Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
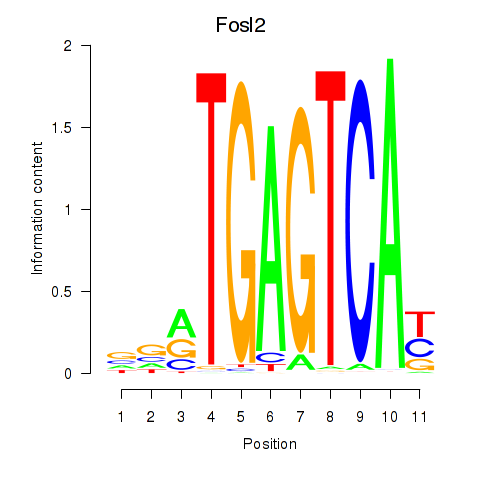
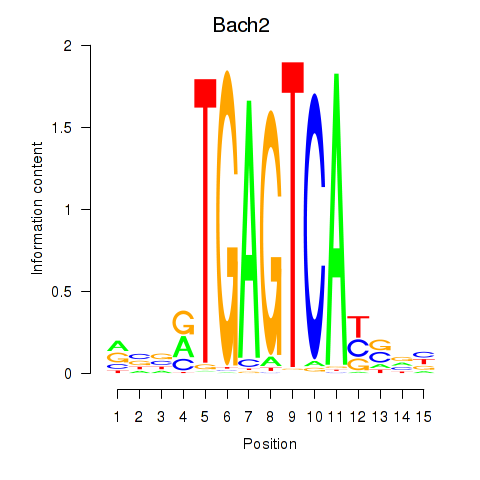
Results for Fosl2_Bach2
Z-value: 0.92
Transcription factors associated with Fosl2_Bach2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Fosl2
|
ENSRNOG00000052357 | FOS like 2, AP-1 transcription factor subunit |
|
Bach2
|
ENSRNOG00000006170 | BTB domain and CNC homolog 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Bach2 | rn6_v1_chr5_+_47546014_47546014 | -0.72 | 1.7e-01 | Click! |
| Fosl2 | rn6_v1_chr6_-_25616995_25616995 | -0.62 | 2.7e-01 | Click! |
Activity profile of Fosl2_Bach2 motif
Sorted Z-values of Fosl2_Bach2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_187863503 | 0.85 |
ENSRNOT00000093036
ENSRNOT00000026705 ENSRNOT00000082174 |
Lmna
|
lamin A/C |
| chr10_-_82229140 | 0.70 |
ENSRNOT00000004400
|
Epn3
|
epsin 3 |
| chr10_-_90312386 | 0.65 |
ENSRNOT00000028445
|
Slc4a1
|
solute carrier family 4 member 1 |
| chr7_+_28654733 | 0.55 |
ENSRNOT00000006174
|
Pmch
|
pro-melanin-concentrating hormone |
| chr1_+_198690794 | 0.53 |
ENSRNOT00000023999
|
Zfp771
|
zinc finger protein 771 |
| chr1_-_24191908 | 0.51 |
ENSRNOT00000061157
|
Sgk1
|
serum/glucocorticoid regulated kinase 1 |
| chr10_-_104575890 | 0.50 |
ENSRNOT00000050223
|
H3f3b
|
H3 histone family member 3B |
| chr17_+_6909728 | 0.48 |
ENSRNOT00000061231
|
LOC681410
|
hypothetical protein LOC681410 |
| chr3_+_147609095 | 0.47 |
ENSRNOT00000041456
|
Srxn1
|
sulfiredoxin 1 |
| chr1_+_218466289 | 0.46 |
ENSRNOT00000017948
|
Mrgprf
|
MAS related GPR family member F |
| chr8_+_48571323 | 0.44 |
ENSRNOT00000059776
|
Ccdc153
|
coiled-coil domain containing 153 |
| chr20_+_5008508 | 0.43 |
ENSRNOT00000001153
|
Vwa7
|
von Willebrand factor A domain containing 7 |
| chr7_+_59200918 | 0.41 |
ENSRNOT00000085073
|
Ptprr
|
protein tyrosine phosphatase, receptor type, R |
| chr1_+_199941161 | 0.41 |
ENSRNOT00000027616
|
Bag3
|
Bcl2-associated athanogene 3 |
| chr4_+_33638709 | 0.40 |
ENSRNOT00000009888
ENSRNOT00000034719 ENSRNOT00000052333 |
Tac1
|
tachykinin, precursor 1 |
| chr1_-_87191588 | 0.39 |
ENSRNOT00000027998
|
LOC100361913
|
rCG54286-like |
| chr1_+_220826560 | 0.35 |
ENSRNOT00000027891
|
Fosl1
|
FOS like 1, AP-1 transcription factor subunit |
| chr8_+_117117430 | 0.35 |
ENSRNOT00000073247
|
Gpx1
|
glutathione peroxidase 1 |
| chr8_+_75687100 | 0.35 |
ENSRNOT00000038677
|
Anxa2
|
annexin A2 |
| chr13_+_89480058 | 0.32 |
ENSRNOT00000050804
|
Cfap126
|
cilia and flagella associated protein 126 |
| chr20_+_5040337 | 0.32 |
ENSRNOT00000068435
|
Clic1
|
chloride intracellular channel 1 |
| chr10_+_35537977 | 0.31 |
ENSRNOT00000071917
|
Rnf130
|
ring finger protein 130 |
| chr11_+_87435185 | 0.29 |
ENSRNOT00000002558
|
P2rx6
|
purinergic receptor P2X 6 |
| chr13_-_50499060 | 0.29 |
ENSRNOT00000065347
ENSRNOT00000076924 |
Etnk2
|
ethanolamine kinase 2 |
| chr3_+_1452644 | 0.26 |
ENSRNOT00000007949
|
Il1rn
|
interleukin 1 receptor antagonist |
| chr3_-_105512939 | 0.25 |
ENSRNOT00000011773
|
Actc1
|
actin, alpha, cardiac muscle 1 |
| chr1_+_105284753 | 0.25 |
ENSRNOT00000041950
|
Slc6a5
|
solute carrier family 6 member 5 |
| chr15_-_61564695 | 0.24 |
ENSRNOT00000068216
|
Rgcc
|
regulator of cell cycle |
| chr1_+_89008117 | 0.24 |
ENSRNOT00000028401
|
Hspb6
|
heat shock protein family B (small) member 6 |
| chr20_-_3818045 | 0.24 |
ENSRNOT00000091622
|
Hsd17b8
|
hydroxysteroid (17-beta) dehydrogenase 8 |
| chr8_+_55178289 | 0.24 |
ENSRNOT00000059127
|
Cryab
|
crystallin, alpha B |
| chr1_+_72636959 | 0.22 |
ENSRNOT00000023489
|
Il11
|
interleukin 11 |
| chr4_+_169147243 | 0.22 |
ENSRNOT00000011580
|
Emp1
|
epithelial membrane protein 1 |
| chr7_+_3355116 | 0.21 |
ENSRNOT00000030453
ENSRNOT00000061657 |
Itga7
|
integrin subunit alpha 7 |
| chr2_-_187854363 | 0.21 |
ENSRNOT00000092993
|
Lmna
|
lamin A/C |
| chr1_-_88193346 | 0.21 |
ENSRNOT00000078600
|
LOC103690068
|
immortalization up-regulated protein-like |
| chr7_+_144647587 | 0.21 |
ENSRNOT00000022398
|
Hoxc4
|
homeo box C4 |
| chr4_+_71740532 | 0.20 |
ENSRNOT00000023537
|
Zyx
|
zyxin |
| chr15_-_34444244 | 0.20 |
ENSRNOT00000027612
|
Cideb
|
cell death-inducing DFFA-like effector b |
| chr2_-_187863349 | 0.20 |
ENSRNOT00000084455
|
Lmna
|
lamin A/C |
| chr1_-_101514547 | 0.20 |
ENSRNOT00000079633
|
Ppp1r15a
|
protein phosphatase 1, regulatory subunit 15A |
| chr12_+_24761210 | 0.20 |
ENSRNOT00000002003
|
Cldn4
|
claudin 4 |
| chr7_-_115981731 | 0.19 |
ENSRNOT00000036549
|
RGD1308195
|
similar to secreted Ly6/uPAR related protein 2 |
| chr14_-_10395047 | 0.19 |
ENSRNOT00000002936
|
Gpat3
|
glycerol-3-phosphate acyltransferase 3 |
| chr1_-_124803363 | 0.18 |
ENSRNOT00000066380
|
Klf13
|
Kruppel-like factor 13 |
| chr16_+_23317953 | 0.18 |
ENSRNOT00000075287
|
AABR07024972.1
|
|
| chr5_+_59008933 | 0.18 |
ENSRNOT00000023060
|
Car9
|
carbonic anhydrase 9 |
| chr11_+_27364916 | 0.18 |
ENSRNOT00000002151
|
Bach1
|
BTB domain and CNC homolog 1 |
| chr18_+_31094965 | 0.17 |
ENSRNOT00000026526
|
Rell2
|
RELT-like 2 |
| chr4_+_97657671 | 0.17 |
ENSRNOT00000075400
|
Gng12
|
G protein subunit gamma 12 |
| chr3_-_2444281 | 0.17 |
ENSRNOT00000013863
|
Tubb4b
|
tubulin, beta 4B class IVb |
| chrM_+_7758 | 0.17 |
ENSRNOT00000046201
|
Mt-atp8
|
mitochondrially encoded ATP synthase 8 |
| chr1_-_170318935 | 0.17 |
ENSRNOT00000024119
|
Prkcdbp
|
protein kinase C, delta binding protein |
| chr10_+_95706685 | 0.17 |
ENSRNOT00000004283
|
Psmd12
|
proteasome 26S subunit, non-ATPase 12 |
| chr2_-_196207560 | 0.16 |
ENSRNOT00000028589
|
Psmd4
|
proteasome 26S subunit, non-ATPase 4 |
| chr5_-_16706909 | 0.16 |
ENSRNOT00000011314
|
Rps20
|
ribosomal protein S20 |
| chr19_+_55381565 | 0.16 |
ENSRNOT00000018923
|
Cdt1
|
chromatin licensing and DNA replication factor 1 |
| chr4_+_66276835 | 0.16 |
ENSRNOT00000007544
|
Fmc1
|
formation of mitochondrial complex V assembly factor 1 |
| chr2_-_26135340 | 0.15 |
ENSRNOT00000074626
|
F2r
|
coagulation factor II (thrombin) receptor |
| chr3_+_2648885 | 0.15 |
ENSRNOT00000020339
|
Abca2
|
ATP binding cassette subfamily A member 2 |
| chr9_+_15166118 | 0.15 |
ENSRNOT00000020432
|
Mdfi
|
MyoD family inhibitor |
| chr1_-_89068348 | 0.15 |
ENSRNOT00000033621
|
Upk1a
|
uroplakin 1A |
| chr1_-_165680176 | 0.15 |
ENSRNOT00000025245
ENSRNOT00000082697 |
Plekhb1
|
pleckstrin homology domain containing B1 |
| chr3_-_10371240 | 0.15 |
ENSRNOT00000012075
|
Ass1
|
argininosuccinate synthase 1 |
| chr7_+_121480723 | 0.15 |
ENSRNOT00000065304
|
Atf4
|
activating transcription factor 4 |
| chrX_-_10413984 | 0.15 |
ENSRNOT00000039551
ENSRNOT00000091448 |
Ddx3x
|
DEAD-box helicase 3, X-linked |
| chr14_+_34354503 | 0.15 |
ENSRNOT00000002941
|
Nmu
|
neuromedin U |
| chr10_-_87232723 | 0.15 |
ENSRNOT00000015150
|
Krt25
|
keratin 25 |
| chr1_-_89560469 | 0.14 |
ENSRNOT00000079091
|
Scn1b
|
sodium voltage-gated channel beta subunit 1 |
| chr7_+_70580198 | 0.14 |
ENSRNOT00000083472
ENSRNOT00000008941 |
Ddit3
|
DNA-damage inducible transcript 3 |
| chr2_+_211337271 | 0.14 |
ENSRNOT00000045155
|
Cox6b1
|
cytochrome c oxidase subunit 6B1 |
| chr2_-_19808937 | 0.13 |
ENSRNOT00000044237
|
Atp6ap1l
|
ATPase H+ transporting accessory protein 1 like |
| chr7_+_125576327 | 0.13 |
ENSRNOT00000016679
|
Prr5
|
proline rich 5 |
| chr2_+_93792601 | 0.13 |
ENSRNOT00000014701
ENSRNOT00000077311 |
Fabp4
|
fatty acid binding protein 4 |
| chr4_-_155401480 | 0.13 |
ENSRNOT00000020735
|
Apobec1
|
apolipoprotein B mRNA editing enzyme catalytic subunit 1 |
| chr19_-_37938857 | 0.13 |
ENSRNOT00000026730
|
Slc12a4
|
solute carrier family 12 member 4 |
| chr4_+_169161585 | 0.13 |
ENSRNOT00000079785
|
Emp1
|
epithelial membrane protein 1 |
| chr1_+_72635267 | 0.13 |
ENSRNOT00000090636
|
Il11
|
interleukin 11 |
| chr7_-_93280009 | 0.13 |
ENSRNOT00000064877
|
Samd12
|
sterile alpha motif domain containing 12 |
| chr8_-_128665988 | 0.13 |
ENSRNOT00000050543
|
Csrnp1
|
cysteine and serine rich nuclear protein 1 |
| chr15_+_45834302 | 0.13 |
ENSRNOT00000051307
|
Serpine3
|
serpin family E member 3 |
| chr1_+_91152635 | 0.13 |
ENSRNOT00000073438
|
LOC100912070
|
dermokine-like |
| chr9_-_82154266 | 0.12 |
ENSRNOT00000024283
|
Cryba2
|
crystallin, beta A2 |
| chr17_-_35037076 | 0.12 |
ENSRNOT00000090946
|
Dusp22
|
dual specificity phosphatase 22 |
| chr4_-_157294047 | 0.12 |
ENSRNOT00000005601
|
Eno2
|
enolase 2 |
| chr20_+_3556975 | 0.12 |
ENSRNOT00000089417
|
Ddr1
|
discoidin domain receptor tyrosine kinase 1 |
| chr10_+_10889488 | 0.12 |
ENSRNOT00000071746
|
Ubald1
|
UBA-like domain containing 1 |
| chr18_-_27206777 | 0.12 |
ENSRNOT00000090569
|
AABR07031691.1
|
|
| chr3_+_22964230 | 0.12 |
ENSRNOT00000041813
|
LOC100362149
|
ribosomal protein S20-like |
| chr5_+_161889342 | 0.12 |
ENSRNOT00000040481
|
LOC100362684
|
ribosomal protein S20-like |
| chr1_+_89215266 | 0.12 |
ENSRNOT00000093612
ENSRNOT00000084799 |
Dmkn
|
dermokine |
| chr10_-_35716260 | 0.11 |
ENSRNOT00000004308
ENSRNOT00000059255 |
Sqstm1
|
sequestosome 1 |
| chr18_-_81539065 | 0.11 |
ENSRNOT00000021400
|
Cndp2
|
CNDP dipeptidase 2 (metallopeptidase M20 family) |
| chr18_-_74688551 | 0.11 |
ENSRNOT00000078016
|
Slc14a2
|
solute carrier family 14 member 2 |
| chr3_+_2642531 | 0.11 |
ENSRNOT00000081798
|
Fut7
|
fucosyltransferase 7 |
| chr4_-_97784842 | 0.11 |
ENSRNOT00000007698
|
Gadd45a
|
growth arrest and DNA-damage-inducible, alpha |
| chr15_-_44860604 | 0.11 |
ENSRNOT00000018637
|
Nefm
|
neurofilament medium |
| chrX_+_28593405 | 0.11 |
ENSRNOT00000071708
|
Tmsb4x
|
thymosin beta 4, X-linked |
| chr1_-_221917901 | 0.11 |
ENSRNOT00000092270
|
Slc22a12
|
solute carrier family 22 member 12 |
| chr17_+_12261102 | 0.11 |
ENSRNOT00000015525
|
Nfil3
|
nuclear factor, interleukin 3 regulated |
| chr7_-_144993652 | 0.11 |
ENSRNOT00000087748
|
Itga5
|
integrin subunit alpha 5 |
| chr11_-_83944763 | 0.11 |
ENSRNOT00000002358
|
Psmd2
|
proteasome 26S subunit, non-ATPase 2 |
| chr9_+_20251521 | 0.11 |
ENSRNOT00000005535
|
LOC100911625
|
gamma-enolase-like |
| chr11_-_84037938 | 0.11 |
ENSRNOT00000002327
|
Abcf3
|
ATP binding cassette subfamily F member 3 |
| chr2_-_149417212 | 0.11 |
ENSRNOT00000018573
|
Gpr87
|
G protein-coupled receptor 87 |
| chr17_+_10537365 | 0.10 |
ENSRNOT00000023651
|
Cltb
|
clathrin, light chain B |
| chr5_+_137357674 | 0.10 |
ENSRNOT00000092813
|
RGD1305347
|
similar to RIKEN cDNA 2610528J11 |
| chr2_-_198339130 | 0.10 |
ENSRNOT00000028767
|
Bola1
|
bolA family member 1 |
| chr3_+_23290819 | 0.10 |
ENSRNOT00000018921
|
Wdr38
|
WD repeat domain 38 |
| chr7_-_11648322 | 0.10 |
ENSRNOT00000026871
|
Gadd45b
|
growth arrest and DNA-damage-inducible, beta |
| chr9_+_82436458 | 0.10 |
ENSRNOT00000026467
ENSRNOT00000081958 |
Dnajb2
|
DnaJ heat shock protein family (Hsp40) member B2 |
| chr1_+_225048149 | 0.10 |
ENSRNOT00000036454
|
Lrrn4cl
|
LRRN4 C-terminal like |
| chr18_+_27923572 | 0.10 |
ENSRNOT00000008041
|
Ctnna1
|
catenin alpha 1 |
| chr5_+_153845094 | 0.09 |
ENSRNOT00000041600
|
Stpg1
|
sperm-tail PG-rich repeat containing 1 |
| chr2_+_46980976 | 0.09 |
ENSRNOT00000083668
ENSRNOT00000082990 |
Mocs2
|
molybdenum cofactor synthesis 2 |
| chr10_+_55712043 | 0.09 |
ENSRNOT00000010141
|
Aloxe3
|
arachidonate lipoxygenase 3 |
| chr2_-_148891900 | 0.09 |
ENSRNOT00000018488
|
Siah2
|
siah E3 ubiquitin protein ligase 2 |
| chr10_+_11338306 | 0.09 |
ENSRNOT00000033695
|
LOC100910875
|
protein FAM100A-like |
| chr2_-_227207584 | 0.09 |
ENSRNOT00000065361
ENSRNOT00000080215 |
Myoz2
|
myozenin 2 |
| chr3_-_91370167 | 0.09 |
ENSRNOT00000006291
|
Prr5l
|
proline rich 5 like |
| chr9_-_37231291 | 0.09 |
ENSRNOT00000016237
|
Ptp4a1
|
protein tyrosine phosphatase type IVA, member 1 |
| chr4_+_176994129 | 0.09 |
ENSRNOT00000018734
|
Cmas
|
cytidine monophosphate N-acetylneuraminic acid synthetase |
| chr1_-_198232344 | 0.09 |
ENSRNOT00000080988
|
Aldoa
|
aldolase, fructose-bisphosphate A |
| chr5_-_151824633 | 0.09 |
ENSRNOT00000043959
|
Sfn
|
stratifin |
| chr8_+_69971778 | 0.09 |
ENSRNOT00000058007
ENSRNOT00000037941 ENSRNOT00000050649 |
Megf11
|
multiple EGF-like-domains 11 |
| chr1_+_78671121 | 0.09 |
ENSRNOT00000021310
|
Ap2s1
|
adaptor-related protein complex 2, sigma 1 subunit |
| chr10_-_45923829 | 0.08 |
ENSRNOT00000035285
|
Olr1462
|
olfactory receptor 1462 |
| chr13_+_105408179 | 0.08 |
ENSRNOT00000003378
|
Ddx3y
|
DEAD (Asp-Glu-Ala-Asp) box helicase 3, Y-linked |
| chr14_+_89314176 | 0.08 |
ENSRNOT00000006765
|
Upp1
|
uridine phosphorylase 1 |
| chr1_+_212359177 | 0.08 |
ENSRNOT00000046314
|
Zfp511
|
zinc finger protein 511 |
| chr14_-_7054548 | 0.08 |
ENSRNOT00000002973
|
Nudt9
|
nudix hydrolase 9 |
| chr1_+_125124743 | 0.08 |
ENSRNOT00000021530
|
Mtmr10
|
myotubularin related protein 10 |
| chr10_-_29367865 | 0.08 |
ENSRNOT00000005304
|
Ttc1
|
tetratricopeptide repeat domain 1 |
| chr2_-_197860699 | 0.08 |
ENSRNOT00000028740
|
Ecm1
|
extracellular matrix protein 1 |
| chr1_-_225128740 | 0.08 |
ENSRNOT00000026897
|
Rom1
|
retinal outer segment membrane protein 1 |
| chr12_+_16988136 | 0.08 |
ENSRNOT00000078925
ENSRNOT00000036744 |
Micall2
|
MICAL-like 2 |
| chr10_+_49299125 | 0.08 |
ENSRNOT00000074853
|
Tvp23b
|
trans-golgi network vesicle protein 23 homolog B |
| chr13_-_99287887 | 0.08 |
ENSRNOT00000004780
|
Ephx1
|
epoxide hydrolase 1 |
| chr9_+_17817721 | 0.08 |
ENSRNOT00000086986
ENSRNOT00000026920 |
Hsp90ab1
|
heat shock protein 90 alpha family class B member 1 |
| chr13_+_92264231 | 0.08 |
ENSRNOT00000066509
ENSRNOT00000004716 |
Spta1
|
spectrin, alpha, erythrocytic 1 |
| chr1_-_266086299 | 0.08 |
ENSRNOT00000026609
|
Cuedc2
|
CUE domain containing 2 |
| chr11_+_73015387 | 0.08 |
ENSRNOT00000090978
|
Gp5
|
glycoprotein V (platelet) |
| chr8_+_65733400 | 0.08 |
ENSRNOT00000089126
|
Uaca
|
uveal autoantigen with coiled-coil domains and ankyrin repeats |
| chr8_-_13513337 | 0.07 |
ENSRNOT00000071532
|
LOC108348070
|
lysine-specific demethylase 4D |
| chr11_+_68198709 | 0.07 |
ENSRNOT00000003048
|
Dirc2
|
disrupted in renal carcinoma 2 |
| chr5_+_168019058 | 0.07 |
ENSRNOT00000077202
|
Tnfrsf9
|
TNF receptor superfamily member 9 |
| chr12_+_7865938 | 0.07 |
ENSRNOT00000061229
|
Ubl3
|
ubiquitin-like 3 |
| chr2_+_198040536 | 0.07 |
ENSRNOT00000028744
|
Anp32e
|
acidic nuclear phosphoprotein 32 family member E |
| chr11_+_82415995 | 0.07 |
ENSRNOT00000051772
|
AABR07034641.1
|
|
| chr13_-_100928811 | 0.07 |
ENSRNOT00000045326
|
Capn2
|
calpain 2 |
| chr13_+_88557860 | 0.07 |
ENSRNOT00000058547
|
Sh2d1b2
|
SH2 domain containing 1B2 |
| chr11_+_46179940 | 0.07 |
ENSRNOT00000088152
|
Tfg
|
Trk-fused gene |
| chr19_+_52077109 | 0.07 |
ENSRNOT00000020225
|
Osgin1
|
oxidative stress induced growth inhibitor 1 |
| chr11_+_30363280 | 0.07 |
ENSRNOT00000002885
|
Sod1
|
superoxide dismutase 1, soluble |
| chr11_+_87374690 | 0.07 |
ENSRNOT00000086117
|
Aifm3
|
apoptosis inducing factor, mitochondria associated 3 |
| chr10_-_89958805 | 0.07 |
ENSRNOT00000084266
|
Mpp3
|
membrane palmitoylated protein 3 |
| chr19_-_39646693 | 0.07 |
ENSRNOT00000019104
|
Psmd7
|
proteasome 26S subunit, non-ATPase 7 |
| chr4_-_147742114 | 0.07 |
ENSRNOT00000041464
|
Efcab12
|
EF-hand calcium binding domain 12 |
| chr3_-_143192408 | 0.07 |
ENSRNOT00000006885
|
LOC689081
|
similar to cystatin E2 |
| chr12_-_22006533 | 0.07 |
ENSRNOT00000059593
|
LOC100362783
|
Uncharacterized protein C7orf61 homolog |
| chr8_+_5522739 | 0.07 |
ENSRNOT00000011507
|
Mmp13
|
matrix metallopeptidase 13 |
| chr1_-_206394346 | 0.07 |
ENSRNOT00000038122
|
LOC100302465
|
hypothetical LOC100302465 |
| chrX_+_43625169 | 0.07 |
ENSRNOT00000086311
|
Sat1
|
spermidine/spermine N1-acetyl transferase 1 |
| chr13_+_83681322 | 0.07 |
ENSRNOT00000004206
|
Mpc2
|
mitochondrial pyruvate carrier 2 |
| chr12_-_21746236 | 0.07 |
ENSRNOT00000001869
|
LOC102550456
|
TSC22 domain family protein 4-like |
| chr9_-_19613360 | 0.07 |
ENSRNOT00000029593
|
Rcan2
|
regulator of calcineurin 2 |
| chr1_-_219294147 | 0.07 |
ENSRNOT00000024601
|
Gstp1
|
glutathione S-transferase pi 1 |
| chr4_+_78618737 | 0.07 |
ENSRNOT00000043790
|
Doxl2
|
diamine oxidase-like protein 2 |
| chr20_+_295250 | 0.06 |
ENSRNOT00000000955
|
Clic2
|
chloride intracellular channel 2 |
| chr20_-_1099336 | 0.06 |
ENSRNOT00000071766
|
LOC100362856
|
olfactory receptor-like protein-like |
| chr2_+_9526209 | 0.06 |
ENSRNOT00000021818
|
Lysmd3
|
LysM domain containing 3 |
| chr5_+_153835821 | 0.06 |
ENSRNOT00000077610
|
Stpg1
|
sperm-tail PG-rich repeat containing 1 |
| chr6_+_72461977 | 0.06 |
ENSRNOT00000007887
ENSRNOT00000090716 |
Ap4s1
|
adaptor-related protein complex 4, sigma 1 subunit |
| chr8_-_85840818 | 0.06 |
ENSRNOT00000013608
|
Eef1a1
|
eukaryotic translation elongation factor 1 alpha 1 |
| chr10_-_14703668 | 0.06 |
ENSRNOT00000024767
|
Tpsab1
|
tryptase alpha/beta 1 |
| chr4_+_45000973 | 0.06 |
ENSRNOT00000084736
ENSRNOT00000080880 |
ST7
|
suppression of tumorigenicity 7 |
| chr13_-_90075144 | 0.06 |
ENSRNOT00000088884
|
Slamf7
|
SLAM family member 7 |
| chr4_+_44573264 | 0.06 |
ENSRNOT00000080271
|
Cav2
|
caveolin 2 |
| chr9_+_71398710 | 0.06 |
ENSRNOT00000049411
|
Ccnyl1
|
cyclin Y-like 1 |
| chr1_-_162533893 | 0.06 |
ENSRNOT00000016783
|
Aamdc
|
adipogenesis associated, Mth938 domain containing |
| chr3_-_71893600 | 0.06 |
ENSRNOT00000006787
ENSRNOT00000057685 |
Tfpi
|
tissue factor pathway inhibitor |
| chr10_+_59831241 | 0.05 |
ENSRNOT00000026596
|
Trpv3
|
transient receptor potential cation channel, subfamily V, member 3 |
| chr1_+_87066289 | 0.05 |
ENSRNOT00000027645
|
Capn12
|
calpain 12 |
| chr5_+_135687538 | 0.05 |
ENSRNOT00000091664
|
AC126292.2
|
|
| chr2_-_45518502 | 0.05 |
ENSRNOT00000014627
|
Hspb3
|
heat shock protein family B (small) member 3 |
| chr17_+_72160735 | 0.05 |
ENSRNOT00000038817
|
Itih2
|
inter-alpha-trypsin inhibitor heavy chain 2 |
| chr11_-_33003021 | 0.05 |
ENSRNOT00000084134
|
Runx1
|
runt-related transcription factor 1 |
| chr10_-_13168217 | 0.05 |
ENSRNOT00000087768
|
Elob
|
elongin B |
| chr15_+_36898195 | 0.05 |
ENSRNOT00000076266
|
Parp4
|
poly (ADP-ribose) polymerase family, member 4 |
| chr2_+_203200427 | 0.05 |
ENSRNOT00000020566
|
Vtcn1
|
V-set domain containing T cell activation inhibitor 1 |
| chr12_+_23463157 | 0.05 |
ENSRNOT00000044841
|
Cux1
|
cut-like homeobox 1 |
| chr11_+_47200029 | 0.05 |
ENSRNOT00000079598
|
Nxpe3
|
neurexophilin and PC-esterase domain family, member 3 |
| chr1_+_72810545 | 0.05 |
ENSRNOT00000092117
|
Ptprh
|
protein tyrosine phosphatase, receptor type, H |
| chr1_+_226258417 | 0.05 |
ENSRNOT00000081839
|
AABR07006258.2
|
|
| chrX_-_71121034 | 0.05 |
ENSRNOT00000076590
|
Snx12
|
sorting nexin 12 |
| chr12_+_30165694 | 0.05 |
ENSRNOT00000001211
|
Asl
|
argininosuccinate lyase |
| chr3_-_82756953 | 0.05 |
ENSRNOT00000044121
|
Accs
|
1-aminocyclopropane-1-carboxylate synthase homolog |
| chr3_-_22951711 | 0.05 |
ENSRNOT00000016876
ENSRNOT00000085981 |
Psmb7
|
proteasome subunit beta 7 |
| chr7_+_13913929 | 0.05 |
ENSRNOT00000009833
|
Ccdc105
|
coiled-coil domain containing 105 |
| chr20_+_21316826 | 0.05 |
ENSRNOT00000000785
|
RGD1306739
|
similar to RIKEN cDNA 1700040L02 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Fosl2_Bach2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:1904178 | sterol regulatory element binding protein import into nucleus(GO:0035105) negative regulation of adipose tissue development(GO:1904178) |
| 0.1 | 0.6 | GO:0010037 | response to carbon dioxide(GO:0010037) |
| 0.1 | 0.4 | GO:2000645 | negative regulation of receptor catabolic process(GO:2000645) |
| 0.1 | 0.4 | GO:0009609 | response to symbiotic bacterium(GO:0009609) |
| 0.1 | 0.5 | GO:0032227 | negative regulation of synaptic transmission, dopaminergic(GO:0032227) |
| 0.1 | 0.4 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.1 | 0.5 | GO:0060623 | pericentric heterochromatin assembly(GO:0031508) regulation of chromosome condensation(GO:0060623) |
| 0.1 | 0.3 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.1 | 0.4 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.1 | 0.2 | GO:0003331 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 0.1 | 0.4 | GO:0046878 | positive regulation of saliva secretion(GO:0046878) positive regulation of glucocorticoid secretion(GO:2000851) |
| 0.1 | 0.1 | GO:1990737 | response to manganese-induced endoplasmic reticulum stress(GO:1990737) |
| 0.1 | 0.2 | GO:1990790 | response to glial cell derived neurotrophic factor(GO:1990790) cellular response to glial cell derived neurotrophic factor(GO:1990792) |
| 0.1 | 0.2 | GO:0000053 | argininosuccinate metabolic process(GO:0000053) |
| 0.1 | 0.2 | GO:0072703 | regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:0060734) response to methyl methanesulfonate(GO:0072702) cellular response to methyl methanesulfonate(GO:0072703) response to environmental enrichment(GO:0090648) positive regulation of peptidyl-serine dephosphorylation(GO:1902310) response to methamphetamine hydrochloride(GO:1904313) |
| 0.1 | 0.3 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 0.1 | 0.2 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.1 | 0.4 | GO:1903659 | regulation of complement-dependent cytotoxicity(GO:1903659) |
| 0.1 | 0.5 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.1 | 0.2 | GO:0071163 | DNA replication preinitiation complex assembly(GO:0071163) regulation of DNA replication origin binding(GO:1902595) |
| 0.1 | 0.2 | GO:0099553 | trans-synaptic signaling by lipid, modulating synaptic transmission(GO:0099552) trans-synaptic signaling by endocannabinoid, modulating synaptic transmission(GO:0099553) |
| 0.1 | 0.2 | GO:0032377 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.1 | 0.3 | GO:0036233 | glycine import(GO:0036233) |
| 0.0 | 0.1 | GO:0033693 | neurofilament bundle assembly(GO:0033693) |
| 0.0 | 0.2 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 0.1 | GO:2000015 | regulation of determination of dorsal identity(GO:2000015) |
| 0.0 | 0.2 | GO:0046133 | pyrimidine ribonucleoside catabolic process(GO:0046133) |
| 0.0 | 0.5 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.0 | 0.2 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.1 | GO:2000468 | regulation of peroxidase activity(GO:2000468) |
| 0.0 | 0.1 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.0 | 0.3 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 0.1 | GO:0021966 | corticospinal neuron axon guidance(GO:0021966) |
| 0.0 | 0.1 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.0 | 0.2 | GO:0061621 | NADH regeneration(GO:0006735) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.0 | 0.1 | GO:2001183 | negative regulation of interleukin-12 secretion(GO:2001183) |
| 0.0 | 0.1 | GO:0051121 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.0 | 0.4 | GO:0097201 | negative regulation of transcription from RNA polymerase II promoter in response to stress(GO:0097201) |
| 0.0 | 0.1 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.0 | 0.1 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 | 0.4 | GO:0035590 | purinergic nucleotide receptor signaling pathway(GO:0035590) |
| 0.0 | 0.1 | GO:0042256 | mature ribosome assembly(GO:0042256) protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.0 | 0.2 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.0 | 0.1 | GO:0075509 | receptor-mediated endocytosis of virus by host cell(GO:0019065) endocytosis involved in viral entry into host cell(GO:0075509) |
| 0.0 | 0.3 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.1 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.0 | 0.1 | GO:2000872 | positive regulation of progesterone secretion(GO:2000872) |
| 0.0 | 0.1 | GO:0090187 | positive regulation of pancreatic juice secretion(GO:0090187) |
| 0.0 | 0.1 | GO:0032919 | spermine acetylation(GO:0032919) |
| 0.0 | 0.1 | GO:0009181 | purine nucleoside diphosphate catabolic process(GO:0009137) purine ribonucleoside diphosphate catabolic process(GO:0009181) |
| 0.0 | 0.1 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.0 | 0.0 | GO:0061152 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.0 | 0.1 | GO:0010482 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.0 | 0.1 | GO:0097332 | response to antipsychotic drug(GO:0097332) |
| 0.0 | 0.1 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 0.0 | 0.0 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.0 | 0.2 | GO:0061418 | regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061418) |
| 0.0 | 0.2 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.1 | GO:0072602 | interleukin-4 secretion(GO:0072602) |
| 0.0 | 0.2 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.0 | 0.2 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 | 0.2 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.0 | 0.0 | GO:1901526 | positive regulation of macromitophagy(GO:1901526) positive regulation of mitophagy in response to mitochondrial depolarization(GO:1904925) |
| 0.0 | 0.1 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.0 | 0.1 | GO:1903336 | negative regulation of vacuolar transport(GO:1903336) |
| 0.0 | 0.1 | GO:0015747 | urate transport(GO:0015747) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.3 | GO:0005638 | lamin filament(GO:0005638) |
| 0.1 | 0.5 | GO:0097413 | Lewy body(GO:0097413) |
| 0.1 | 0.5 | GO:0001740 | Barr body(GO:0001740) |
| 0.0 | 0.1 | GO:0036488 | CHOP-C/EBP complex(GO:0036488) CHOP-ATF3 complex(GO:1990622) |
| 0.0 | 0.2 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.0 | 0.3 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 0.0 | 0.2 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.2 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.0 | 0.2 | GO:0031094 | platelet dense tubular network(GO:0031094) |
| 0.0 | 0.5 | GO:0005838 | proteasome regulatory particle(GO:0005838) |
| 0.0 | 0.1 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.2 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.1 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 0.1 | GO:0043293 | apoptosome(GO:0043293) |
| 0.0 | 0.2 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.2 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.1 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.0 | 0.2 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.1 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.0 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.0 | 0.0 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.1 | 0.3 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.1 | 0.2 | GO:0047025 | 3-oxoacyl-[acyl-carrier-protein] reductase (NADH) activity(GO:0047025) |
| 0.1 | 0.5 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.1 | 0.3 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.1 | 1.5 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.3 | GO:0048019 | receptor antagonist activity(GO:0048019) |
| 0.0 | 0.4 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.2 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.3 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 0.2 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 1.1 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.0 | 0.6 | GO:0015106 | bicarbonate transmembrane transporter activity(GO:0015106) |
| 0.0 | 0.1 | GO:0002134 | UTP binding(GO:0002134) sulfonylurea receptor binding(GO:0017098) pyrimidine ribonucleoside binding(GO:0032551) |
| 0.0 | 0.1 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 0.1 | GO:0086062 | voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 0.0 | 0.3 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.5 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.1 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.0 | 0.1 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.0 | 0.5 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.1 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.0 | 0.5 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.1 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.0 | 0.1 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.0 | 0.1 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.1 | GO:0043262 | adenosine-diphosphatase activity(GO:0043262) ADP-ribose diphosphatase activity(GO:0047631) |
| 0.0 | 0.1 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 0.2 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.0 | 0.1 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.0 | 0.2 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.0 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 0.0 | 0.5 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.3 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.1 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.2 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.1 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.0 | 0.2 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.0 | 0.2 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.1 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.0 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.1 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.0 | 0.1 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.1 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 0.0 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.0 | 0.2 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.3 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.1 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.4 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.7 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 0.3 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 1.3 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.3 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.5 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.0 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.0 | 0.3 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.7 | REACTOME CDT1 ASSOCIATION WITH THE CDC6 ORC ORIGIN COMPLEX | Genes involved in CDT1 association with the CDC6:ORC:origin complex |
| 0.0 | 0.1 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |