Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
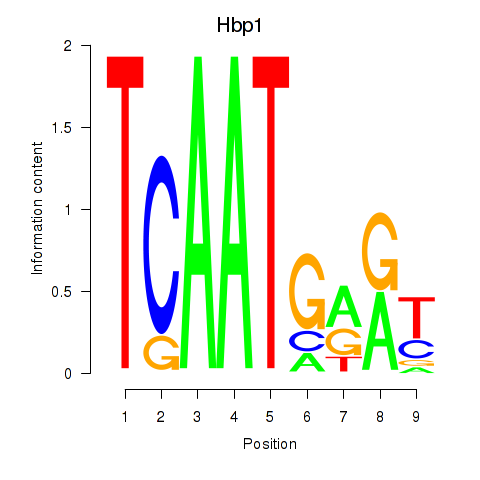
Results for Hbp1
Z-value: 0.66
Transcription factors associated with Hbp1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hbp1
|
ENSRNOG00000008927 | HMG-box transcription factor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hbp1 | rn6_v1_chr6_-_51257625_51257625 | -0.20 | 7.4e-01 | Click! |
Activity profile of Hbp1 motif
Sorted Z-values of Hbp1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_20262680 | 0.29 |
ENSRNOT00000046378
|
LOC300308
|
similar to hypothetical protein 4930509O22 |
| chr8_-_104155775 | 0.28 |
ENSRNOT00000042885
|
LOC102550734
|
60S ribosomal protein L31-like |
| chr14_-_46153212 | 0.23 |
ENSRNOT00000079269
|
Nwd2
|
NACHT and WD repeat domain containing 2 |
| chr17_-_32783427 | 0.22 |
ENSRNOT00000059921
|
Serpinb6b
|
serine (or cysteine) peptidase inhibitor, clade B, member 6b |
| chr2_+_149899836 | 0.21 |
ENSRNOT00000086481
|
RGD1560324
|
similar to hypothetical protein C130079G13 |
| chr7_-_120882392 | 0.21 |
ENSRNOT00000056179
ENSRNOT00000056178 |
Fam227a
|
family with sequence similarity 227, member A |
| chr14_+_63405408 | 0.20 |
ENSRNOT00000086658
|
AABR07015559.1
|
|
| chr16_+_35116440 | 0.19 |
ENSRNOT00000074353
|
AABR07025358.1
|
|
| chr6_+_127941526 | 0.19 |
ENSRNOT00000033897
|
LOC500712
|
Ab1-233 |
| chr12_+_24148567 | 0.19 |
ENSRNOT00000077378
ENSRNOT00000045157 |
Ccl24
|
C-C motif chemokine ligand 24 |
| chr9_-_30844199 | 0.18 |
ENSRNOT00000017169
|
Col19a1
|
collagen type XIX alpha 1 chain |
| chr9_-_98597359 | 0.18 |
ENSRNOT00000027506
|
Per2
|
period circadian clock 2 |
| chr6_-_87427153 | 0.17 |
ENSRNOT00000071999
|
AABR07064622.1
|
|
| chr12_-_51965779 | 0.16 |
ENSRNOT00000056733
|
LOC100362927
|
replication protein A3-like |
| chr8_-_22336794 | 0.16 |
ENSRNOT00000066340
|
Ap1m2
|
adaptor-related protein complex 1, mu 2 subunit |
| chr13_-_111948753 | 0.16 |
ENSRNOT00000048074
|
Hsd11b1
|
hydroxysteroid 11-beta dehydrogenase 1 |
| chrX_-_10031167 | 0.16 |
ENSRNOT00000060988
|
Gpr34
|
G protein-coupled receptor 34 |
| chr1_+_65851060 | 0.15 |
ENSRNOT00000036880
|
Zscan18
|
zinc finger and SCAN domain containing 18 |
| chr8_-_64572850 | 0.15 |
ENSRNOT00000015415
|
Senp8
|
SUMO/sentrin peptidase family member, NEDD8 specific |
| chrX_+_135187468 | 0.15 |
ENSRNOT00000006805
|
Bcorl1
|
BCL6 co-repressor-like 1 |
| chrX_+_65040775 | 0.14 |
ENSRNOT00000081354
|
Zc3h12b
|
zinc finger CCCH-type containing 12B |
| chr2_+_71753816 | 0.14 |
ENSRNOT00000025809
|
AABR07008724.1
|
|
| chr7_+_20462081 | 0.13 |
ENSRNOT00000088383
|
AABR07056131.1
|
|
| chr20_-_26589209 | 0.13 |
ENSRNOT00000049437
|
Ctnna3
|
catenin alpha 3 |
| chr8_-_84506328 | 0.13 |
ENSRNOT00000064754
|
Mlip
|
muscular LMNA-interacting protein |
| chr1_-_112811936 | 0.13 |
ENSRNOT00000093339
|
Gabrg3
|
gamma-aminobutyric acid type A receptor gamma 3 subunit |
| chr5_+_159734838 | 0.13 |
ENSRNOT00000079905
|
Rsg1
|
REM2 and RAB-like small GTPase 1 |
| chr8_-_76579387 | 0.12 |
ENSRNOT00000090747
|
Fam81a
|
family with sequence similarity 81, member A |
| chr8_-_76579099 | 0.12 |
ENSRNOT00000088628
|
Fam81a
|
family with sequence similarity 81, member A |
| chr9_+_61738471 | 0.12 |
ENSRNOT00000090305
|
AABR07067762.1
|
|
| chr2_+_180012414 | 0.12 |
ENSRNOT00000082273
|
Pdgfc
|
platelet derived growth factor C |
| chrX_-_142248369 | 0.12 |
ENSRNOT00000091330
|
Fgf13
|
fibroblast growth factor 13 |
| chr6_+_78567970 | 0.12 |
ENSRNOT00000032743
|
Ttc6
|
tetratricopeptide repeat domain 6 |
| chr4_-_161681660 | 0.12 |
ENSRNOT00000039086
|
Rhno1
|
RAD9-HUS1-RAD1 interacting nuclear orphan 1 |
| chr9_-_15274917 | 0.12 |
ENSRNOT00000019650
|
Pgc
|
progastricsin |
| chr8_-_48762342 | 0.12 |
ENSRNOT00000049125
|
Foxr1
|
forkhead box R1 |
| chr8_-_84632817 | 0.12 |
ENSRNOT00000076942
|
Mlip
|
muscular LMNA-interacting protein |
| chr16_+_22979444 | 0.12 |
ENSRNOT00000017822
|
Csgalnact1
|
chondroitin sulfate N-acetylgalactosaminyltransferase 1 |
| chr5_+_147476221 | 0.12 |
ENSRNOT00000010730
|
Sync
|
syncoilin, intermediate filament protein |
| chr6_+_30038777 | 0.12 |
ENSRNOT00000072340
|
Fam228b
|
family with sequence similarity 228, member B |
| chr17_+_45175121 | 0.12 |
ENSRNOT00000080417
|
Nkapl
|
NFKB activating protein-like |
| chr3_-_148407778 | 0.11 |
ENSRNOT00000011262
|
Foxs1
|
forkhead box S1 |
| chr6_-_95502775 | 0.11 |
ENSRNOT00000074990
ENSRNOT00000034289 |
Dhrs7l1
|
dehydrogenase/reductase (SDR family) member 7-like 1 |
| chr14_-_71973419 | 0.11 |
ENSRNOT00000079895
|
Cc2d2a
|
coiled-coil and C2 domain containing 2A |
| chr13_+_51795867 | 0.11 |
ENSRNOT00000006747
|
Ube2t
|
ubiquitin-conjugating enzyme E2T |
| chr8_+_26652121 | 0.11 |
ENSRNOT00000009342
|
Eepd1
|
endonuclease/exonuclease/phosphatase family domain containing 1 |
| chrX_-_17708388 | 0.11 |
ENSRNOT00000048887
|
LOC302576
|
similar to mgclh |
| chr10_+_73868943 | 0.11 |
ENSRNOT00000081012
|
Tubd1
|
tubulin, delta 1 |
| chr5_+_10125070 | 0.11 |
ENSRNOT00000085321
|
AABR07046866.1
|
|
| chr10_+_29606748 | 0.10 |
ENSRNOT00000080720
|
AABR07029467.2
|
|
| chr14_+_71542057 | 0.10 |
ENSRNOT00000082592
ENSRNOT00000083701 ENSRNOT00000084322 |
Prom1
|
prominin 1 |
| chr6_-_138508753 | 0.10 |
ENSRNOT00000006888
|
Ighm
|
immunoglobulin heavy constant mu |
| chr3_+_72385666 | 0.10 |
ENSRNOT00000011168
|
Prg2
|
proteoglycan 2 |
| chr13_+_101790865 | 0.10 |
ENSRNOT00000087784
|
Taf1a
|
TATA-box binding protein associated factor, RNA polymerase I subunit A |
| chr2_-_186333805 | 0.10 |
ENSRNOT00000022150
|
Cd1d1
|
CD1d1 molecule |
| chr2_-_208225888 | 0.10 |
ENSRNOT00000054860
|
AABR07012775.1
|
|
| chr19_+_39067363 | 0.10 |
ENSRNOT00000083515
|
Has3
|
hyaluronan synthase 3 |
| chr2_-_57600820 | 0.10 |
ENSRNOT00000083247
|
Nipbl
|
NIPBL, cohesin loading factor |
| chr7_-_116106368 | 0.10 |
ENSRNOT00000035678
|
Ly6k
|
lymphocyte antigen 6 complex, locus K |
| chr12_-_19254527 | 0.10 |
ENSRNOT00000089349
|
AABR07035541.3
|
|
| chr7_-_120380200 | 0.10 |
ENSRNOT00000014878
|
RGD1359634
|
similar to RIKEN cDNA 1700088E04 |
| chr1_+_67340809 | 0.10 |
ENSRNOT00000045266
|
LOC100362054
|
mCG114696-like |
| chr9_+_111249351 | 0.10 |
ENSRNOT00000076729
|
Ppip5k2
|
diphosphoinositol pentakisphosphate kinase 2 |
| chr1_+_101517714 | 0.10 |
ENSRNOT00000028423
|
Plekha4
|
pleckstrin homology domain containing A4 |
| chr1_+_199555722 | 0.09 |
ENSRNOT00000054983
|
Itgax
|
integrin subunit alpha X |
| chr15_+_58016238 | 0.09 |
ENSRNOT00000075786
|
Kctd4
|
potassium channel tetramerization domain containing 4 |
| chr6_-_108976489 | 0.09 |
ENSRNOT00000007350
|
Rps6kl1
|
ribosomal protein S6 kinase-like 1 |
| chr1_-_100993269 | 0.09 |
ENSRNOT00000027777
|
Bcl2l12
|
BCL2 like 12 |
| chr8_+_115151627 | 0.09 |
ENSRNOT00000064252
|
Abhd14b
|
abhydrolase domain containing 14b |
| chr4_-_118342176 | 0.09 |
ENSRNOT00000032477
|
AC139642.2
|
|
| chr16_-_32868680 | 0.09 |
ENSRNOT00000015974
ENSRNOT00000082392 |
Aadat
|
aminoadipate aminotransferase |
| chr1_+_98398660 | 0.09 |
ENSRNOT00000047473
|
Cd33
|
CD33 molecule |
| chr8_-_39907478 | 0.09 |
ENSRNOT00000064141
|
Robo3
|
roundabout guidance receptor 3 |
| chr1_+_188288738 | 0.09 |
ENSRNOT00000055103
|
Tmc5
|
transmembrane channel-like 5 |
| chr7_+_28414350 | 0.09 |
ENSRNOT00000085680
|
Igf1
|
insulin-like growth factor 1 |
| chr7_+_71157664 | 0.09 |
ENSRNOT00000005919
|
Sdr9c7
|
short chain dehydrogenase/reductase family 9C, member 7 |
| chr13_-_104284068 | 0.09 |
ENSRNOT00000005407
|
LOC100912365
|
uncharacterized LOC100912365 |
| chr10_+_1622573 | 0.09 |
ENSRNOT00000040132
ENSRNOT00000090207 ENSRNOT00000089118 |
AABR07028989.1
|
|
| chr4_+_98457810 | 0.09 |
ENSRNOT00000074175
|
AABR07060872.1
|
|
| chr10_-_103687425 | 0.09 |
ENSRNOT00000039284
|
Cd300lf
|
Cd300 molecule-like family member F |
| chr20_+_14578605 | 0.09 |
ENSRNOT00000041165
|
Rtdr1
|
rhabdoid tumor deletion region gene 1 |
| chr5_+_14415841 | 0.08 |
ENSRNOT00000010682
|
Rgs20
|
regulator of G-protein signaling 20 |
| chr12_+_14092541 | 0.08 |
ENSRNOT00000033998
|
Radil
|
Rap associating with DIL domain |
| chr9_+_70059683 | 0.08 |
ENSRNOT00000016038
|
Zdbf2
|
zinc finger, DBF-type containing 2 |
| chr1_-_261446570 | 0.08 |
ENSRNOT00000020182
|
Sfrp5
|
secreted frizzled-related protein 5 |
| chr8_+_28352772 | 0.08 |
ENSRNOT00000012391
|
Igsf9b
|
immunoglobulin superfamily, member 9B |
| chr7_+_15422479 | 0.08 |
ENSRNOT00000066520
|
Zfp563
|
zinc finger protein 563 |
| chr20_-_11644166 | 0.08 |
ENSRNOT00000073009
|
LOC100361739
|
keratin associated protein 12-1-like |
| chr10_-_85049331 | 0.08 |
ENSRNOT00000012538
|
Tbx21
|
T-box 21 |
| chr7_-_15821927 | 0.08 |
ENSRNOT00000050658
|
LOC691422
|
similar to zinc finger protein 101 |
| chr15_-_29548400 | 0.08 |
ENSRNOT00000078176
|
AABR07017639.2
|
|
| chr2_+_58448917 | 0.08 |
ENSRNOT00000082562
|
Ranbp3l
|
RAN binding protein 3-like |
| chr11_+_74050166 | 0.08 |
ENSRNOT00000002348
|
Lrrc15
|
leucine rich repeat containing 15 |
| chr3_+_147042944 | 0.08 |
ENSRNOT00000012608
|
Fkbp1a
|
FK506 binding protein 1a |
| chr9_+_47281961 | 0.08 |
ENSRNOT00000065234
|
Slc9a4
|
solute carrier family 9 member A4 |
| chr19_-_17494516 | 0.08 |
ENSRNOT00000078786
ENSRNOT00000076138 |
Chd9
|
chromodomain helicase DNA binding protein 9 |
| chr7_+_41475163 | 0.08 |
ENSRNOT00000037844
|
Dusp6
|
dual specificity phosphatase 6 |
| chr2_+_22909569 | 0.08 |
ENSRNOT00000073871
|
Homer1
|
homer scaffolding protein 1 |
| chr14_-_45859908 | 0.08 |
ENSRNOT00000086994
|
Pgm2
|
phosphoglucomutase 2 |
| chr13_-_67206688 | 0.08 |
ENSRNOT00000003630
ENSRNOT00000090693 |
Pla2g4a
|
phospholipase A2 group IVA |
| chr9_-_13877130 | 0.08 |
ENSRNOT00000015705
|
Lrfn2
|
leucine rich repeat and fibronectin type III domain containing 2 |
| chr2_+_58534476 | 0.08 |
ENSRNOT00000077646
|
Lmbrd2
|
LMBR1 domain containing 2 |
| chr14_+_7630482 | 0.08 |
ENSRNOT00000091581
|
LOC498330
|
similar to hypothetical protein MGC26744 |
| chr8_+_44136496 | 0.08 |
ENSRNOT00000087022
|
Scn3b
|
sodium voltage-gated channel beta subunit 3 |
| chr1_+_252944103 | 0.08 |
ENSRNOT00000025770
|
Ifit1
|
interferon-induced protein with tetratricopeptide repeats 1 |
| chrX_+_908044 | 0.08 |
ENSRNOT00000072087
|
Zfp300
|
zinc finger protein 300 |
| chr1_-_198900375 | 0.08 |
ENSRNOT00000024969
|
Zfp689
|
zinc finger protein 689 |
| chr10_+_62191656 | 0.07 |
ENSRNOT00000029866
|
Smyd4
|
SET and MYND domain containing 4 |
| chr5_-_151895016 | 0.07 |
ENSRNOT00000081841
|
Pigv
|
phosphatidylinositol glycan anchor biosynthesis, class V |
| chr20_-_19468848 | 0.07 |
ENSRNOT00000000756
|
Fam13c
|
family with sequence similarity 13, member C |
| chr7_-_97605113 | 0.07 |
ENSRNOT00000089734
|
AABR07058000.1
|
|
| chr13_-_88061108 | 0.07 |
ENSRNOT00000003774
|
Rgs4
|
regulator of G-protein signaling 4 |
| chr4_+_163358009 | 0.07 |
ENSRNOT00000082064
|
Klrd1
|
killer cell lectin like receptor D1 |
| chr15_-_34693034 | 0.07 |
ENSRNOT00000083314
|
Mcpt8
|
mast cell protease 8 |
| chr2_+_251200686 | 0.07 |
ENSRNOT00000019210
|
Col24a1
|
collagen type XXIV alpha 1 chain |
| chr14_-_10446909 | 0.07 |
ENSRNOT00000002962
|
Mrps18c
|
mitochondrial ribosomal protein S18C |
| chr2_+_188528979 | 0.07 |
ENSRNOT00000087934
|
Thbs3
|
thrombospondin 3 |
| chr6_+_52702544 | 0.07 |
ENSRNOT00000014252
|
Efcab10
|
EF-hand calcium binding domain 10 |
| chr20_+_28572242 | 0.07 |
ENSRNOT00000072485
|
Sh3rf3
|
SH3 domain containing ring finger 3 |
| chr9_+_8399632 | 0.07 |
ENSRNOT00000092165
|
LOC100360856
|
hypothetical protein LOC100360856 |
| chr16_+_75465710 | 0.07 |
ENSRNOT00000039969
|
Defa9
|
defensin alpha 9 |
| chr3_+_101899068 | 0.07 |
ENSRNOT00000079600
ENSRNOT00000006256 |
Muc15
|
mucin 15, cell surface associated |
| chr16_-_6404578 | 0.07 |
ENSRNOT00000051371
|
Cacna1d
|
calcium voltage-gated channel subunit alpha1 D |
| chr6_+_137953545 | 0.07 |
ENSRNOT00000006804
|
Crip2
|
cysteine-rich protein 2 |
| chr7_+_70946228 | 0.07 |
ENSRNOT00000039306
|
Stat6
|
signal transducer and activator of transcription 6 |
| chr1_-_228755866 | 0.07 |
ENSRNOT00000083283
|
Dtx4
|
deltex E3 ubiquitin ligase 4 |
| chr12_-_6078411 | 0.07 |
ENSRNOT00000001197
|
Rxfp2
|
relaxin/insulin-like family peptide receptor 2 |
| chr9_+_14560504 | 0.07 |
ENSRNOT00000091532
|
Nfya
|
nuclear transcription factor Y subunit alpha |
| chr8_-_62386264 | 0.07 |
ENSRNOT00000026165
|
Cplx3
|
complexin 3 |
| chr10_-_91661558 | 0.07 |
ENSRNOT00000043156
|
AABR07030521.1
|
|
| chr14_-_114047527 | 0.07 |
ENSRNOT00000083199
|
AABR07016779.1
|
|
| chr3_+_122816924 | 0.07 |
ENSRNOT00000045604
|
RGD1561317
|
similar to ribosomal protein L31 |
| chr1_-_73733788 | 0.07 |
ENSRNOT00000025338
|
Leng8
|
leukocyte receptor cluster member 8 |
| chr18_+_30909490 | 0.07 |
ENSRNOT00000026967
|
Pcdhgb8
|
protocadherin gamma subfamily B, 8 |
| chr5_-_8864578 | 0.07 |
ENSRNOT00000008480
|
Cspp1
|
centrosome and spindle pole associated protein 1 |
| chr5_+_25349928 | 0.06 |
ENSRNOT00000020826
|
Gem
|
GTP binding protein overexpressed in skeletal muscle |
| chr18_+_53088994 | 0.06 |
ENSRNOT00000091921
|
AC104053.2
|
|
| chr8_-_54998864 | 0.06 |
ENSRNOT00000059191
|
Tex12
|
testis expressed 12 |
| chr2_-_157066781 | 0.06 |
ENSRNOT00000001187
|
LOC304239
|
similar to RalA binding protein 1 |
| chr8_+_32018560 | 0.06 |
ENSRNOT00000007358
|
Adamts8
|
ADAM metallopeptidase with thrombospondin type 1 motif, 8 |
| chr3_-_146470293 | 0.06 |
ENSRNOT00000009627
|
Acss1
|
acyl-CoA synthetase short-chain family member 1 |
| chr6_-_94834908 | 0.06 |
ENSRNOT00000006284
|
L3hypdh
|
trans-L-3-hydroxyproline dehydratase |
| chrX_-_13601069 | 0.06 |
ENSRNOT00000004686
|
Otc
|
ornithine carbamoyltransferase |
| chr1_-_37726151 | 0.06 |
ENSRNOT00000071842
|
RGD1308544
|
LOC361192 |
| chr14_+_100373129 | 0.06 |
ENSRNOT00000029980
|
Wdr92
|
WD repeat domain 92 |
| chr1_-_189911571 | 0.06 |
ENSRNOT00000080996
ENSRNOT00000088536 |
Zp2
|
zona pellucida glycoprotein 2 |
| chr14_+_39368530 | 0.06 |
ENSRNOT00000084367
|
Cox7b2
|
cytochrome c oxidase subunit 7B2 |
| chr2_+_248398917 | 0.06 |
ENSRNOT00000045855
|
Gbp1
|
guanylate binding protein 1 |
| chrX_+_68752597 | 0.06 |
ENSRNOT00000077039
|
Stard8
|
StAR-related lipid transfer domain containing 8 |
| chr3_+_121408146 | 0.06 |
ENSRNOT00000023708
|
Fbln7
|
fibulin 7 |
| chr6_-_25211494 | 0.06 |
ENSRNOT00000009634
|
Xdh
|
xanthine dehydrogenase |
| chr15_+_42659371 | 0.06 |
ENSRNOT00000091612
|
Clu
|
clusterin |
| chr1_-_167698263 | 0.06 |
ENSRNOT00000093046
|
Trim21
|
tripartite motif-containing 21 |
| chr15_+_31948035 | 0.06 |
ENSRNOT00000071627
|
AABR07017868.1
|
|
| chr14_+_69800156 | 0.06 |
ENSRNOT00000072746
|
Lcorl
|
ligand dependent nuclear receptor corepressor-like |
| chr1_+_72889270 | 0.06 |
ENSRNOT00000058843
ENSRNOT00000034957 |
Tnnt1
|
troponin T1, slow skeletal type |
| chr8_-_22150005 | 0.06 |
ENSRNOT00000041678
|
Tyk2
|
tyrosine kinase 2 |
| chr2_+_35977828 | 0.06 |
ENSRNOT00000076712
|
LOC108348103
|
serine protease inhibitor Kazal-type 5-like |
| chr13_-_51784639 | 0.06 |
ENSRNOT00000089068
|
Ppp1r12b
|
protein phosphatase 1, regulatory subunit 12B |
| chr15_+_41643541 | 0.06 |
ENSRNOT00000019646
|
Arl11
|
ADP-ribosylation factor like GTPase 11 |
| chr8_-_115149419 | 0.06 |
ENSRNOT00000016056
ENSRNOT00000077514 |
Abhd14a
|
abhydrolase domain containing 14A |
| chr15_+_87722221 | 0.06 |
ENSRNOT00000082688
|
Scel
|
sciellin |
| chrM_+_11736 | 0.06 |
ENSRNOT00000048767
|
Mt-nd5
|
mitochondrially encoded NADH dehydrogenase 5 |
| chr4_-_28310178 | 0.06 |
ENSRNOT00000084021
|
RGD1563091
|
similar to OEF2 |
| chr3_-_8659102 | 0.06 |
ENSRNOT00000050908
|
Zdhhc12
|
zinc finger, DHHC-type containing 12 |
| chr9_-_27761733 | 0.06 |
ENSRNOT00000040034
|
Kcnq5
|
potassium voltage-gated channel subfamily Q member 5 |
| chr8_-_123829749 | 0.06 |
ENSRNOT00000090467
|
AABR07071598.1
|
|
| chrX_+_31984612 | 0.06 |
ENSRNOT00000005181
|
Bmx
|
BMX non-receptor tyrosine kinase |
| chr3_+_172550258 | 0.06 |
ENSRNOT00000075148
|
Tubb1
|
tubulin, beta 1 class VI |
| chrX_+_1311121 | 0.06 |
ENSRNOT00000038909
|
Cfp
|
complement factor properdin |
| chr1_-_257498844 | 0.06 |
ENSRNOT00000019056
|
Noc3l
|
NOC3-like DNA replication regulator |
| chr13_-_90839411 | 0.06 |
ENSRNOT00000010594
|
Slamf9
|
SLAM family member 9 |
| chr14_-_108658371 | 0.06 |
ENSRNOT00000008919
|
Papolg
|
poly(A) polymerase gamma |
| chr14_+_84417320 | 0.06 |
ENSRNOT00000008788
|
Tbc1d10a
|
TBC1 domain family, member 10a |
| chr5_+_152680407 | 0.06 |
ENSRNOT00000076864
|
Stmn1
|
stathmin 1 |
| chr1_-_143392532 | 0.06 |
ENSRNOT00000026089
|
Fsd2
|
fibronectin type III and SPRY domain containing 2 |
| chr18_-_37096132 | 0.06 |
ENSRNOT00000041188
|
Ppp2r2b
|
protein phosphatase 2, regulatory subunit B, beta |
| chr18_-_7081356 | 0.06 |
ENSRNOT00000021253
|
Chst9
|
carbohydrate sulfotransferase 9 |
| chrX_-_14331486 | 0.06 |
ENSRNOT00000067603
|
Rpgr
|
retinitis pigmentosa GTPase regulator |
| chr7_-_54778848 | 0.06 |
ENSRNOT00000005399
|
Glipr1
|
GLI pathogenesis-related 1 |
| chr7_+_42304534 | 0.06 |
ENSRNOT00000085097
|
Kitlg
|
KIT ligand |
| chr1_-_13915594 | 0.06 |
ENSRNOT00000015927
|
Arfgef3
|
ARFGEF family member 3 |
| chrX_+_123751089 | 0.06 |
ENSRNOT00000092384
|
Nkap
|
NFKB activating protein |
| chr4_-_172063391 | 0.05 |
ENSRNOT00000010158
|
Slc15a5
|
solute carrier family 15, member 5 |
| chr11_-_11585078 | 0.05 |
ENSRNOT00000088878
|
Robo2
|
roundabout guidance receptor 2 |
| chr8_+_71591360 | 0.05 |
ENSRNOT00000090096
|
Csnk1g1
|
casein kinase 1, gamma 1 |
| chr4_-_119568736 | 0.05 |
ENSRNOT00000041234
|
Aplf
|
aprataxin and PNKP like factor |
| chr18_+_65388685 | 0.05 |
ENSRNOT00000080844
|
Tcf4
|
transcription factor 4 |
| chr2_+_220298245 | 0.05 |
ENSRNOT00000022625
|
Plppr4
|
phospholipid phosphatase related 4 |
| chr8_+_115546712 | 0.05 |
ENSRNOT00000079801
|
Dcaf1
|
DDB1 and CUL4 associated factor 1 |
| chr2_-_40386669 | 0.05 |
ENSRNOT00000014074
|
Elovl7
|
ELOVL fatty acid elongase 7 |
| chr4_-_66899914 | 0.05 |
ENSRNOT00000011481
|
Parp12
|
poly (ADP-ribose) polymerase family, member 12 |
| chr4_-_65780799 | 0.05 |
ENSRNOT00000079883
|
Atp6v0a4
|
ATPase H+ transporting V0 subunit a4 |
| chr2_+_192538899 | 0.05 |
ENSRNOT00000045691
ENSRNOT00000085931 |
LOC102550416
|
small proline-rich protein 2I-like |
| chr14_+_113867209 | 0.05 |
ENSRNOT00000067591
|
Ccdc88a
|
coiled coil domain containing 88A |
| chr1_-_84145916 | 0.05 |
ENSRNOT00000081796
|
Ltbp4
|
latent transforming growth factor beta binding protein 4 |
| chr10_-_34242985 | 0.05 |
ENSRNOT00000046438
|
RGD1559575
|
similar to novel protein |
| chr3_+_56355431 | 0.05 |
ENSRNOT00000037188
|
Myo3b
|
myosin IIIB |
| chr19_-_23554594 | 0.05 |
ENSRNOT00000004590
|
Il15
|
interleukin 15 |
| chr1_-_98046770 | 0.05 |
ENSRNOT00000056528
|
Grb2
|
growth factor receptor bound protein 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hbp1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0051933 | amino acid neurotransmitter reuptake(GO:0051933) glutamate reuptake(GO:0051935) |
| 0.1 | 0.2 | GO:0006713 | glucocorticoid catabolic process(GO:0006713) |
| 0.0 | 0.2 | GO:2000418 | positive regulation of eosinophil migration(GO:2000418) |
| 0.0 | 0.1 | GO:0002215 | defense response to nematode(GO:0002215) |
| 0.0 | 0.1 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.0 | 0.1 | GO:0002296 | T-helper 1 cell lineage commitment(GO:0002296) |
| 0.0 | 0.1 | GO:0048003 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.0 | 0.1 | GO:0035261 | external genitalia morphogenesis(GO:0035261) gall bladder development(GO:0061010) |
| 0.0 | 0.1 | GO:0033512 | L-lysine catabolic process to acetyl-CoA via saccharopine(GO:0033512) |
| 0.0 | 0.1 | GO:0002803 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.0 | 0.1 | GO:0061643 | chemoattraction of axon(GO:0061642) chemorepulsion of axon(GO:0061643) |
| 0.0 | 0.1 | GO:1902214 | regulation of interleukin-4-mediated signaling pathway(GO:1902214) negative regulation of apoptotic cell clearance(GO:2000426) |
| 0.0 | 0.1 | GO:0044333 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) regulation of planar cell polarity pathway involved in axis elongation(GO:2000040) negative regulation of planar cell polarity pathway involved in axis elongation(GO:2000041) |
| 0.0 | 0.1 | GO:0072139 | glomerular parietal epithelial cell differentiation(GO:0072139) |
| 0.0 | 0.1 | GO:0045226 | hyaluranon cable assembly(GO:0036118) extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 0.0 | 0.1 | GO:0070318 | positive regulation of G0 to G1 transition(GO:0070318) |
| 0.0 | 0.1 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 0.0 | 0.1 | GO:0001834 | trophectodermal cell proliferation(GO:0001834) |
| 0.0 | 0.1 | GO:0006083 | acetate metabolic process(GO:0006083) propionate metabolic process(GO:0019541) |
| 0.0 | 0.1 | GO:0060467 | negative regulation of fertilization(GO:0060467) prevention of polyspermy(GO:0060468) |
| 0.0 | 0.1 | GO:0009115 | xanthine catabolic process(GO:0009115) |
| 0.0 | 0.1 | GO:1902996 | regulation of neuronal signal transduction(GO:1902847) positive regulation of tau-protein kinase activity(GO:1902949) neurofibrillary tangle assembly(GO:1902988) regulation of neurofibrillary tangle assembly(GO:1902996) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 0.0 | 0.1 | GO:0045196 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.0 | 0.1 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.0 | 0.1 | GO:0019563 | glycerol catabolic process(GO:0019563) |
| 0.0 | 0.1 | GO:0060315 | negative regulation of ryanodine-sensitive calcium-release channel activity(GO:0060315) |
| 0.0 | 0.1 | GO:1902623 | negative regulation of neutrophil migration(GO:1902623) |
| 0.0 | 0.1 | GO:0051106 | regulation of DNA ligation(GO:0051105) positive regulation of DNA ligation(GO:0051106) |
| 0.0 | 0.1 | GO:1990164 | histone H2A phosphorylation(GO:1990164) |
| 0.0 | 0.1 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.0 | 0.1 | GO:0071350 | interleukin-15-mediated signaling pathway(GO:0035723) cellular response to interleukin-15(GO:0071350) regulation of protein O-linked glycosylation(GO:1904098) positive regulation of protein O-linked glycosylation(GO:1904100) |
| 0.0 | 0.1 | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.0 | 0.0 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.0 | 0.0 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.0 | 0.1 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.0 | 0.1 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.0 | 0.0 | GO:0001905 | activation of membrane attack complex(GO:0001905) regulation of activation of membrane attack complex(GO:0001969) |
| 0.0 | 0.0 | GO:0043606 | formate metabolic process(GO:0015942) histidine catabolic process to glutamate and formamide(GO:0019556) formamide metabolic process(GO:0043606) |
| 0.0 | 0.1 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.1 | GO:0001554 | luteolysis(GO:0001554) |
| 0.0 | 0.0 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.0 | 0.1 | GO:0070494 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 0.0 | 0.0 | GO:0021678 | third ventricle development(GO:0021678) |
| 0.0 | 0.1 | GO:0070668 | negative regulation of mast cell apoptotic process(GO:0033026) mast cell proliferation(GO:0070662) regulation of mast cell proliferation(GO:0070666) positive regulation of mast cell proliferation(GO:0070668) |
| 0.0 | 0.1 | GO:0046813 | receptor-mediated virion attachment to host cell(GO:0046813) |
| 0.0 | 0.1 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.0 | 0.0 | GO:0006480 | N-terminal protein amino acid methylation(GO:0006480) |
| 0.0 | 0.1 | GO:0045359 | positive regulation of interferon-beta biosynthetic process(GO:0045359) |
| 0.0 | 0.1 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 0.0 | 0.1 | GO:0098912 | membrane depolarization during atrial cardiac muscle cell action potential(GO:0098912) |
| 0.0 | 0.1 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.0 | 0.0 | GO:0033386 | geranylgeranyl diphosphate metabolic process(GO:0033385) geranylgeranyl diphosphate biosynthetic process(GO:0033386) |
| 0.0 | 0.1 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.0 | 0.0 | GO:2000118 | dentinogenesis(GO:0097187) regulation of sodium-dependent phosphate transport(GO:2000118) |
| 0.0 | 0.1 | GO:0034626 | fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.1 | GO:0048690 | regulation of sprouting of injured axon(GO:0048686) regulation of axon extension involved in regeneration(GO:0048690) |
| 0.0 | 0.0 | GO:0071963 | establishment or maintenance of cell polarity regulating cell shape(GO:0071963) |
| 0.0 | 0.1 | GO:0052697 | xenobiotic glucuronidation(GO:0052697) |
| 0.0 | 0.0 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.0 | 0.1 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) |
| 0.0 | 0.0 | GO:0060830 | ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0036117 | hyaluranon cable(GO:0036117) |
| 0.0 | 0.1 | GO:0071914 | prominosome(GO:0071914) |
| 0.0 | 0.1 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.0 | 0.1 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.1 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.0 | 0.1 | GO:0072534 | perineuronal net(GO:0072534) |
| 0.0 | 0.1 | GO:0020005 | symbiont-containing vacuole membrane(GO:0020005) |
| 0.0 | 0.2 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.1 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.0 | 0.1 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.0 | GO:0005607 | laminin-2 complex(GO:0005607) |
| 0.0 | 0.0 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.0 | 0.1 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 0.1 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 0.0 | GO:0008275 | gamma-tubulin small complex(GO:0008275) |
| 0.0 | 0.1 | GO:0072557 | IPAF inflammasome complex(GO:0072557) |
| 0.0 | 0.0 | GO:0097232 | lamellar body membrane(GO:0097232) alveolar lamellar body membrane(GO:0097233) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.0 | 0.1 | GO:0047237 | glucuronylgalactosylproteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047237) |
| 0.0 | 0.1 | GO:0052591 | sn-glycerol-3-phosphate:ubiquinone oxidoreductase activity(GO:0052590) sn-glycerol-3-phosphate:ubiquinone-8 oxidoreductase activity(GO:0052591) |
| 0.0 | 0.2 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 0.0 | GO:0002134 | UTP binding(GO:0002134) pyrimidine ribonucleoside binding(GO:0032551) |
| 0.0 | 0.1 | GO:0016212 | kynurenine-oxoglutarate transaminase activity(GO:0016212) kynurenine aminotransferase activity(GO:0036137) |
| 0.0 | 0.1 | GO:0000829 | inositol heptakisphosphate kinase activity(GO:0000829) |
| 0.0 | 0.1 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 0.0 | 0.1 | GO:0023030 | MHC class Ib protein complex binding(GO:0023025) MHC class Ib protein binding, via antigen binding groove(GO:0023030) |
| 0.0 | 0.1 | GO:0016726 | xanthine dehydrogenase activity(GO:0004854) oxidoreductase activity, acting on CH or CH2 groups, NAD or NADP as acceptor(GO:0016726) |
| 0.0 | 0.1 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.1 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.0 | 0.1 | GO:0086059 | voltage-gated calcium channel activity involved SA node cell action potential(GO:0086059) |
| 0.0 | 0.1 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.1 | GO:0071723 | lipopeptide binding(GO:0071723) |
| 0.0 | 0.1 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.0 | GO:0001605 | adrenomedullin receptor activity(GO:0001605) |
| 0.0 | 0.1 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.0 | 0.1 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.1 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 0.1 | GO:0019002 | GMP binding(GO:0019002) |
| 0.0 | 0.1 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.0 | 0.0 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.0 | 0.0 | GO:0071885 | N-terminal protein N-methyltransferase activity(GO:0071885) |
| 0.0 | 0.1 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.0 | 0.0 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.0 | 0.0 | GO:0015265 | urea channel activity(GO:0015265) |
| 0.0 | 0.1 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.0 | 0.1 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.0 | 0.0 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.0 | 0.0 | GO:0016433 | rRNA (adenine) methyltransferase activity(GO:0016433) |
| 0.0 | 0.0 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.0 | 0.2 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 0.0 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.1 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.1 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |