Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
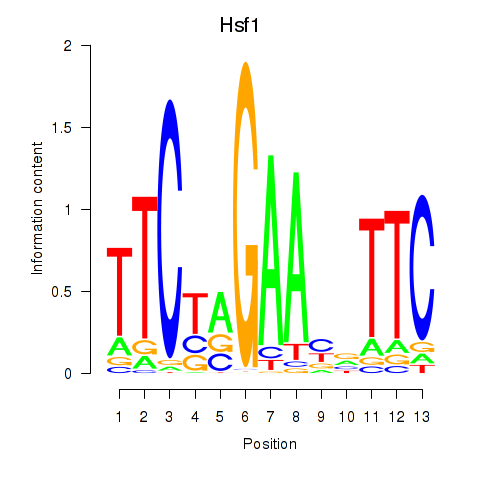
Results for Hsf1
Z-value: 0.65
Transcription factors associated with Hsf1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hsf1
|
ENSRNOG00000021732 | heat shock transcription factor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hsf1 | rn6_v1_chr7_+_117538523_117538523 | 0.15 | 8.1e-01 | Click! |
Activity profile of Hsf1 motif
Sorted Z-values of Hsf1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_23841049 | 0.28 |
ENSRNOT00000031555
|
Hspb1
|
heat shock protein family B (small) member 1 |
| chr1_+_80135391 | 0.22 |
ENSRNOT00000021893
|
Gpr4
|
G protein-coupled receptor 4 |
| chr7_+_11737293 | 0.18 |
ENSRNOT00000046059
|
Lingo3
|
leucine rich repeat and Ig domain containing 3 |
| chr3_+_154043873 | 0.15 |
ENSRNOT00000072502
ENSRNOT00000034166 |
Nnat
|
neuronatin |
| chr7_-_59763219 | 0.15 |
ENSRNOT00000041210
|
RGD1564883
|
similar to 60S ribosomal protein L12 |
| chr1_-_164308317 | 0.15 |
ENSRNOT00000022983
|
Serpinh1
|
serpin family H member 1 |
| chr20_+_295250 | 0.15 |
ENSRNOT00000000955
|
Clic2
|
chloride intracellular channel 2 |
| chr7_-_125497691 | 0.15 |
ENSRNOT00000049445
|
AABR07058578.1
|
|
| chr20_+_3149114 | 0.14 |
ENSRNOT00000084770
|
RT1-N2
|
RT1 class Ib, locus N2 |
| chr1_-_154170409 | 0.13 |
ENSRNOT00000089014
|
Hikeshi
|
Hikeshi, heat shock protein nuclear import factor |
| chr4_-_161757447 | 0.13 |
ENSRNOT00000008737
|
Fkbp4
|
FK506 binding protein 4 |
| chr12_-_6341902 | 0.13 |
ENSRNOT00000001201
|
Hsph1
|
heat shock protein family H (Hsp110) member 1 |
| chr10_-_105787803 | 0.13 |
ENSRNOT00000064671
|
Jmjd6
|
jumonji domain containing 6 |
| chr5_-_139864393 | 0.13 |
ENSRNOT00000030016
|
Zfp69
|
zinc finger protein 69 |
| chr5_+_57028467 | 0.13 |
ENSRNOT00000010061
|
Dnaja1
|
DnaJ heat shock protein family (Hsp40) member A1 |
| chr2_-_187668677 | 0.12 |
ENSRNOT00000056898
ENSRNOT00000092563 |
Tsacc
|
TSSK6 activating co-chaperone |
| chr1_-_222293148 | 0.11 |
ENSRNOT00000028743
|
Stip1
|
stress-induced phosphoprotein 1 |
| chr20_+_28920616 | 0.11 |
ENSRNOT00000070897
|
P4ha1
|
prolyl 4-hydroxylase subunit alpha 1 |
| chr11_+_37798370 | 0.11 |
ENSRNOT00000002679
|
Bace2
|
beta-site APP-cleaving enzyme 2 |
| chr20_-_12938891 | 0.11 |
ENSRNOT00000017141
|
RGD1564149
|
similar to Protein C21orf58 |
| chr5_-_151768123 | 0.10 |
ENSRNOT00000079380
|
Nudc
|
nuclear distribution C, dynein complex regulator |
| chr5_+_58393233 | 0.09 |
ENSRNOT00000000142
|
Dnajb5
|
DnaJ heat shock protein family (Hsp40) member B5 |
| chr5_+_169357517 | 0.09 |
ENSRNOT00000000079
|
Acot7
|
acyl-CoA thioesterase 7 |
| chr2_-_251532312 | 0.09 |
ENSRNOT00000019501
|
Cyr61
|
cysteine-rich, angiogenic inducer, 61 |
| chr1_-_12952906 | 0.08 |
ENSRNOT00000078193
|
LOC100360362
|
hypothetical protein LOC100360362 |
| chr11_+_64472072 | 0.08 |
ENSRNOT00000042756
|
RGD1563835
|
similar to ribosomal protein L27 |
| chrX_-_112328642 | 0.08 |
ENSRNOT00000083150
|
Psmd10
|
proteasome 26S subunit, non-ATPase 10 |
| chr20_-_32133431 | 0.08 |
ENSRNOT00000000443
|
Srgn
|
serglycin |
| chr14_-_17225389 | 0.07 |
ENSRNOT00000052035
|
Art3
|
ADP-ribosyltransferase 3 |
| chr8_-_111965889 | 0.07 |
ENSRNOT00000032376
|
Bfsp2
|
beaded filament structural protein 2 |
| chr1_+_105094411 | 0.07 |
ENSRNOT00000036258
|
Htatip2
|
HIV-1 Tat interactive protein 2 |
| chr2_-_84678790 | 0.07 |
ENSRNOT00000015886
|
Cct5
|
chaperonin containing TCP1 subunit 5 |
| chr10_+_31324512 | 0.06 |
ENSRNOT00000008559
|
Fndc9
|
fibronectin type III domain containing 9 |
| chr6_-_127248372 | 0.06 |
ENSRNOT00000085517
|
Asb2
|
ankyrin repeat and SOCS box-containing 2 |
| chr14_+_107767392 | 0.06 |
ENSRNOT00000012847
|
Cct4
|
chaperonin containing TCP1 subunit 4 |
| chr7_-_122635731 | 0.06 |
ENSRNOT00000025925
|
St13
|
suppression of tumorigenicity 13 |
| chr14_-_43143973 | 0.05 |
ENSRNOT00000003248
|
Uchl1
|
ubiquitin C-terminal hydrolase L1 |
| chr8_+_17421557 | 0.05 |
ENSRNOT00000030569
|
Chordc1
|
cysteine and histidine rich domain containing 1 |
| chr11_-_61530567 | 0.05 |
ENSRNOT00000076277
|
Naa50
|
N(alpha)-acetyltransferase 50, NatE catalytic subunit |
| chr1_-_252461461 | 0.05 |
ENSRNOT00000026093
|
Ankrd22
|
ankyrin repeat domain 22 |
| chr14_-_85350948 | 0.05 |
ENSRNOT00000083464
|
LOC100912481
|
RNA-binding protein EWS-like |
| chr9_-_61690956 | 0.04 |
ENSRNOT00000066589
|
Hspd1
|
heat shock protein family D member 1 |
| chr4_+_24612205 | 0.04 |
ENSRNOT00000057382
|
Ewsr1
|
EWS RNA-binding protein 1 |
| chr11_-_81379640 | 0.04 |
ENSRNOT00000002484
|
Eif4a2
|
eukaryotic translation initiation factor 4A2 |
| chrX_+_157095937 | 0.04 |
ENSRNOT00000091792
|
Bcap31
|
B-cell receptor-associated protein 31 |
| chr6_-_94980004 | 0.04 |
ENSRNOT00000006373
|
Rtn1
|
reticulon 1 |
| chr12_-_31323810 | 0.04 |
ENSRNOT00000001247
|
Ran
|
RAN, member RAS oncogene family |
| chr14_+_88543630 | 0.04 |
ENSRNOT00000060515
|
Hnrnpc
|
heterogeneous nuclear ribonucleoprotein C |
| chr19_+_24747178 | 0.04 |
ENSRNOT00000038879
ENSRNOT00000079106 |
Dnajb1
|
DnaJ heat shock protein family (Hsp40) member B1 |
| chrX_+_45213228 | 0.04 |
ENSRNOT00000088538
|
LOC103690319
|
odorant-binding protein |
| chr18_-_77579969 | 0.04 |
ENSRNOT00000034896
|
Sall3
|
spalt-like transcription factor 3 |
| chr17_+_36771639 | 0.04 |
ENSRNOT00000015147
|
Gpr50
|
G protein-coupled receptor 50 |
| chr14_-_85350786 | 0.03 |
ENSRNOT00000012634
|
LOC100912481
|
RNA-binding protein EWS-like |
| chr3_+_122928964 | 0.03 |
ENSRNOT00000074724
|
LOC100365450
|
rCG26795-like |
| chrX_+_15321713 | 0.03 |
ENSRNOT00000071835
|
LOC108348138
|
GTPase ERas |
| chr16_+_20336638 | 0.03 |
ENSRNOT00000080467
|
Kcnn1
|
potassium calcium-activated channel subfamily N member 1 |
| chr10_-_56008885 | 0.03 |
ENSRNOT00000067677
|
Kdm6b
|
lysine demethylase 6B |
| chr16_+_50049828 | 0.03 |
ENSRNOT00000034448
|
Fam149a
|
family with sequence similarity 149, member A |
| chr1_-_89045586 | 0.03 |
ENSRNOT00000063808
|
Zbtb32
|
zinc finger and BTB domain containing 32 |
| chr2_+_187668796 | 0.03 |
ENSRNOT00000025824
|
Cct3
|
chaperonin containing TCP1 subunit 3 |
| chr12_-_6956914 | 0.03 |
ENSRNOT00000072129
ENSRNOT00000001210 |
Uspl1
|
ubiquitin specific peptidase like 1 |
| chr7_+_40318490 | 0.03 |
ENSRNOT00000081374
|
LOC500827
|
similar to hypothetical protein FLJ35821 |
| chr10_+_105787935 | 0.03 |
ENSRNOT00000000266
|
Mettl23
|
methyltransferase like 23 |
| chr8_+_55178289 | 0.03 |
ENSRNOT00000059127
|
Cryab
|
crystallin, alpha B |
| chr1_+_192025357 | 0.03 |
ENSRNOT00000025072
|
Ubfd1
|
ubiquitin family domain containing 1 |
| chr12_-_30501860 | 0.02 |
ENSRNOT00000001227
|
Cct6a
|
chaperonin containing TCP1 subunit 6A |
| chrX_-_112159458 | 0.02 |
ENSRNOT00000087403
|
Tex13b
|
testis expressed 13B |
| chr2_+_178354890 | 0.02 |
ENSRNOT00000037890
|
Ppid
|
peptidylprolyl isomerase D |
| chr8_-_49301125 | 0.02 |
ENSRNOT00000091190
|
Cd3e
|
CD3e molecule |
| chrX_-_1346181 | 0.02 |
ENSRNOT00000042736
|
LOC102555453
|
60S ribosomal protein L12-like |
| chr8_-_58293102 | 0.02 |
ENSRNOT00000011913
|
Rab39a
|
RAB39A, member RAS oncogene family |
| chr1_-_48033343 | 0.02 |
ENSRNOT00000019531
ENSRNOT00000076422 |
Tcp1
|
t-complex 1 |
| chr1_+_221443896 | 0.02 |
ENSRNOT00000073942
|
Tmem262
|
transmembrane protein 262 |
| chr20_+_3148665 | 0.02 |
ENSRNOT00000086026
|
RT1-N2
|
RT1 class Ib, locus N2 |
| chr20_+_4118461 | 0.02 |
ENSRNOT00000083085
|
RT1-Db2
|
RT1 class II, locus Db2 |
| chr15_+_87886783 | 0.02 |
ENSRNOT00000065710
|
Slain1
|
SLAIN motif family, member 1 |
| chr19_-_41161765 | 0.01 |
ENSRNOT00000023117
|
Hydin
|
Hydin, axonemal central pair apparatus protein |
| chr14_-_64535170 | 0.01 |
ENSRNOT00000082338
|
Gba3
|
glucosidase, beta, acid 3 |
| chr11_+_83855753 | 0.01 |
ENSRNOT00000030686
|
RGD1563956
|
similar to 60S ribosomal protein L12 |
| chr9_+_64643281 | 0.01 |
ENSRNOT00000081497
|
Spats2l
|
spermatogenesis associated, serine-rich 2-like |
| chr3_+_94035905 | 0.01 |
ENSRNOT00000014767
|
RGD1563222
|
similar to RIKEN cDNA A930018P22 |
| chr11_-_27141881 | 0.01 |
ENSRNOT00000002169
|
Cct8
|
chaperonin containing TCP1 subunit 8 |
| chrX_-_83864150 | 0.01 |
ENSRNOT00000073734
|
Obp1f
|
odorant binding protein I f |
| chr6_+_93539271 | 0.01 |
ENSRNOT00000078791
|
Tomm20l
|
translocase of outer mitochondrial membrane 20 homolog (yeast)-like |
| chr9_+_10172832 | 0.01 |
ENSRNOT00000074555
|
Acsbg2
|
acyl-CoA synthetase bubblegum family member 2 |
| chr8_+_128790348 | 0.00 |
ENSRNOT00000025142
|
Slc25a38
|
solute carrier family 25, member 38 |
| chr16_-_930527 | 0.00 |
ENSRNOT00000014787
|
Spin1
|
spindlin 1 |
| chr14_+_7169519 | 0.00 |
ENSRNOT00000003026
|
Klhl8
|
kelch-like family member 8 |
| chr2_-_173087648 | 0.00 |
ENSRNOT00000091079
|
Iqcj
|
IQ motif containing J |
| chr8_-_76239995 | 0.00 |
ENSRNOT00000071321
|
LOC100912027
|
60S ribosomal protein L27-like |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hsf1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0071348 | cellular response to interleukin-11(GO:0071348) response to butyrate(GO:1903544) |
| 0.0 | 0.2 | GO:0072144 | glomerular mesangial cell development(GO:0072144) |
| 0.0 | 0.1 | GO:0017185 | peptidyl-lysine hydroxylation(GO:0017185) |
| 0.0 | 0.1 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.0 | 0.1 | GO:0036112 | medium-chain fatty-acyl-CoA metabolic process(GO:0036112) fatty-acyl-CoA catabolic process(GO:0036115) |
| 0.0 | 0.1 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.0 | 0.1 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.0 | 0.1 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.0 | 0.2 | GO:1904871 | protein localization to nuclear body(GO:1903405) protein localization to Cajal body(GO:1904867) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) protein localization to nucleoplasm(GO:1990173) |
| 0.0 | 0.1 | GO:1903753 | negative regulation of p38MAPK cascade(GO:1903753) |
| 0.0 | 0.1 | GO:0060315 | negative regulation of ryanodine-sensitive calcium-release channel activity(GO:0060315) |
| 0.0 | 0.1 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.0 | 0.1 | GO:0003433 | chondrocyte development involved in endochondral bone morphogenesis(GO:0003433) |
| 0.0 | 0.0 | GO:2000298 | regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 0.0 | 0.1 | GO:2000304 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.0 | 0.0 | GO:0045041 | T cell mediated immune response to tumor cell(GO:0002424) regulation of T cell mediated immune response to tumor cell(GO:0002840) protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.0 | 0.1 | GO:0061084 | negative regulation of protein refolding(GO:0061084) |
| 0.0 | 0.0 | GO:1904154 | positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.0 | 0.1 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.0 | 0.2 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.0 | 0.0 | GO:1990826 | nucleoplasmic periphery of the nuclear pore complex(GO:1990826) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0070815 | peptidyl-lysine 5-dioxygenase activity(GO:0070815) |
| 0.0 | 0.1 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.0 | 0.1 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 0.0 | 0.1 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.0 | 0.5 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.1 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.0 | 0.1 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.1 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 0.0 | 0.0 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.0 | 0.1 | GO:0032564 | dATP binding(GO:0032564) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |