154606490 | 0.09 |
ENSRNOT00000024095
|
Ccdc89
|
coiled-coil domain containing 89 |
| chr3_+_3788583 | 0.09 |
ENSRNOT00000084567
|
Gpsm1
|
G-protein signaling modulator 1 |
| chr8_-_130429132 | 0.09 |
ENSRNOT00000026261
|
Hhatl
|
hedgehog acyltransferase-like |
| chr8_+_116096458 | 0.09 |
ENSRNOT00000021188
|
RGD1307461
|
similar to RIKEN cDNA 6430571L13 gene; similar to g20 protein |
| chr5_+_59008933 | 0.09 |
ENSRNOT00000023060
|
Car9
|
carbonic anhydrase 9 |
| chr4_-_148845267 | 0.09 |
ENSRNOT00000037397
|
Tmem72
|
transmembrane protein 72 |
| chr3_-_3574787 | 0.09 |
ENSRNOT00000087651
|
Nacc2
|
NACC family member 2 |
| chr8_+_95968652 | 0.08 |
ENSRNOT00000015057
|
Nt5e
|
5' nucleotidase, ecto |
| chr1_+_86938138 | 0.08 |
ENSRNOT00000075601
|
Ccer2
|
coiled-coil glutamate-rich protein 2 |
| chr1_-_101514547 | 0.08 |
ENSRNOT00000079633
|
Ppp1r15a
|
protein phosphatase 1, regulatory subunit 15A |
| chr17_-_32558180 | 0.08 |
ENSRNOT00000022681
|
Serpinb1b
|
serine (or cysteine) peptidase inhibitor, clade B, member 1b |
| chr7_-_15225309 | 0.08 |
ENSRNOT00000075589
ENSRNOT00000066095 |
Cyp4f6
|
cytochrome P450, family 4, subfamily f, polypeptide 6 |
| chr1_-_156327352 | 0.08 |
ENSRNOT00000074282
|
LOC100911776
|
coiled-coil domain-containing protein 89-like |
| chr1_+_89008117 | 0.08 |
ENSRNOT00000028401
|
Hspb6
|
heat shock protein family B (small) member 6 |
| chrX_+_106823491 | 0.08 |
ENSRNOT00000045997
|
Bex3
|
brain expressed X-linked 3 |
| chr14_+_80403001 | 0.08 |
ENSRNOT00000012109
|
Cpz
|
carboxypeptidase Z |
| chr15_-_52210746 | 0.08 |
ENSRNOT00000046054
|
Bmp1
|
bone morphogenetic protein 1 |
| chr2_+_226900619 | 0.08 |
ENSRNOT00000019638
|
Pde5a
|
phosphodiesterase 5A |
| chr2_-_187854363 | 0.08 |
ENSRNOT00000092993
|
Lmna
|
lamin A/C |
| chr18_-_25997555 | 0.08 |
ENSRNOT00000027755
|
Stard4
|
StAR-related lipid transfer domain containing 4 |
| chr6_-_45669148 | 0.08 |
ENSRNOT00000010092
|
Rsad2
|
radical S-adenosyl methionine domain containing 2 |
| chr5_-_144345531 | 0.08 |
ENSRNOT00000014721
|
Tekt2
|
tektin 2 |
| chr3_+_11679530 | 0.08 |
ENSRNOT00000074562
ENSRNOT00000071801 |
Eng
|
endoglin |
| chr9_-_9985630 | 0.08 |
ENSRNOT00000071780
|
Crb3
|
crumbs 3, cell polarity complex component |
| chr12_-_21761487 | 0.08 |
ENSRNOT00000082910
|
LOC102550456
|
TSC22 domain family protein 4-like |
| chr15_+_28287024 | 0.08 |
ENSRNOT00000064907
|
Mettl17
|
methyltransferase like 17 |
| chr4_+_55720010 | 0.08 |
ENSRNOT00000063987
|
Fscn3
|
fascin actin-bundling protein 3 |
| chr13_+_89919667 | 0.07 |
ENSRNOT00000006196
|
Itln1
|
intelectin 1 |
| chr1_+_12823363 | 0.07 |
ENSRNOT00000086790
|
Cited2
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2 |
| chr17_-_70548071 | 0.07 |
ENSRNOT00000073144
|
Il2ra
|
interleukin 2 receptor subunit alpha |
| chr1_-_142020525 | 0.07 |
ENSRNOT00000042558
|
Cib1
|
calcium and integrin binding 1 |
| chr1_-_162385575 | 0.07 |
ENSRNOT00000016540
|
Thrsp
|
thyroid hormone responsive |
| chr10_-_18942540 | 0.07 |
ENSRNOT00000007187
|
Kcnip1
|
potassium voltage-gated channel interacting protein 1 |
| chr14_+_10534423 | 0.07 |
ENSRNOT00000002983
|
Hpse
|
heparanase |
| chr9_+_10941613 | 0.07 |
ENSRNOT00000070794
|
Sema6b
|
semaphorin 6B |
| chr14_+_46001849 | 0.07 |
ENSRNOT00000076611
|
Rell1
|
RELT-like 1 |
| chr7_-_70842405 | 0.07 |
ENSRNOT00000047449
|
Nxph4
|
neurexophilin 4 |
| chr13_+_90723092 | 0.07 |
ENSRNOT00000010146
|
Kcnj10
|
ATP-sensitive inward rectifier potassium channel 10 |
| chr5_-_58950373 | 0.07 |
ENSRNOT00000060442
|
Cd72
|
Cd72 molecule |
| chr1_+_219833299 | 0.06 |
ENSRNOT00000087432
|
Pc
|
pyruvate carboxylase |
| chr10_+_58875826 | 0.06 |
ENSRNOT00000020071
|
Fbxo39
|
F-box protein 39 |
| chr13_+_89622632 | 0.06 |
ENSRNOT00000004713
|
Adamts4
|
ADAM metallopeptidase with thrombospondin type 1 motif, 4 |
| chr5_-_126395668 | 0.06 |
ENSRNOT00000010396
|
Acot11
|
acyl-CoA thioesterase 11 |
| chr8_+_64481172 | 0.06 |
ENSRNOT00000015332
|
Pkm
|
pyruvate kinase, muscle |
| chr7_+_120465130 | 0.06 |
ENSRNOT00000016077
|
Pick1
|
protein interacting with PRKCA 1 |
| chr9_+_81655629 | 0.06 |
ENSRNOT00000088679
ENSRNOT00000057472 |
Slc11a1
|
solute carrier family 11 member 1 |
| chr2_+_233615739 | 0.06 |
ENSRNOT00000051009
|
Pitx2
|
paired-like homeodomain 2 |
| chr7_+_18409147 | 0.06 |
ENSRNOT00000086336
ENSRNOT00000011849 |
Adamts10
|
ADAM metallopeptidase with thrombospondin type 1 motif, 10 |
| chr20_-_2224463 | 0.06 |
ENSRNOT00000001019
|
Trim26
|
tripartite motif-containing 26 |
| chr12_+_22449985 | 0.06 |
ENSRNOT00000075951
|
Slc12a9
|
solute carrier family 12, member 9 |
| chr1_+_226993250 | 0.06 |
ENSRNOT00000054808
|
Ptgdr2
|
prostaglandin D2 receptor 2 |
| chr4_+_144382945 | 0.06 |
ENSRNOT00000007601
|
Cav3
|
caveolin 3 |
| chr10_+_17486579 | 0.06 |
ENSRNOT00000080404
|
Stk10
|
serine/threonine kinase 10 |
| chr16_+_21282467 | 0.06 |
ENSRNOT00000065345
|
Yjefn3
|
YjeF N-terminal domain containing 3 |
| chr5_-_156781291 | 0.06 |
ENSRNOT00000075128
|
Cda
|
cytidine deaminase |
| chr13_+_112031594 | 0.06 |
ENSRNOT00000008440
|
Lamb3
|
laminin subunit beta 3 |
| chr3_-_151224123 | 0.05 |
ENSRNOT00000026091
|
Trpc4ap
|
transient receptor potential cation channel, subfamily C, member 4 associated protein |
| chrX_+_33895769 | 0.05 |
ENSRNOT00000085993
ENSRNOT00000079365 |
Reps2
|
RALBP1 associated Eps domain containing protein 2 |
| chr16_-_83132785 | 0.05 |
ENSRNOT00000043449
|
Arhgef7
|
Rho guanine nucleotide exchange factor 7 |
| chr15_-_55277713 | 0.05 |
ENSRNOT00000023037
|
Itm2b
|
integral membrane protein 2B |
| chr20_-_5212624 | 0.05 |
ENSRNOT00000074261
|
LOC103689996
|
antigen peptide transporter 2 |
| chr5_-_155143539 | 0.05 |
ENSRNOT00000016983
|
Ephb2
|
Eph receptor B2 |
| chr1_+_164380577 | 0.05 |
ENSRNOT00000055321
|
Gdpd5
|
glycerophosphodiester phosphodiesterase domain containing 5 |
| chr16_+_49266903 | 0.05 |
ENSRNOT00000014704
|
Slc25a4
|
solute carrier family 25 member 4 |
| chr3_+_3389612 | 0.05 |
ENSRNOT00000041984
|
Rpl8
|
ribosomal protein L8 |
| chr12_-_22021851 | 0.05 |
ENSRNOT00000039280
|
Tsc22d4
|
TSC22 domain family, member 4 |
| chr5_+_138300107 | 0.05 |
ENSRNOT00000047151
|
Cldn19
|
claudin 19 |
| chr1_-_222177421 | 0.05 |
ENSRNOT00000078393
|
Esrra
|
estrogen related receptor, alpha |
| chr20_+_40769586 | 0.05 |
ENSRNOT00000001079
|
Fabp7
|
fatty acid binding protein 7 |
| chr5_+_152466331 | 0.05 |
ENSRNOT00000082841
|
LOC108351043
|
uncharacterized LOC108351043 |
| chr7_+_76980040 | 0.05 |
ENSRNOT00000050726
|
LOC108350501
|
40S ribosomal protein S29 |
| chr16_-_82100222 | 0.05 |
ENSRNOT00000023163
|
Atp11a
|
ATPase phospholipid transporting 11A |
| chr16_+_21178138 | 0.05 |
ENSRNOT00000027943
|
Gatad2a
|
GATA zinc finger domain containing 2A |
| chr20_+_7136007 | 0.05 |
ENSRNOT00000000580
|
Hmga1
|
high mobility group AT-hook 1 |
| chr18_-_62965538 | 0.05 |
ENSRNOT00000025164
|
Mppe1
|
metallophosphoesterase 1 |
| chr1_-_101514974 | 0.05 |
ENSRNOT00000044788
|
Ppp1r15a
|
protein phosphatase 1, regulatory subunit 15A |
| chr1_-_252550394 | 0.05 |
ENSRNOT00000083468
|
Acta2
|
actin, alpha 2, smooth muscle, aorta |
| chr10_-_13051154 | 0.05 |
ENSRNOT00000033457
|
AABR07029202.1
|
|
| chr3_+_14889510 | 0.05 |
ENSRNOT00000080760
|
Dab2ip
|
DAB2 interacting protein |
| chr4_+_88328061 | 0.05 |
ENSRNOT00000084775
|
Vom1r87
|
vomeronasal 1 receptor 87 |
| chr6_+_111076351 | 0.05 |
ENSRNOT00000089644
|
Tmem63c
|
transmembrane protein 63c |
| chr20_+_1749716 | 0.05 |
ENSRNOT00000048856
|
Olr1735
|
olfactory receptor 1735 |
| chr20_-_2020537 | 0.05 |
ENSRNOT00000011704
|
Zfp57
|
zinc finger protein 57 |
| chr5_-_61077627 | 0.05 |
ENSRNOT00000015338
|
Shb
|
SH2 domain containing adaptor protein B |
| chr7_+_12487361 | 0.05 |
ENSRNOT00000077146
|
Sbno2
|
strawberry notch homolog 2 |
| chr2_+_221823687 | 0.04 |
ENSRNOT00000072735
|
Dpyd
|
dihydropyrimidine dehydrogenase |
| chr2_+_61991742 | 0.04 |
ENSRNOT00000046715
|
LOC499544
|
LRRGT00154 |
| chr1_-_174350196 | 0.04 |
ENSRNOT00000055158
|
LOC499240
|
similar to predicted gene ICRFP703B1614Q5.5 |
| chr9_+_46962288 | 0.04 |
ENSRNOT00000082146
|
Il1r1
|
interleukin 1 receptor type 1 |
| chr10_+_90988732 | 0.04 |
ENSRNOT00000003892
|
FAM187A
|
family with sequence similarity 187, member A |
| chr1_-_170318935 | 0.04 |
ENSRNOT00000024119
|
Prkcdbp
|
protein kinase C, delta binding protein |
| chr14_+_18983853 | 0.04 |
ENSRNOT00000003836
|
Rassf6
|
Ras association domain family member 6 |
| chr4_+_55715744 | 0.04 |
ENSRNOT00000010429
|
Arf5
|
ADP-ribosylation factor 5 |
| chr17_+_32904119 | 0.04 |
ENSRNOT00000059854
|
Serpinb1a
|
serpin family B member 1A |
| chr1_-_8751198 | 0.04 |
ENSRNOT00000030511
|
Adgrg6
|
adhesion G protein-coupled receptor G6 |
| chr5_-_33182147 | 0.04 |
ENSRNOT00000080358
|
Maged2
|
MAGE family member D2 |
| chr1_-_222197252 | 0.04 |
ENSRNOT00000028707
|
Gpr137
|
G protein-coupled receptor 137 |
| chr8_-_55177818 | 0.04 |
ENSRNOT00000013960
|
Hspb2
|
heat shock protein family B (small) member 2 |
| chr5_+_153269526 | 0.04 |
ENSRNOT00000091494
|
Syf2
|
SYF2 pre-mRNA-splicing factor |
| chr7_+_139698148 | 0.04 |
ENSRNOT00000078579
|
Pfkm
|
phosphofructokinase, muscle |
| chr7_-_143420027 | 0.04 |
ENSRNOT00000082863
|
Krt2
|
keratin 2 |
| chr6_-_62798384 | 0.04 |
ENSRNOT00000093200
|
Ndufa10l1
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex 10-like 1 |
| chr8_+_32188617 | 0.04 |
ENSRNOT00000081145
|
Zbtb44
|
zinc finger and BTB domain containing 44 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Nfic_Nfib
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.2 | GO:0060155 | platelet dense granule organization(GO:0060155) |
| 0.2 | 0.5 | GO:1902303 | negative regulation of potassium ion export(GO:1902303) |
| 0.1 | 0.5 | GO:1901423 | response to benzene(GO:1901423) |
| 0.1 | 0.6 | GO:1990086 | lens fiber cell apoptotic process(GO:1990086) |
| 0.1 | 0.6 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.1 | 0.3 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.1 | 0.4 | GO:0046087 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 0.1 | 0.3 | GO:0010986 | positive regulation of lipoprotein particle clearance(GO:0010986) |
| 0.1 | 0.3 | GO:1904209 | regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) positive regulation of metalloendopeptidase activity(GO:1904685) |
| 0.1 | 0.1 | GO:1905072 | cardiac jelly development(GO:1905072) |
| 0.1 | 0.2 | GO:2000820 | negative regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000820) |
| 0.1 | 0.2 | GO:0034769 | basement membrane disassembly(GO:0034769) |
| 0.1 | 0.4 | GO:0018879 | biphenyl metabolic process(GO:0018879) |
| 0.1 | 0.2 | GO:1903903 | protein processing in phagocytic vesicle(GO:1900756) regulation of establishment of T cell polarity(GO:1903903) regulation of protein processing in phagocytic vesicle(GO:1903921) positive regulation of protein processing in phagocytic vesicle(GO:1903923) |
| 0.1 | 0.2 | GO:1900147 | Schwann cell migration(GO:0036135) regulation of Schwann cell migration(GO:1900147) |
| 0.1 | 0.2 | GO:0007468 | regulation of rhodopsin gene expression(GO:0007468) |
| 0.0 | 0.2 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.0 | 0.5 | GO:0019372 | lipoxygenase pathway(GO:0019372) |
| 0.0 | 0.1 | GO:0090648 | regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:0060734) response to methyl methanesulfonate(GO:0072702) cellular response to methyl methanesulfonate(GO:0072703) response to environmental enrichment(GO:0090648) positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 0.0 | 0.1 | GO:0071298 | cellular response to L-ascorbic acid(GO:0071298) |
| 0.0 | 0.2 | GO:0019074 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.0 | 0.1 | GO:0010752 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) negative regulation of nitric oxide mediated signal transduction(GO:0010751) regulation of cGMP-mediated signaling(GO:0010752) negative regulation of endothelial cell chemotaxis(GO:2001027) |
| 0.0 | 0.1 | GO:1902462 | regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 0.0 | 0.2 | GO:0071224 | cellular response to peptidoglycan(GO:0071224) |
| 0.0 | 0.1 | GO:0060279 | positive regulation of ovulation(GO:0060279) |
| 0.0 | 0.1 | GO:0097069 | cellular response to thyroxine stimulus(GO:0097069) |
| 0.0 | 0.1 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.0 | 0.1 | GO:0046086 | adenosine biosynthetic process(GO:0046086) |
| 0.0 | 0.2 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.0 | 0.2 | GO:0097116 | gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 0.0 | 0.1 | GO:1904178 | sterol regulatory element binding protein import into nucleus(GO:0035105) negative regulation of adipose tissue development(GO:1904178) |
| 0.0 | 0.1 | GO:1904253 | positive regulation of bile acid biosynthetic process(GO:0070859) positive regulation of bile acid metabolic process(GO:1904253) |
| 0.0 | 0.1 | GO:0034165 | positive regulation of toll-like receptor 9 signaling pathway(GO:0034165) |
| 0.0 | 0.1 | GO:0006114 | glycerol biosynthetic process(GO:0006114) |
| 0.0 | 0.1 | GO:0038163 | thrombopoietin-mediated signaling pathway(GO:0038163) |
| 0.0 | 0.1 | GO:0030200 | proteoglycan catabolic process(GO:0030167) heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.0 | 0.1 | GO:0097052 | L-kynurenine metabolic process(GO:0097052) |
| 0.0 | 0.3 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.0 | 0.1 | GO:0060623 | pericentric heterochromatin assembly(GO:0031508) regulation of chromosome condensation(GO:0060623) |
| 0.0 | 0.1 | GO:0051935 | amino acid neurotransmitter reuptake(GO:0051933) glutamate reuptake(GO:0051935) |
| 0.0 | 0.3 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 | 0.4 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.0 | 0.2 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 0.0 | 0.1 | GO:0055073 | cadmium ion homeostasis(GO:0055073) |
| 0.0 | 0.1 | GO:0003350 | pulmonary myocardium development(GO:0003350) subthalamus development(GO:0021539) deltoid tuberosity development(GO:0035993) left lung morphogenesis(GO:0060460) pulmonary vein morphogenesis(GO:0060577) |
| 0.0 | 0.1 | GO:0055118 | negative regulation of cardiac muscle contraction(GO:0055118) |
| 0.0 | 0.1 | GO:0075509 | receptor-mediated endocytosis of virus by host cell(GO:0019065) endocytosis involved in viral entry into host cell(GO:0075509) |
| 0.0 | 0.1 | GO:0044691 | tooth eruption(GO:0044691) |
| 0.0 | 0.1 | GO:0035995 | detection of muscle stretch(GO:0035995) |
| 0.0 | 0.1 | GO:0071630 | nucleus-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071630) |
| 0.0 | 0.1 | GO:0061156 | embryonic heart tube left/right pattern formation(GO:0060971) pulmonary artery morphogenesis(GO:0061156) |
| 0.0 | 0.1 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.0 | 0.2 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.0 | 0.0 | GO:0035986 | senescence-associated heterochromatin focus assembly(GO:0035986) |
| 0.0 | 0.1 | GO:1904714 | positive regulation of Schwann cell proliferation(GO:0010625) regulation of chaperone-mediated autophagy(GO:1904714) |
| 0.0 | 0.0 | GO:0071348 | cellular response to interleukin-11(GO:0071348) |
| 0.0 | 0.0 | GO:0046127 | thymidine catabolic process(GO:0006214) pyrimidine deoxyribonucleoside catabolic process(GO:0046127) |
| 0.0 | 0.1 | GO:0050748 | negative regulation of lipoprotein metabolic process(GO:0050748) regulation of protein lipidation(GO:1903059) |
| 0.0 | 0.1 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.0 | 0.1 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.0 | 0.1 | GO:0099545 | trans-synaptic signaling by trans-synaptic complex(GO:0099545) |
| 0.0 | 0.1 | GO:2000277 | positive regulation of oxidative phosphorylation uncoupler activity(GO:2000277) |
| 0.0 | 0.1 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.1 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.0 | 0.0 | GO:0072144 | glomerular mesangial cell development(GO:0072144) |
| 0.0 | 0.0 | GO:0036324 | vascular endothelial growth factor receptor-2 signaling pathway(GO:0036324) |
| 0.0 | 0.3 | GO:0010758 | regulation of macrophage chemotaxis(GO:0010758) |
| 0.0 | 0.1 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.0 | 0.1 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.0 | 0.1 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.1 | GO:0009624 | response to nematode(GO:0009624) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.2 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.1 | 0.3 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.3 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.2 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.0 | 0.1 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.0 | 0.1 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 0.0 | 0.1 | GO:0070821 | tertiary granule membrane(GO:0070821) |
| 0.0 | 0.1 | GO:0001740 | Barr body(GO:0001740) |
| 0.0 | 0.0 | GO:1990032 | parallel fiber(GO:1990032) |
| 0.0 | 0.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.3 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.2 | GO:0042581 | specific granule(GO:0042581) |
| 0.0 | 0.0 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.0 | 0.0 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.3 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.1 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.0 | 0.0 | GO:0097233 | lamellar body membrane(GO:0097232) alveolar lamellar body membrane(GO:0097233) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.1 | 1.5 | GO:0008320 | protein transmembrane transporter activity(GO:0008320) |
| 0.1 | 0.4 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.1 | 0.3 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) |
| 0.1 | 0.2 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.1 | 0.5 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.1 | 0.2 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 0.1 | 0.6 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.2 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.0 | 0.2 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.1 | GO:0005163 | nerve growth factor receptor binding(GO:0005163) |
| 0.0 | 0.1 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.0 | 0.1 | GO:0070548 | L-glutamine aminotransferase activity(GO:0070548) |
| 0.0 | 0.3 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.0 | 0.2 | GO:0010853 | cyclase activator activity(GO:0010853) guanylate cyclase activator activity(GO:0030250) |
| 0.0 | 0.1 | GO:0008900 | hydrogen:potassium-exchanging ATPase activity(GO:0008900) |
| 0.0 | 0.1 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 0.0 | 0.3 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.1 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.0 | 0.2 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.0 | 0.2 | GO:1901612 | phosphatidylglycerol binding(GO:1901611) cardiolipin binding(GO:1901612) |
| 0.0 | 0.1 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.0 | 0.1 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.1 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.0 | 0.4 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.1 | GO:0004956 | prostaglandin D receptor activity(GO:0004956) |
| 0.0 | 0.2 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.0 | 0.1 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 0.0 | 0.2 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.0 | 0.1 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.1 | GO:0098519 | nucleotide phosphatase activity, acting on free nucleotides(GO:0098519) |
| 0.0 | 0.0 | GO:0004159 | dihydrouracil dehydrogenase (NAD+) activity(GO:0004159) dihydropyrimidine dehydrogenase (NADP+) activity(GO:0017113) |
| 0.0 | 0.5 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.0 | 0.0 | GO:0005333 | norepinephrine transmembrane transporter activity(GO:0005333) |
| 0.0 | 0.1 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.0 | 0.1 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.2 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.3 | GO:0042923 | neuropeptide binding(GO:0042923) |
| 0.0 | 0.3 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.0 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.0 | 0.0 | GO:0052723 | inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 0.0 | 0.2 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.0 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 0.1 | GO:0015440 | peptide-transporting ATPase activity(GO:0015440) |
| 0.0 | 0.1 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.2 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.0 | 0.5 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 0.5 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 1.2 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.4 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 0.4 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.3 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.3 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.1 | REACTOME FGFR2C LIGAND BINDING AND ACTIVATION | Genes involved in FGFR2c ligand binding and activation |
| 0.0 | 0.2 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |