Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
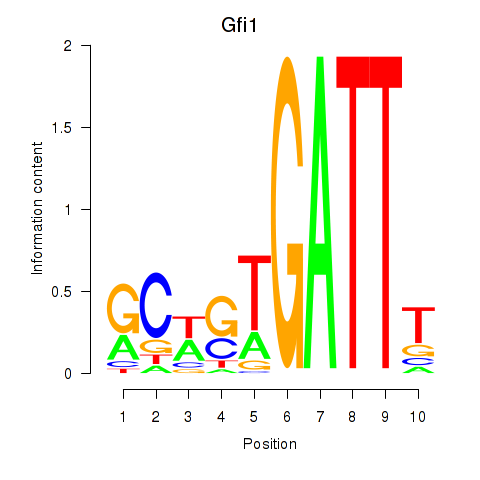
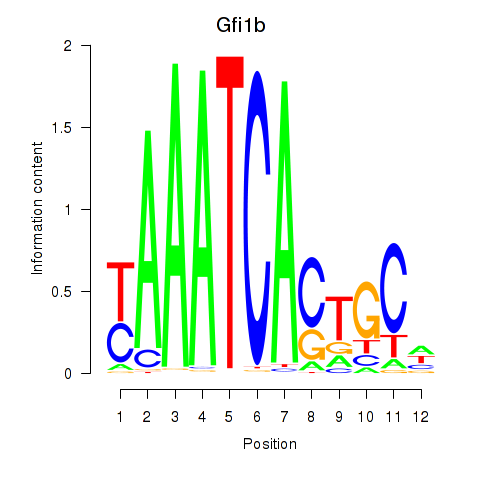
Results for Gfi1_Gfi1b
Z-value: 0.44
Transcription factors associated with Gfi1_Gfi1b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Gfi1
|
ENSRNOG00000002042 | growth factor independent 1 transcriptional repressor |
|
Gfi1b
|
ENSRNOG00000011353 | growth factor independent 1B transcriptional repressor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Gfi1b | rn6_v1_chr3_-_7203420_7203420 | -0.32 | 6.0e-01 | Click! |
| Gfi1 | rn6_v1_chr14_+_3058993_3058993 | -0.11 | 8.6e-01 | Click! |
Activity profile of Gfi1_Gfi1b motif
Sorted Z-values of Gfi1_Gfi1b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_+_43694183 | 0.45 |
ENSRNOT00000046342
|
RGD1563570
|
similar to ribosomal protein S23 |
| chr14_-_18745457 | 0.34 |
ENSRNOT00000003778
|
Cxcl1
|
C-X-C motif chemokine ligand 1 |
| chr3_-_123119460 | 0.32 |
ENSRNOT00000028833
|
Avp
|
arginine vasopressin |
| chr17_-_22143324 | 0.29 |
ENSRNOT00000019361
|
Edn1
|
endothelin 1 |
| chr1_-_227047290 | 0.26 |
ENSRNOT00000037226
|
Ms4a15
|
membrane spanning 4-domains A15 |
| chr4_-_44136815 | 0.18 |
ENSRNOT00000086810
|
Tfec
|
transcription factor EC |
| chr10_-_70871066 | 0.15 |
ENSRNOT00000015139
|
Ccl3
|
C-C motif chemokine ligand 3 |
| chr14_-_42221225 | 0.15 |
ENSRNOT00000036103
|
Shisa3
|
shisa family member 3 |
| chr8_-_55491152 | 0.15 |
ENSRNOT00000014965
|
LOC100359479
|
rCG58364-like |
| chr8_-_55194692 | 0.14 |
ENSRNOT00000068366
|
RGD1564937
|
similar to RIKEN cDNA 1110032A03 |
| chr13_-_50514151 | 0.14 |
ENSRNOT00000003951
|
Ren
|
renin |
| chr7_-_145174771 | 0.13 |
ENSRNOT00000055270
|
Glycam1
|
glycosylation dependent cell adhesion molecule 1 |
| chr1_-_88690534 | 0.13 |
ENSRNOT00000072570
|
Ovol3
|
ovo-like zinc finger 3 |
| chr6_+_8669722 | 0.13 |
ENSRNOT00000048550
|
Camkmt
|
calmodulin-lysine N-methyltransferase |
| chr6_-_123577695 | 0.13 |
ENSRNOT00000006604
|
Foxn3
|
forkhead box N3 |
| chr1_+_81372650 | 0.13 |
ENSRNOT00000088829
|
Zfp428
|
zinc finger protein 428 |
| chr4_+_170958196 | 0.13 |
ENSRNOT00000007905
|
Pde6h
|
phosphodiesterase 6H |
| chr10_+_70884531 | 0.13 |
ENSRNOT00000015199
|
Ccl4
|
C-C motif chemokine ligand 4 |
| chr14_-_6900733 | 0.12 |
ENSRNOT00000061224
|
Dmp1
|
dentin matrix acidic phosphoprotein 1 |
| chr14_-_8510138 | 0.12 |
ENSRNOT00000080758
|
Arhgap24
|
Rho GTPase activating protein 24 |
| chrX_+_124894466 | 0.12 |
ENSRNOT00000080894
|
Mcts1
|
MCTS1, re-initiation and release factor |
| chr8_+_53678777 | 0.12 |
ENSRNOT00000045944
|
Drd2
|
dopamine receptor D2 |
| chr5_-_136721379 | 0.12 |
ENSRNOT00000026704
|
Atp6v0b
|
ATPase H+ transporting V0 subunit B |
| chr1_+_91038886 | 0.11 |
ENSRNOT00000041301
|
Ovol3
|
ovo-like zinc finger 3 |
| chr6_-_76270457 | 0.11 |
ENSRNOT00000009894
|
Nfkbia
|
NFKB inhibitor alpha |
| chrX_+_124894706 | 0.11 |
ENSRNOT00000066677
|
Mcts1
|
MCTS1, re-initiation and release factor |
| chr5_-_130085838 | 0.10 |
ENSRNOT00000035252
|
Elavl4
|
ELAV like RNA binding protein 4 |
| chr20_+_4593389 | 0.10 |
ENSRNOT00000001174
|
Slc44a4
|
solute carrier family 44, member 4 |
| chr14_-_43694584 | 0.10 |
ENSRNOT00000041866
|
AABR07072400.1
|
|
| chr4_+_33638709 | 0.10 |
ENSRNOT00000009888
ENSRNOT00000034719 ENSRNOT00000052333 |
Tac1
|
tachykinin, precursor 1 |
| chr12_-_22021851 | 0.09 |
ENSRNOT00000039280
|
Tsc22d4
|
TSC22 domain family, member 4 |
| chr1_-_179010257 | 0.09 |
ENSRNOT00000017199
|
Rras2
|
related RAS viral (r-ras) oncogene homolog 2 |
| chr8_+_118378059 | 0.09 |
ENSRNOT00000043247
|
AABR07071482.1
|
|
| chr16_-_82439441 | 0.09 |
ENSRNOT00000040315
|
AABR07026539.1
|
|
| chr17_-_27969433 | 0.09 |
ENSRNOT00000073967
|
Nrn1
|
neuritin 1 |
| chrX_+_15049462 | 0.09 |
ENSRNOT00000007015
|
Ebp
|
emopamil binding protein (sterol isomerase) |
| chr6_+_50725085 | 0.08 |
ENSRNOT00000009473
|
Slc26a3
|
solute carrier family 26 member 3 |
| chr2_-_148807813 | 0.08 |
ENSRNOT00000047649
|
AC112531.1
|
|
| chr15_+_4475051 | 0.08 |
ENSRNOT00000008461
|
Kcnk16
|
potassium two pore domain channel subfamily K member 16 |
| chr8_-_13420439 | 0.08 |
ENSRNOT00000051471
|
Izumo1r
|
IZUMO1 receptor, JUNO |
| chr3_+_170252901 | 0.08 |
ENSRNOT00000005871
|
Mc3r
|
melanocortin 3 receptor |
| chr1_-_166189741 | 0.08 |
ENSRNOT00000025728
|
AABR07004891.1
|
|
| chr17_+_72429618 | 0.08 |
ENSRNOT00000026187
|
Gata3
|
GATA binding protein 3 |
| chr7_-_29152442 | 0.08 |
ENSRNOT00000079774
|
Mybpc1
|
myosin binding protein C, slow type |
| chr2_+_154580558 | 0.08 |
ENSRNOT00000076976
|
Gmps
|
guanine monophosphate synthase |
| chr11_-_60679555 | 0.08 |
ENSRNOT00000059735
|
Ccdc80
|
coiled-coil domain containing 80 |
| chr1_-_25839198 | 0.07 |
ENSRNOT00000090388
ENSRNOT00000092757 ENSRNOT00000042072 |
Trdn
|
triadin |
| chr16_-_49453394 | 0.07 |
ENSRNOT00000041617
|
Lrp2bp
|
Lrp2 binding protein |
| chr4_+_114854458 | 0.07 |
ENSRNOT00000013312
|
LOC500227
|
hypothetical gene supported by BC079424 |
| chr10_-_13780283 | 0.07 |
ENSRNOT00000084200
|
AC103090.1
|
|
| chr11_-_88092756 | 0.07 |
ENSRNOT00000002542
|
Ydjc
|
YdjC homolog (bacterial) |
| chr4_-_114853868 | 0.07 |
ENSRNOT00000090828
|
Wdr54
|
WD repeat domain 54 |
| chr10_+_69423086 | 0.07 |
ENSRNOT00000000256
|
Ccl7
|
C-C motif chemokine ligand 7 |
| chr19_-_26243223 | 0.07 |
ENSRNOT00000037766
|
Zfp791
|
zinc finger protein 791 |
| chr7_-_96464049 | 0.07 |
ENSRNOT00000006517
|
Has2
|
hyaluronan synthase 2 |
| chr1_-_12952906 | 0.07 |
ENSRNOT00000078193
|
LOC100360362
|
hypothetical protein LOC100360362 |
| chr5_+_58393233 | 0.07 |
ENSRNOT00000000142
|
Dnajb5
|
DnaJ heat shock protein family (Hsp40) member B5 |
| chrX_-_136807885 | 0.07 |
ENSRNOT00000010325
|
Igsf1
|
immunoglobulin superfamily, member 1 |
| chr11_+_32655616 | 0.07 |
ENSRNOT00000084412
ENSRNOT00000034383 |
Clic6
|
chloride intracellular channel 6 |
| chr10_-_47059216 | 0.07 |
ENSRNOT00000007092
|
Shmt1
|
serine hydroxymethyltransferase 1 |
| chr14_-_6813945 | 0.07 |
ENSRNOT00000002938
|
Ibsp
|
integrin-binding sialoprotein |
| chr2_-_257484587 | 0.07 |
ENSRNOT00000083344
|
Nexn
|
nexilin (F actin binding protein) |
| chr1_-_124803363 | 0.06 |
ENSRNOT00000066380
|
Klf13
|
Kruppel-like factor 13 |
| chr5_-_139864393 | 0.06 |
ENSRNOT00000030016
|
Zfp69
|
zinc finger protein 69 |
| chr5_+_107712579 | 0.06 |
ENSRNOT00000090739
|
Mtap
|
methylthioadenosine phosphorylase |
| chr5_-_126782726 | 0.06 |
ENSRNOT00000048784
|
Tceanc2
|
transcription elongation factor A N-terminal and central domain containing 2 |
| chr3_+_66673246 | 0.06 |
ENSRNOT00000081338
|
Ppp1r1c
|
protein phosphatase 1, regulatory (inhibitor) subunit 1C |
| chr13_+_89386023 | 0.06 |
ENSRNOT00000086223
|
Fcgr3a
|
Fc fragment of IgG receptor IIIa |
| chr4_-_16130848 | 0.06 |
ENSRNOT00000042914
|
Cacna2d1
|
calcium voltage-gated channel auxiliary subunit alpha2delta 1 |
| chr9_-_100888107 | 0.06 |
ENSRNOT00000024794
|
Thap4
|
THAP domain containing 4 |
| chr5_-_68139199 | 0.06 |
ENSRNOT00000009987
|
Plppr1
|
phospholipid phosphatase related 1 |
| chr2_-_187915596 | 0.06 |
ENSRNOT00000093008
|
Lamtor2
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 2 |
| chrX_-_1369384 | 0.06 |
ENSRNOT00000013745
|
Timp1
|
TIMP metallopeptidase inhibitor 1 |
| chr18_-_27520295 | 0.06 |
ENSRNOT00000027544
|
Gfra3
|
GDNF family receptor alpha 3 |
| chr2_-_28799266 | 0.06 |
ENSRNOT00000089293
|
Tmem171
|
transmembrane protein 171 |
| chr13_+_63526486 | 0.06 |
ENSRNOT00000003788
|
Brinp3
|
BMP/retinoic acid inducible neural specific 3 |
| chrX_-_72133692 | 0.06 |
ENSRNOT00000004263
|
Cited1
|
Cbp/p300-interacting transactivator with Glu/Asp-rich carboxy-terminal domain 1 |
| chr1_+_167320797 | 0.05 |
ENSRNOT00000076923
|
Pgap2
|
post-GPI attachment to proteins 2 |
| chr20_+_55594676 | 0.05 |
ENSRNOT00000057010
|
Sim1
|
single-minded family bHLH transcription factor 1 |
| chr13_+_89385859 | 0.05 |
ENSRNOT00000047434
|
Fcgr3a
|
Fc fragment of IgG receptor IIIa |
| chr18_+_56044369 | 0.05 |
ENSRNOT00000059501
|
Rps14
|
ribosomal protein S14 |
| chr8_+_91464229 | 0.05 |
ENSRNOT00000013249
|
Bckdhb
|
branched chain keto acid dehydrogenase E1 subunit beta |
| chr16_-_62294769 | 0.05 |
ENSRNOT00000020663
|
Ppp2cb
|
protein phosphatase 2 catalytic subunit beta |
| chr4_-_129430251 | 0.05 |
ENSRNOT00000067881
|
Fam19a4
|
family with sequence similarity 19 member A4, C-C motif chemokine like |
| chr5_-_172307431 | 0.05 |
ENSRNOT00000018453
|
Fam213b
|
family with sequence similarity 213, member B |
| chr18_-_4371899 | 0.05 |
ENSRNOT00000051207
|
LOC108349606
|
60S ribosomal protein L7a-like |
| chr4_-_168884886 | 0.05 |
ENSRNOT00000011141
|
Gprc5d
|
G protein-coupled receptor, class C, group 5, member D |
| chr9_+_41045363 | 0.05 |
ENSRNOT00000066284
|
Cfc1
|
cripto, FRL-1, cryptic family 1 |
| chrX_+_104684676 | 0.05 |
ENSRNOT00000083229
|
Tnmd
|
tenomodulin |
| chr2_+_205160405 | 0.05 |
ENSRNOT00000035605
|
Tspan2
|
tetraspanin 2 |
| chr1_+_77541359 | 0.05 |
ENSRNOT00000049028
|
LOC103690015
|
40S ribosomal protein S19-like |
| chr5_+_131257127 | 0.05 |
ENSRNOT00000010628
|
Bend5
|
BEN domain containing 5 |
| chr6_+_34030620 | 0.05 |
ENSRNOT00000091651
|
Laptm4a
|
lysosomal protein transmembrane 4 alpha |
| chr1_-_89473904 | 0.05 |
ENSRNOT00000089474
|
Fxyd5
|
FXYD domain-containing ion transport regulator 5 |
| chr10_-_57653359 | 0.05 |
ENSRNOT00000089638
|
Derl2
|
derlin 2 |
| chr1_+_201429771 | 0.05 |
ENSRNOT00000027836
|
Plekha1
|
pleckstrin homology domain containing A1 |
| chr17_-_2705123 | 0.05 |
ENSRNOT00000024940
|
Olr1652
|
olfactory receptor 1652 |
| chr2_+_208323882 | 0.05 |
ENSRNOT00000085178
|
Tmigd3
|
transmembrane and immunoglobulin domain containing 3 |
| chr9_-_71900044 | 0.05 |
ENSRNOT00000020322
|
Idh1
|
isocitrate dehydrogenase (NADP(+)) 1, cytosolic |
| chr2_-_218658761 | 0.05 |
ENSRNOT00000018318
|
S1pr1
|
sphingosine-1-phosphate receptor 1 |
| chr9_+_81816872 | 0.05 |
ENSRNOT00000041407
|
Plcd4
|
phospholipase C, delta 4 |
| chr16_-_49574314 | 0.05 |
ENSRNOT00000017568
ENSRNOT00000085535 ENSRNOT00000017054 |
Pdlim3
|
PDZ and LIM domain 3 |
| chr3_+_67849966 | 0.05 |
ENSRNOT00000057826
|
Dusp19
|
dual specificity phosphatase 19 |
| chr17_-_43640387 | 0.05 |
ENSRNOT00000087731
|
Hist1h1c
|
histone cluster 1 H1 family member c |
| chr19_+_56024903 | 0.05 |
ENSRNOT00000021607
|
Cdk10
|
cyclin-dependent kinase 10 |
| chr10_+_86819929 | 0.05 |
ENSRNOT00000038228
|
Cdc6
|
cell division cycle 6 |
| chr6_+_70289464 | 0.05 |
ENSRNOT00000085187
|
AABR07064232.1
|
|
| chr17_-_43675934 | 0.05 |
ENSRNOT00000081345
|
Hist1h1t
|
histone cluster 1 H1 family member t |
| chr10_+_96661696 | 0.05 |
ENSRNOT00000077567
|
AABR07030630.1
|
|
| chr7_-_49741540 | 0.05 |
ENSRNOT00000006523
|
Myf6
|
myogenic factor 6 |
| chr1_-_187779675 | 0.05 |
ENSRNOT00000024648
|
Arl6ip1
|
ADP-ribosylation factor like GTPase 6 interacting protein 1 |
| chr1_+_51619875 | 0.05 |
ENSRNOT00000023319
|
Pabpc6
|
poly(A) binding protein, cytoplasmic 6 |
| chr1_+_48033768 | 0.04 |
ENSRNOT00000020064
|
Mrpl18
|
mitochondrial ribosomal protein L18 |
| chr12_-_12025549 | 0.04 |
ENSRNOT00000001331
|
Nptx2
|
neuronal pentraxin 2 |
| chrX_-_71169865 | 0.04 |
ENSRNOT00000050415
|
Il2rg
|
interleukin 2 receptor subunit gamma |
| chr2_+_196277978 | 0.04 |
ENSRNOT00000028625
|
Vps72
|
vacuolar protein sorting 72 homolog |
| chr7_-_123586919 | 0.04 |
ENSRNOT00000011484
|
Ndufa6
|
NADH:ubiquinone oxidoreductase subunit A6 |
| chr2_-_251532312 | 0.04 |
ENSRNOT00000019501
|
Cyr61
|
cysteine-rich, angiogenic inducer, 61 |
| chr4_+_14039977 | 0.04 |
ENSRNOT00000091249
ENSRNOT00000075878 |
Cd36
|
CD36 molecule |
| chr2_+_224851383 | 0.04 |
ENSRNOT00000015354
|
Alg14
|
ALG14, UDP-N-acetylglucosaminyltransferase subunit |
| chr10_-_8498422 | 0.04 |
ENSRNOT00000082332
|
Rbfox1
|
RNA binding protein, fox-1 homolog 1 |
| chr2_-_96032722 | 0.04 |
ENSRNOT00000015746
|
LOC100360846
|
proteasome subunit beta type 6-like |
| chr4_-_179700130 | 0.04 |
ENSRNOT00000021306
|
Lmntd1
|
lamin tail domain containing 1 |
| chr1_-_100887853 | 0.04 |
ENSRNOT00000027675
|
Med25
|
mediator complex subunit 25 |
| chr2_-_210738378 | 0.04 |
ENSRNOT00000025746
|
Gstm6
|
glutathione S-transferase, mu 6 |
| chr4_-_40385349 | 0.04 |
ENSRNOT00000039005
|
Gpr85
|
G protein-coupled receptor 85 |
| chr8_+_118926478 | 0.04 |
ENSRNOT00000028426
|
Ccdc12
|
coiled-coil domain containing 12 |
| chr1_+_88620317 | 0.04 |
ENSRNOT00000075116
|
Cox7a1
|
cytochrome c oxidase subunit 7A1 |
| chr17_-_70325855 | 0.04 |
ENSRNOT00000024952
|
Gdi2
|
GDP dissociation inhibitor 2 |
| chr9_+_95161157 | 0.04 |
ENSRNOT00000071200
|
Ugt1a5
|
UDP glucuronosyltransferase family 1 member A5 |
| chr10_-_81653717 | 0.04 |
ENSRNOT00000003611
|
Nme2
|
NME/NM23 nucleoside diphosphate kinase 2 |
| chr17_+_82066152 | 0.04 |
ENSRNOT00000083034
|
Arl5b
|
ADP-ribosylation factor like GTPase 5B |
| chr15_-_37830131 | 0.04 |
ENSRNOT00000075861
|
Eef1akmt1
|
eukaryotic translation elongation factor 1 alpha lysine methyltransferase 1 |
| chr20_+_46044892 | 0.04 |
ENSRNOT00000057187
|
Ak9
|
adenylate kinase 9 |
| chr1_+_239266921 | 0.04 |
ENSRNOT00000044486
|
Abhd17b
|
abhydrolase domain containing 17B |
| chr3_-_111422203 | 0.04 |
ENSRNOT00000084290
|
Oip5
|
Opa interacting protein 5 |
| chr19_+_32770655 | 0.04 |
ENSRNOT00000071471
|
LOC100360750
|
Sm protein F-like |
| chr8_+_100260049 | 0.04 |
ENSRNOT00000011090
|
AABR07071101.1
|
|
| chr16_-_38063821 | 0.04 |
ENSRNOT00000077452
ENSRNOT00000089533 |
Glra3
|
glycine receptor, alpha 3 |
| chr1_-_44474674 | 0.04 |
ENSRNOT00000078768
|
Tfb1m
|
transcription factor B1, mitochondrial |
| chr4_-_170932618 | 0.04 |
ENSRNOT00000007779
|
Arhgdib
|
Rho GDP dissociation inhibitor beta |
| chr17_+_36771639 | 0.04 |
ENSRNOT00000015147
|
Gpr50
|
G protein-coupled receptor 50 |
| chr1_+_105349069 | 0.04 |
ENSRNOT00000056030
|
Nell1
|
neural EGFL like 1 |
| chr15_+_12569235 | 0.04 |
ENSRNOT00000083787
|
Thoc7
|
THO complex 7 |
| chrX_-_118998300 | 0.04 |
ENSRNOT00000025575
|
AABR07041089.1
|
|
| chr6_-_47904163 | 0.04 |
ENSRNOT00000011333
|
Rps7
|
ribosomal protein S7 |
| chr4_+_150199627 | 0.04 |
ENSRNOT00000052259
|
AABR07061871.1
|
|
| chrX_+_66314418 | 0.04 |
ENSRNOT00000035405
|
Pgr15l
|
G protein-coupled receptor 15-like |
| chr6_-_92643847 | 0.04 |
ENSRNOT00000009183
|
Pygl
|
glycogen phosphorylase L |
| chr6_+_69971227 | 0.04 |
ENSRNOT00000075349
|
Foxg1
|
forkhead box G1 |
| chrX_+_15679254 | 0.04 |
ENSRNOT00000045256
|
Plp2
|
proteolipid protein 2 |
| chr10_+_64952119 | 0.04 |
ENSRNOT00000012154
|
Pipox
|
pipecolic acid and sarcosine oxidase |
| chr1_+_243589607 | 0.04 |
ENSRNOT00000021792
|
Dmrt3
|
doublesex and mab-3 related transcription factor 3 |
| chr2_+_198726118 | 0.04 |
ENSRNOT00000056237
|
Lix1l
|
limb and CNS expressed 1 like |
| chrX_-_72077590 | 0.04 |
ENSRNOT00000004278
|
Rps4x
|
ribosomal protein S4, X-linked |
| chr10_-_83655182 | 0.04 |
ENSRNOT00000007897
|
Abi3
|
ABI family, member 3 |
| chr1_-_80835701 | 0.04 |
ENSRNOT00000064305
|
PVR
|
poliovirus receptor |
| chr1_-_146823762 | 0.03 |
ENSRNOT00000018098
|
Zfand6
|
zinc finger AN1-type containing 6 |
| chr1_+_102396538 | 0.03 |
ENSRNOT00000015109
|
Myod1
|
myogenic differentiation 1 |
| chr19_+_54314865 | 0.03 |
ENSRNOT00000024069
|
Irf8
|
interferon regulatory factor 8 |
| chr1_-_154170409 | 0.03 |
ENSRNOT00000089014
|
Hikeshi
|
Hikeshi, heat shock protein nuclear import factor |
| chr15_-_5510289 | 0.03 |
ENSRNOT00000085883
|
Spetex-2F
|
Spetex-2F protein |
| chr10_+_110893232 | 0.03 |
ENSRNOT00000072190
|
Metrnl
|
meteorin-like, glial cell differentiation regulator |
| chr9_+_111327402 | 0.03 |
ENSRNOT00000045749
|
RGD1562136
|
similar to D1Ertd622e protein |
| chr4_+_8066737 | 0.03 |
ENSRNOT00000063994
|
Srpk2
|
SRSF protein kinase 2 |
| chr17_-_89780691 | 0.03 |
ENSRNOT00000091830
|
LOC108348042
|
ankyrin repeat domain-containing protein 7-like |
| chr3_-_72895740 | 0.03 |
ENSRNOT00000012568
|
Fads2l1
|
fatty acid desaturase 2-like 1 |
| chr6_-_47904437 | 0.03 |
ENSRNOT00000092867
|
Rps7
|
ribosomal protein S7 |
| chr2_+_187951344 | 0.03 |
ENSRNOT00000027123
|
Ssr2
|
signal sequence receptor, beta |
| chr15_-_27997810 | 0.03 |
ENSRNOT00000034647
|
Olr1637
|
olfactory receptor 1637 |
| chr11_+_60072727 | 0.03 |
ENSRNOT00000090230
|
Tagln3
|
transgelin 3 |
| chr5_-_9035811 | 0.03 |
ENSRNOT00000082023
|
Mcmdc2
|
minichromosome maintenance domain containing 2 |
| chr18_+_60745035 | 0.03 |
ENSRNOT00000045098
|
LOC502176
|
hypothetical protein LOC502176 |
| chr5_-_115387377 | 0.03 |
ENSRNOT00000036030
ENSRNOT00000077492 |
RGD1560146
|
similar to hypothetical protein MGC34837 |
| chr4_+_136512201 | 0.03 |
ENSRNOT00000051645
|
Cntn6
|
contactin 6 |
| chr8_-_118378460 | 0.03 |
ENSRNOT00000047247
|
RGD1563705
|
similar to ribosomal protein S23 |
| chr2_+_208749996 | 0.03 |
ENSRNOT00000086321
|
Chia
|
chitinase, acidic |
| chr4_+_8066907 | 0.03 |
ENSRNOT00000080811
|
Srpk2
|
SRSF protein kinase 2 |
| chr10_-_34333305 | 0.03 |
ENSRNOT00000071365
|
Olr1384
|
olfactory receptor gene Olr1384 |
| chr8_-_72714664 | 0.03 |
ENSRNOT00000024286
|
Rab8b
|
RAB8B, member RAS oncogene family |
| chr5_+_126783061 | 0.03 |
ENSRNOT00000013224
|
Tmem59
|
transmembrane protein 59 |
| chr9_+_46657922 | 0.03 |
ENSRNOT00000019180
|
Map4k4
|
mitogen-activated protein kinase kinase kinase kinase 4 |
| chr11_-_62067655 | 0.03 |
ENSRNOT00000093382
|
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr15_-_27981663 | 0.03 |
ENSRNOT00000064936
|
Rnase12
|
ribonuclease A family member 12 |
| chr2_+_188478082 | 0.03 |
ENSRNOT00000085817
|
Clk2
|
CDC-like kinase 2 |
| chr4_-_10269748 | 0.03 |
ENSRNOT00000074662
|
Fam185a
|
family with sequence similarity 185, member A |
| chr3_+_163552166 | 0.03 |
ENSRNOT00000049082
|
Tp53rk
|
TP53 regulating kinase |
| chr9_-_113246545 | 0.03 |
ENSRNOT00000034759
|
Tmem232
|
transmembrane protein 232 |
| chr1_-_64099277 | 0.03 |
ENSRNOT00000084846
|
Tsen34
|
tRNA splicing endonuclease subunit 34 |
| chr1_-_156327352 | 0.03 |
ENSRNOT00000074282
|
LOC100911776
|
coiled-coil domain-containing protein 89-like |
| chr17_+_47302272 | 0.03 |
ENSRNOT00000090021
|
Nme8
|
NME/NM23 family member 8 |
| chr11_+_57505005 | 0.03 |
ENSRNOT00000002942
|
LOC103693564
|
transgelin-3 |
| chr3_+_170996193 | 0.03 |
ENSRNOT00000008959
|
Spo11
|
SPO11, initiator of meiotic double stranded breaks |
| chr1_+_86996093 | 0.03 |
ENSRNOT00000027425
ENSRNOT00000027494 |
Hnrnpl
|
heterogeneous nuclear ribonucleoprotein L |
| chr1_-_53563032 | 0.03 |
ENSRNOT00000044831
|
Ttll2
|
tubulin tyrosine ligase like 2 |
| chr1_-_84353725 | 0.03 |
ENSRNOT00000057177
|
Pld3
|
phospholipase D family, member 3 |
| chr1_+_81499821 | 0.03 |
ENSRNOT00000027100
|
Lypd3
|
Ly6/Plaur domain containing 3 |
| chr12_+_2213705 | 0.03 |
ENSRNOT00000031653
|
Mcemp1
|
mast cell-expressed membrane protein 1 |
| chr14_-_1505085 | 0.03 |
ENSRNOT00000090361
|
LOC102554799
|
uncharacterized LOC102554799 |
| chr16_-_14210449 | 0.03 |
ENSRNOT00000016494
|
Ccser2
|
coiled-coil serine-rich protein 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Gfi1_Gfi1b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0070948 | regulation of neutrophil mediated cytotoxicity(GO:0070948) regulation of neutrophil mediated killing of symbiont cell(GO:0070949) |
| 0.1 | 0.3 | GO:0031583 | phospholipase D-activating G-protein coupled receptor signaling pathway(GO:0031583) |
| 0.1 | 0.3 | GO:0002125 | maternal aggressive behavior(GO:0002125) |
| 0.1 | 0.2 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.1 | 0.3 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.0 | 0.1 | GO:0070173 | regulation of enamel mineralization(GO:0070173) |
| 0.0 | 0.1 | GO:0090158 | endoplasmic reticulum membrane organization(GO:0090158) |
| 0.0 | 0.1 | GO:0060160 | negative regulation of dopamine receptor signaling pathway(GO:0060160) |
| 0.0 | 0.1 | GO:0002018 | angiotensin maturation(GO:0002003) renin-angiotensin regulation of aldosterone production(GO:0002018) |
| 0.0 | 0.1 | GO:0002572 | pro-T cell differentiation(GO:0002572) |
| 0.0 | 0.1 | GO:1901367 | response to L-cysteine(GO:1901367) |
| 0.0 | 0.1 | GO:1901164 | negative regulation of trophoblast cell migration(GO:1901164) |
| 0.0 | 0.1 | GO:0009177 | pyrimidine deoxyribonucleoside monophosphate biosynthetic process(GO:0009177) |
| 0.0 | 0.1 | GO:0046495 | nicotinamide riboside catabolic process(GO:0006738) nicotinamide riboside metabolic process(GO:0046495) pyridine nucleoside metabolic process(GO:0070637) pyridine nucleoside catabolic process(GO:0070638) |
| 0.0 | 0.1 | GO:1900620 | acetylcholine biosynthetic process(GO:0008292) acetate ester biosynthetic process(GO:1900620) |
| 0.0 | 0.1 | GO:0046878 | positive regulation of saliva secretion(GO:0046878) positive regulation of glucocorticoid secretion(GO:2000851) |
| 0.0 | 0.0 | GO:0043179 | rhythmic excitation(GO:0043179) |
| 0.0 | 0.1 | GO:0070427 | nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070427) |
| 0.0 | 0.1 | GO:0051454 | pH elevation(GO:0045852) intracellular pH elevation(GO:0051454) |
| 0.0 | 0.1 | GO:1900623 | positive regulation of hyaluronan biosynthetic process(GO:1900127) regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.0 | 0.0 | GO:0060460 | left lung morphogenesis(GO:0060460) |
| 0.0 | 0.0 | GO:0009216 | purine deoxyribonucleoside triphosphate biosynthetic process(GO:0009216) |
| 0.0 | 0.0 | GO:0035928 | rRNA import into mitochondrion(GO:0035928) |
| 0.0 | 0.1 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.0 | 0.1 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.0 | 0.1 | GO:1990918 | double-strand break repair involved in meiotic recombination(GO:1990918) |
| 0.0 | 0.1 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.0 | 0.0 | GO:0046487 | glyoxylate metabolic process(GO:0046487) |
| 0.0 | 0.0 | GO:0060370 | susceptibility to T cell mediated cytotoxicity(GO:0060370) |
| 0.0 | 0.0 | GO:0002584 | negative regulation of antigen processing and presentation of peptide antigen(GO:0002584) |
| 0.0 | 0.1 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.0 | 0.1 | GO:0071104 | negative regulation of mesenchymal to epithelial transition involved in metanephros morphogenesis(GO:0003340) response to interleukin-9(GO:0071104) |
| 0.0 | 0.0 | GO:0090283 | regulation of protein glycosylation in Golgi(GO:0090283) |
| 0.0 | 0.0 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.1 | GO:0086048 | membrane depolarization during bundle of His cell action potential(GO:0086048) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.0 | 0.1 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.0 | 0.1 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.0 | GO:0098982 | glycine-gated chloride channel complex(GO:0016935) GABA-ergic synapse(GO:0098982) |
| 0.0 | 0.1 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 0.1 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.0 | 0.1 | GO:0042788 | polysomal ribosome(GO:0042788) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0031893 | vasopressin receptor binding(GO:0031893) |
| 0.1 | 0.3 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.1 | 0.3 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.0 | 0.1 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.1 | GO:0019767 | IgE receptor activity(GO:0019767) |
| 0.0 | 0.1 | GO:0070905 | serine binding(GO:0070905) |
| 0.0 | 0.1 | GO:0003922 | GMP synthase activity(GO:0003921) GMP synthase (glutamine-hydrolyzing) activity(GO:0003922) |
| 0.0 | 0.3 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.1 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.0 | 0.1 | GO:0086057 | voltage-gated calcium channel activity involved in bundle of His cell action potential(GO:0086057) |
| 0.0 | 0.1 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.0 | 0.0 | GO:0002060 | purine nucleobase binding(GO:0002060) |
| 0.0 | 0.1 | GO:0043199 | sulfate binding(GO:0043199) |
| 0.0 | 0.1 | GO:0031750 | D3 dopamine receptor binding(GO:0031750) |
| 0.0 | 0.0 | GO:0051990 | (R)-2-hydroxyglutarate dehydrogenase activity(GO:0051990) |
| 0.0 | 0.0 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.1 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 0.0 | GO:0050145 | nucleoside phosphate kinase activity(GO:0050145) |
| 0.0 | 0.0 | GO:0022852 | glycine-gated chloride ion channel activity(GO:0022852) |
| 0.0 | 0.0 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.0 | 0.0 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.0 | 0.1 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.0 | GO:0061505 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.0 | 0.1 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.1 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.0 | 0.1 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.0 | 0.0 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.7 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |