Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
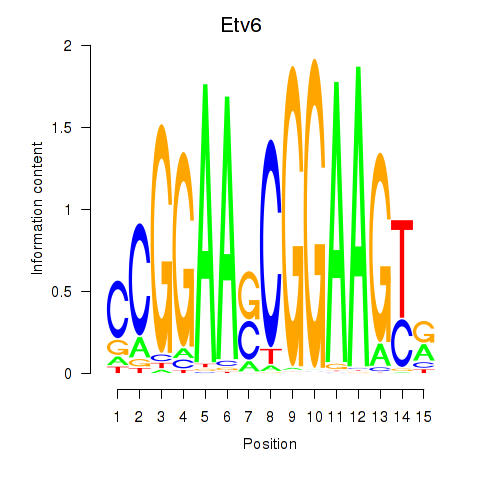
Results for Etv6
Z-value: 0.58
Transcription factors associated with Etv6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Etv6
|
ENSRNOG00000005984 | ets variant 6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Etv6 | rn6_v1_chr4_+_167754525_167754525 | 0.53 | 3.6e-01 | Click! |
Activity profile of Etv6 motif
Sorted Z-values of Etv6 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_+_70522092 | 0.34 |
ENSRNOT00000025873
|
Dpp8
|
dipeptidylpeptidase 8 |
| chr4_-_63039422 | 0.33 |
ENSRNOT00000015808
|
Mtpn
|
myotrophin |
| chr6_-_108796124 | 0.27 |
ENSRNOT00000086545
|
Arel1
|
apoptosis resistant E3 ubiquitin protein ligase 1 |
| chr5_-_137238354 | 0.22 |
ENSRNOT00000039235
|
Szt2
|
seizure threshold 2 homolog (mouse) |
| chr7_-_121783435 | 0.19 |
ENSRNOT00000034912
|
Enthd1
|
ENTH domain containing 1 |
| chr7_+_144647587 | 0.17 |
ENSRNOT00000022398
|
Hoxc4
|
homeo box C4 |
| chr2_-_189096785 | 0.17 |
ENSRNOT00000028200
|
Chrnb2
|
cholinergic receptor nicotinic beta 2 subunit |
| chr8_+_49354115 | 0.17 |
ENSRNOT00000032837
|
Mpzl3
|
myelin protein zero-like 3 |
| chr12_+_46316236 | 0.17 |
ENSRNOT00000001508
|
Prkab1
|
protein kinase AMP-activated non-catalytic subunit beta 1 |
| chr13_+_97838361 | 0.16 |
ENSRNOT00000003641
|
Cnst
|
consortin, connexin sorting protein |
| chr4_-_150471806 | 0.15 |
ENSRNOT00000008741
ENSRNOT00000076557 |
Bms1
|
BMS1 ribosome biogenesis factor |
| chr7_+_130498199 | 0.15 |
ENSRNOT00000092684
ENSRNOT00000092431 |
Shank3
|
SH3 and multiple ankyrin repeat domains 3 |
| chr3_+_8498122 | 0.15 |
ENSRNOT00000020630
|
Gle1
|
GLE1 RNA export mediator |
| chr3_-_12007570 | 0.14 |
ENSRNOT00000060186
|
Lrsam1
|
leucine rich repeat and sterile alpha motif containing 1 |
| chr1_+_29432152 | 0.14 |
ENSRNOT00000019436
ENSRNOT00000083276 |
Trmt11
|
tRNA methyltransferase 11 homolog |
| chr19_-_41433346 | 0.13 |
ENSRNOT00000022952
|
Cmtr2
|
cap methyltransferase 2 |
| chr9_+_81880177 | 0.13 |
ENSRNOT00000022839
|
Stk36
|
serine/threonine kinase 36 |
| chr8_-_47393503 | 0.13 |
ENSRNOT00000059868
|
Arhgef12
|
Rho guanine nucleotide exchange factor 12 |
| chr9_-_65387169 | 0.13 |
ENSRNOT00000051843
|
Orc2
|
origin recognition complex, subunit 2 |
| chr5_-_14588422 | 0.13 |
ENSRNOT00000011312
|
Lypla1
|
lysophospholipase I |
| chr9_-_111220651 | 0.13 |
ENSRNOT00000016028
|
Gin1
|
gypsy retrotransposon integrase 1 |
| chr5_+_173256637 | 0.12 |
ENSRNOT00000025531
|
Ccnl2
|
cyclin L2 |
| chr13_-_53224768 | 0.12 |
ENSRNOT00000091015
|
Camsap2
|
calmodulin regulated spectrin-associated protein family, member 2 |
| chr11_-_27080701 | 0.12 |
ENSRNOT00000002180
|
Ltn1
|
listerin E3 ubiquitin protein ligase 1 |
| chr1_-_173682226 | 0.12 |
ENSRNOT00000020137
|
Ric3
|
RIC3 acetylcholine receptor chaperone |
| chr5_+_173256834 | 0.12 |
ENSRNOT00000089936
|
Ccnl2
|
cyclin L2 |
| chr16_-_81945127 | 0.12 |
ENSRNOT00000023352
|
Mcf2l
|
MCF.2 cell line derived transforming sequence-like |
| chrX_+_128416722 | 0.12 |
ENSRNOT00000009336
ENSRNOT00000085110 |
Xiap
|
X-linked inhibitor of apoptosis |
| chr12_-_2555164 | 0.11 |
ENSRNOT00000084460
ENSRNOT00000061821 |
Map2k7
|
mitogen activated protein kinase kinase 7 |
| chr15_+_24078280 | 0.11 |
ENSRNOT00000015511
ENSRNOT00000063807 |
Mapk1ip1l
|
mitogen-activated protein kinase 1 interacting protein 1-like |
| chr3_-_119405453 | 0.10 |
ENSRNOT00000090355
|
Sppl2a
|
signal peptide peptidase-like 2A |
| chr2_+_165805144 | 0.10 |
ENSRNOT00000073716
|
Rabif
|
RAB interacting factor |
| chr4_+_157523770 | 0.10 |
ENSRNOT00000055985
ENSRNOT00000023240 |
Zfp384
|
zinc finger protein 384 |
| chr3_+_79728796 | 0.10 |
ENSRNOT00000068124
|
Kbtbd4
|
kelch repeat and BTB domain containing 4 |
| chr10_-_37209881 | 0.10 |
ENSRNOT00000090475
|
Sec24a
|
SEC24 homolog A, COPII coat complex component |
| chr3_+_125470551 | 0.10 |
ENSRNOT00000028898
|
Mcm8
|
minichromosome maintenance 8 homologous recombination repair factor |
| chr8_-_21661259 | 0.09 |
ENSRNOT00000068308
|
Fbxl12
|
F-box and leucine-rich repeat protein 12 |
| chr2_-_187786700 | 0.09 |
ENSRNOT00000092257
ENSRNOT00000092612 ENSRNOT00000068360 |
Slc25a44
|
solute carrier family 25, member 44 |
| chr6_+_86713803 | 0.09 |
ENSRNOT00000005861
|
Fam179b
|
family with sequence similarity 179, member B |
| chr19_+_37252843 | 0.09 |
ENSRNOT00000021145
|
E2f4
|
E2F transcription factor 4 |
| chr7_+_74994605 | 0.09 |
ENSRNOT00000013801
|
Spag1
|
sperm associated antigen 1 |
| chr3_-_7422738 | 0.09 |
ENSRNOT00000088339
|
Gtf3c4
|
general transcription factor IIIC subunit 4 |
| chr19_+_45938915 | 0.09 |
ENSRNOT00000065508
|
Mon1b
|
MON1 homolog B, secretory trafficking associated |
| chr1_+_57345577 | 0.09 |
ENSRNOT00000082790
ENSRNOT00000084017 |
Fam120b
|
family with sequence similarity 120B |
| chr1_-_241046249 | 0.09 |
ENSRNOT00000067796
ENSRNOT00000081866 |
Smc5
|
structural maintenance of chromosomes 5 |
| chr10_-_67285617 | 0.09 |
ENSRNOT00000019044
|
Utp6
|
UTP6 small subunit processome component |
| chrX_-_78911601 | 0.09 |
ENSRNOT00000003188
|
RGD1566265
|
similar to RIKEN cDNA 2610002M06 |
| chr1_-_103128743 | 0.08 |
ENSRNOT00000075006
|
Spty2d1
|
SPT2 chromatin protein domain containing 1 |
| chr4_+_157523320 | 0.08 |
ENSRNOT00000023192
|
Zfp384
|
zinc finger protein 384 |
| chr6_-_50786967 | 0.08 |
ENSRNOT00000009566
|
Cbll1
|
Cbl proto-oncogene like 1 |
| chr12_-_37398329 | 0.08 |
ENSRNOT00000083401
|
Tctn2
|
tectonic family member 2 |
| chr7_-_2972521 | 0.08 |
ENSRNOT00000061995
|
Zc3h10
|
zinc finger CCCH type containing 10 |
| chr15_+_34270648 | 0.08 |
ENSRNOT00000026333
|
Rnf31
|
ring finger protein 31 |
| chr15_-_28611946 | 0.08 |
ENSRNOT00000016288
|
Supt16h
|
SPT16 homolog, facilitates chromatin remodeling subunit |
| chr8_-_21968415 | 0.07 |
ENSRNOT00000067325
ENSRNOT00000064932 |
Dnmt1
|
DNA methyltransferase 1 |
| chr3_-_119405300 | 0.07 |
ENSRNOT00000015568
|
Sppl2a
|
signal peptide peptidase-like 2A |
| chr10_+_93354003 | 0.07 |
ENSRNOT00000008140
|
Mettl2b
|
methyltransferase like 2B |
| chr3_-_94419048 | 0.07 |
ENSRNOT00000015775
|
Hipk3
|
homeodomain interacting protein kinase 3 |
| chr17_-_18421861 | 0.07 |
ENSRNOT00000060500
ENSRNOT00000036560 |
Fam8a1
|
family with sequence similarity 8, member A1 |
| chr6_-_86713370 | 0.07 |
ENSRNOT00000005821
|
Klhl28
|
kelch-like family member 28 |
| chr4_-_59014416 | 0.07 |
ENSRNOT00000049811
|
RGD1561636
|
similar to 60S ribosomal protein L38 |
| chr9_+_52687868 | 0.07 |
ENSRNOT00000083864
|
Wdr75
|
WD repeat domain 75 |
| chr13_+_56513286 | 0.07 |
ENSRNOT00000015596
|
Zbtb41
|
zinc finger and BTB domain containing 41 |
| chr3_+_147713821 | 0.07 |
ENSRNOT00000007558
|
Csnk2a1
|
casein kinase 2 alpha 1 |
| chr13_+_89805962 | 0.07 |
ENSRNOT00000074035
|
Tstd1
|
thiosulfate sulfurtransferase like domain containing 1 |
| chr1_-_80515694 | 0.07 |
ENSRNOT00000075144
|
Clasrp
|
CLK4-associating serine/arginine rich protein |
| chr12_-_37468935 | 0.07 |
ENSRNOT00000001387
|
Ddx55
|
DEAD-box helicase 55 |
| chr20_+_2501252 | 0.06 |
ENSRNOT00000079307
ENSRNOT00000084559 |
Trim39
|
tripartite motif-containing 39 |
| chr2_+_58534476 | 0.06 |
ENSRNOT00000077646
|
Lmbrd2
|
LMBR1 domain containing 2 |
| chr7_-_139734568 | 0.06 |
ENSRNOT00000079377
|
Asb8
|
ankyrin repeat and SOCS box-containing 8 |
| chr7_+_97760134 | 0.06 |
ENSRNOT00000086792
|
Tbc1d31
|
TBC1 domain family, member 31 |
| chr8_+_117246376 | 0.06 |
ENSRNOT00000074493
|
Ccdc71
|
coiled-coil domain containing 71 |
| chr5_+_126226291 | 0.06 |
ENSRNOT00000009582
|
Ttc22
|
tetratricopeptide repeat domain 22 |
| chr3_+_21698032 | 0.06 |
ENSRNOT00000013378
|
Rabgap1
|
RAB GTPase activating protein 1 |
| chr3_+_119640298 | 0.06 |
ENSRNOT00000043895
|
Snrnp200
|
small nuclear ribonucleoprotein U5 subunit 200 |
| chr6_-_64170122 | 0.06 |
ENSRNOT00000093248
ENSRNOT00000005363 |
Dnajb9
|
DnaJ heat shock protein family (Hsp40) member B9 |
| chr1_-_199395363 | 0.06 |
ENSRNOT00000090368
ENSRNOT00000026587 |
Prss36
|
protease, serine, 36 |
| chr5_+_118541979 | 0.06 |
ENSRNOT00000013089
|
Efcab7
|
EF-hand calcium binding domain 7 |
| chr5_+_142797366 | 0.06 |
ENSRNOT00000031928
|
Mtf1
|
metal-regulatory transcription factor 1 |
| chr6_+_26537707 | 0.06 |
ENSRNOT00000050102
|
Zfp513
|
zinc finger protein 513 |
| chr1_+_197839430 | 0.06 |
ENSRNOT00000025043
|
Rabep2
|
rabaptin, RAB GTPase binding effector protein 2 |
| chr6_+_126636491 | 0.06 |
ENSRNOT00000089601
|
Ubr7
|
ubiquitin protein ligase E3 component n-recognin 7 (putative) |
| chr8_+_44047592 | 0.06 |
ENSRNOT00000085084
|
Zfp202
|
zinc finger protein 202 |
| chr1_-_218532118 | 0.06 |
ENSRNOT00000086385
ENSRNOT00000018487 |
Ighmbp2
|
immunoglobulin mu binding protein 2 |
| chr15_-_52210746 | 0.06 |
ENSRNOT00000046054
|
Bmp1
|
bone morphogenetic protein 1 |
| chr5_-_134348995 | 0.06 |
ENSRNOT00000015961
|
Faah
|
fatty acid amide hydrolase |
| chr19_+_30668602 | 0.06 |
ENSRNOT00000035084
|
Usp38
|
ubiquitin specific peptidase 38 |
| chr9_+_114022137 | 0.06 |
ENSRNOT00000007657
|
Map3k7
|
mitogen activated protein kinase kinase kinase 7 |
| chr5_+_172488708 | 0.05 |
ENSRNOT00000019930
|
Morn1
|
MORN repeat containing 1 |
| chr12_+_16899025 | 0.05 |
ENSRNOT00000001716
|
Psmg3
|
proteasome assembly chaperone 3 |
| chr10_+_109630099 | 0.05 |
ENSRNOT00000072099
|
Ccdc137
|
coiled-coil domain containing 137 |
| chr14_-_104375649 | 0.05 |
ENSRNOT00000006607
|
Actr2
|
ARP2 actin related protein 2 homolog |
| chr11_+_71151132 | 0.05 |
ENSRNOT00000082594
ENSRNOT00000082435 |
Rubcn
|
RUN and cysteine rich domain containing beclin 1 interacting protein |
| chr6_+_86713604 | 0.05 |
ENSRNOT00000059271
|
Fam179b
|
family with sequence similarity 179, member B |
| chr8_-_62542622 | 0.05 |
ENSRNOT00000080071
|
Clk3
|
CDC-like kinase 3 |
| chr8_-_89130991 | 0.05 |
ENSRNOT00000017411
|
Htr1b
|
5-hydroxytryptamine receptor 1B |
| chr2_-_252382955 | 0.05 |
ENSRNOT00000021705
|
Rpf1
|
ribosome production factor 1 homolog |
| chr9_-_23352668 | 0.05 |
ENSRNOT00000075279
|
Mut
|
methylmalonyl CoA mutase |
| chr6_+_126636662 | 0.05 |
ENSRNOT00000010739
|
Ubr7
|
ubiquitin protein ligase E3 component n-recognin 7 (putative) |
| chr2_-_58534211 | 0.05 |
ENSRNOT00000089178
|
Skp2
|
S-phase kinase associated protein 2 |
| chr20_+_27832932 | 0.05 |
ENSRNOT00000001092
|
Gcc2
|
GRIP and coiled-coil domain containing 2 |
| chr3_-_140141679 | 0.05 |
ENSRNOT00000014632
|
Crnkl1
|
crooked neck pre-mRNA splicing factor 1 |
| chr5_+_142332607 | 0.04 |
ENSRNOT00000072841
|
LOC100909856
|
metal regulatory transcription factor 1-like |
| chr11_+_1896209 | 0.04 |
ENSRNOT00000000909
|
Cggbp1
|
CGG triplet repeat binding protein 1 |
| chr9_+_111220858 | 0.04 |
ENSRNOT00000076669
|
Ppip5k2
|
diphosphoinositol pentakisphosphate kinase 2 |
| chr1_+_198911911 | 0.04 |
ENSRNOT00000055002
|
Prr14
|
proline rich 14 |
| chrX_+_27015884 | 0.04 |
ENSRNOT00000065814
|
Msl3
|
male-specific lethal 3 homolog (Drosophila) |
| chr1_+_220746387 | 0.04 |
ENSRNOT00000027753
|
Eif1ad
|
eukaryotic translation initiation factor 1A domain containing |
| chr3_-_21697945 | 0.04 |
ENSRNOT00000012433
|
Zbtb26
|
zinc finger and BTB domain containing 26 |
| chr1_-_222118459 | 0.04 |
ENSRNOT00000067217
|
Ccdc88b
|
coiled-coil domain containing 88B |
| chr5_-_62001196 | 0.04 |
ENSRNOT00000012602
|
Trmo
|
tRNA methyltransferase O |
| chr5_+_47186558 | 0.04 |
ENSRNOT00000007654
|
LOC100910771
|
mitogen-activated protein kinase kinase kinase 7-like |
| chr1_+_114453275 | 0.04 |
ENSRNOT00000019245
|
Herc2
|
HECT and RLD domain containing E3 ubiquitin protein ligase 2 |
| chr3_+_32947378 | 0.04 |
ENSRNOT00000007404
|
Acvr2a
|
activin A receptor type 2A |
| chr3_+_173955686 | 0.04 |
ENSRNOT00000088447
|
Fam217b
|
family with sequence similarity 217, member B |
| chr5_+_143063099 | 0.04 |
ENSRNOT00000037510
|
Snip1
|
Smad nuclear interacting protein 1 |
| chr8_+_68526093 | 0.04 |
ENSRNOT00000011385
|
Aagab
|
alpha- and gamma-adaptin binding protein |
| chr16_-_21362955 | 0.04 |
ENSRNOT00000039607
|
Gmip
|
Gem-interacting protein |
| chrX_-_105355716 | 0.04 |
ENSRNOT00000015043
|
Timm8a1
|
translocase of inner mitochondrial membrane 8 homolog A1 (yeast) |
| chr13_-_101597570 | 0.04 |
ENSRNOT00000004834
|
Disp1
|
dispatched RND transporter family member 1 |
| chr3_-_125470413 | 0.04 |
ENSRNOT00000028893
|
Trmt6
|
tRNA methyltransferase 6 |
| chr4_-_7113919 | 0.04 |
ENSRNOT00000014082
|
Chpf2
|
chondroitin polymerizing factor 2 |
| chr1_+_197999037 | 0.04 |
ENSRNOT00000091065
|
Apobr
|
apolipoprotein B receptor |
| chr3_-_83306781 | 0.04 |
ENSRNOT00000014088
|
Ttc17
|
tetratricopeptide repeat domain 17 |
| chr20_+_22728208 | 0.04 |
ENSRNOT00000000794
|
LOC100363472
|
nuclear receptor-binding factor 2-like |
| chr10_-_70516421 | 0.04 |
ENSRNOT00000013233
|
Pex12
|
peroxisomal biogenesis factor 12 |
| chr1_+_264309214 | 0.04 |
ENSRNOT00000019070
|
Hif1an
|
hypoxia-inducible factor 1, alpha subunit inhibitor |
| chr13_+_51317460 | 0.03 |
ENSRNOT00000005845
|
RGD1563962
|
similar to Mss4 protein |
| chr2_-_183582553 | 0.03 |
ENSRNOT00000014236
|
Arfip1
|
ADP-ribosylation factor interacting protein 1 |
| chr4_-_87111025 | 0.03 |
ENSRNOT00000018442
|
Kbtbd2
|
kelch repeat and BTB domain containing 2 |
| chr1_+_222250995 | 0.03 |
ENSRNOT00000031458
|
Trpt1
|
tRNA phosphotransferase 1 |
| chr4_+_114776797 | 0.03 |
ENSRNOT00000089635
|
LOC103692167
|
polycomb group RING finger protein 1 |
| chr10_-_94445503 | 0.03 |
ENSRNOT00000013295
|
Ftsj3
|
FtsJ homolog 3 |
| chr20_+_5113339 | 0.03 |
ENSRNOT00000001124
|
Gpank1
|
G patch domain and ankyrin repeats 1 |
| chr1_+_114453054 | 0.03 |
ENSRNOT00000090796
|
Herc2
|
HECT and RLD domain containing E3 ubiquitin protein ligase 2 |
| chr16_+_7082450 | 0.03 |
ENSRNOT00000013291
|
Glt8d1
|
glycosyltransferase 8 domain containing 1 |
| chr2_+_62887267 | 0.03 |
ENSRNOT00000048026
|
Drosha
|
drosha ribonuclease III |
| chr10_+_3321476 | 0.03 |
ENSRNOT00000090166
|
Mpv17l
|
MPV17 mitochondrial inner membrane protein like |
| chr6_-_75996216 | 0.03 |
ENSRNOT00000009180
|
RGD1304624
|
similar to RIKEN cDNA 2700097O09 |
| chr10_+_908806 | 0.03 |
ENSRNOT00000091520
|
Marf1
|
meiosis arrest female 1 |
| chr10_+_28243 | 0.03 |
ENSRNOT00000003193
ENSRNOT00000087156 |
LOC103689943
|
meiosis arrest female protein 1-like |
| chr6_-_111417813 | 0.03 |
ENSRNOT00000016324
ENSRNOT00000085458 |
Sptlc2
|
serine palmitoyltransferase, long chain base subunit 2 |
| chrX_+_115561332 | 0.03 |
ENSRNOT00000076680
ENSRNOT00000075912 ENSRNOT00000007968 |
Alg13
|
ALG13, UDP-N-acetylglucosaminyltransferase subunit |
| chr7_-_141136700 | 0.03 |
ENSRNOT00000091538
|
Bcdin3d
|
BCDIN3 domain containing RNA methyltransferase |
| chr8_-_39830306 | 0.03 |
ENSRNOT00000040901
|
Ccdc15
|
coiled-coil domain containing 15 |
| chr15_-_55424024 | 0.03 |
ENSRNOT00000077733
|
Nudt15
|
nudix hydrolase 15 |
| chr6_+_75996643 | 0.02 |
ENSRNOT00000052076
|
Srp54a
|
signal recognition particle 54A |
| chr5_-_60871705 | 0.02 |
ENSRNOT00000016582
|
Exosc3
|
exosome component 3 |
| chr3_+_11554457 | 0.02 |
ENSRNOT00000073087
|
Fam102a
|
family with sequence similarity 102, member A |
| chr1_-_226255886 | 0.02 |
ENSRNOT00000027842
|
Fen1
|
flap structure-specific endonuclease 1 |
| chr9_+_38544398 | 0.02 |
ENSRNOT00000016828
|
Prim2
|
primase (DNA) subunit 2 |
| chr1_+_197999336 | 0.02 |
ENSRNOT00000023555
|
Apobr
|
apolipoprotein B receptor |
| chr13_-_97838228 | 0.02 |
ENSRNOT00000003618
|
Tfb2m
|
transcription factor B2, mitochondrial |
| chr5_-_118541928 | 0.02 |
ENSRNOT00000012947
|
Itgb3bp
|
integrin subunit beta 3 binding protein |
| chr10_-_84789832 | 0.02 |
ENSRNOT00000071719
|
Cdk5rap3
|
CDK5 regulatory subunit associated protein 3 |
| chr14_+_83226601 | 0.02 |
ENSRNOT00000072368
|
Prr14l
|
proline rich 14-like |
| chr19_+_52032886 | 0.02 |
ENSRNOT00000019923
|
Mlycd
|
malonyl-CoA decarboxylase |
| chr2_-_197971456 | 0.02 |
ENSRNOT00000063866
ENSRNOT00000085971 |
Prpf3
|
pre-mRNA processing factor 3 |
| chr2_+_263864331 | 0.02 |
ENSRNOT00000055259
|
Zranb2
|
zinc finger RANBP2-type containing 2 |
| chr15_-_4057104 | 0.02 |
ENSRNOT00000084350
ENSRNOT00000012270 |
Sec24c
|
SEC24 homolog C, COPII coat complex component |
| chr1_+_201337416 | 0.02 |
ENSRNOT00000067742
|
Btbd16
|
BTB domain containing 16 |
| chr10_-_86355006 | 0.02 |
ENSRNOT00000075076
|
Pgap3
|
post-GPI attachment to proteins 3 |
| chr18_+_70248713 | 0.02 |
ENSRNOT00000032202
|
Mbd1
|
methyl-CpG binding domain protein 1 |
| chrX_-_70461553 | 0.02 |
ENSRNOT00000076110
ENSRNOT00000003872 ENSRNOT00000076812 |
Pdzd11
|
PDZ domain containing 11 |
| chr5_-_57947183 | 0.02 |
ENSRNOT00000060642
|
Fam219a
|
family with sequence similarity 219, member A |
| chr7_+_12256387 | 0.02 |
ENSRNOT00000021939
|
RGD1359127
|
similar to RIKEN cDNA 2310011J03 |
| chr9_-_16581078 | 0.02 |
ENSRNOT00000022582
|
Pex6
|
peroxisomal biogenesis factor 6 |
| chr5_+_137356801 | 0.02 |
ENSRNOT00000027432
|
RGD1305347
|
similar to RIKEN cDNA 2610528J11 |
| chr15_+_51779839 | 0.01 |
ENSRNOT00000024614
ENSRNOT00000077259 |
Bin3
|
bridging integrator 3 |
| chr9_-_10293980 | 0.01 |
ENSRNOT00000072703
|
Dus3l
|
dihydrouridine synthase 3-like |
| chr2_-_62887048 | 0.01 |
ENSRNOT00000039234
|
RGD1306502
|
similar to hypothetical protein FLJ11193 |
| chr10_-_65507581 | 0.01 |
ENSRNOT00000016759
|
Supt6h
|
SPT6 homolog, histone chaperone |
| chr12_+_51937175 | 0.01 |
ENSRNOT00000056780
|
Pus1
|
pseudouridylate synthase 1 |
| chr11_-_72929003 | 0.01 |
ENSRNOT00000071483
|
Ncbp2
|
nuclear cap binding protein subunit 2 |
| chr20_-_5441706 | 0.01 |
ENSRNOT00000000549
|
Vps52
|
VPS52 GARP complex subunit |
| chr3_+_151032952 | 0.01 |
ENSRNOT00000064013
|
Acss2
|
acyl-CoA synthetase short-chain family member 2 |
| chr10_-_109802739 | 0.01 |
ENSRNOT00000054951
|
Sirt7
|
sirtuin 7 |
| chr15_+_28287024 | 0.01 |
ENSRNOT00000064907
|
Mettl17
|
methyltransferase like 17 |
| chr5_-_160070916 | 0.01 |
ENSRNOT00000067517
ENSRNOT00000055791 |
Spen
|
spen family transcriptional repressor |
| chr10_-_3321404 | 0.01 |
ENSRNOT00000093722
ENSRNOT00000003284 |
Pdxdc1
|
pyridoxal-dependent decarboxylase domain containing 1 |
| chr19_+_34041219 | 0.01 |
ENSRNOT00000017226
|
Tmem184c
|
transmembrane protein 184C |
| chr7_-_33584564 | 0.01 |
ENSRNOT00000005463
|
Nedd1
|
neural precursor cell expressed, developmentally down-regulated 1 |
| chr6_+_129609375 | 0.01 |
ENSRNOT00000083732
|
Papola
|
poly (A) polymerase alpha |
| chr10_+_72197977 | 0.01 |
ENSRNOT00000003886
|
Myo19
|
myosin XIX |
| chr8_-_50277797 | 0.01 |
ENSRNOT00000082508
|
Pafah1b2
|
platelet-activating factor acetylhydrolase 1b, catalytic subunit 2 |
| chr13_-_37287458 | 0.01 |
ENSRNOT00000003391
|
Insig2
|
insulin induced gene 2 |
| chr8_+_117280705 | 0.01 |
ENSRNOT00000085038
|
Usp19
|
ubiquitin specific peptidase 19 |
| chr1_+_247084228 | 0.01 |
ENSRNOT00000020475
|
Plpp6
|
phospholipid phosphatase 6 |
| chr10_+_43768708 | 0.01 |
ENSRNOT00000086218
|
Sh3bp5l
|
SH3 binding domain protein 5 like |
| chr19_+_43392085 | 0.01 |
ENSRNOT00000023854
|
Sf3b3
|
splicing factor 3b, subunit 3 |
| chr4_+_113910685 | 0.01 |
ENSRNOT00000011173
|
LOC103692167
|
polycomb group RING finger protein 1 |
| chr1_+_177249226 | 0.01 |
ENSRNOT00000021671
|
Parva
|
parvin, alpha |
| chr9_+_111649631 | 0.01 |
ENSRNOT00000056430
|
Fer
|
FER tyrosine kinase |
| chr8_+_132553218 | 0.01 |
ENSRNOT00000006554
|
Limd1
|
LIM domains containing 1 |
| chr3_+_7422820 | 0.01 |
ENSRNOT00000064323
|
Ddx31
|
DEAD-box helicase 31 |
| chr14_+_78939903 | 0.00 |
ENSRNOT00000078562
ENSRNOT00000088221 |
Man2b2
Mrfap1
|
mannosidase, alpha, class 2B, member 2 Morf4 family associated protein 1 |
| chr9_+_82718709 | 0.00 |
ENSRNOT00000027256
ENSRNOT00000080524 |
Stk11ip
|
serine/threonine kinase 11 interacting protein |
| chr2_-_189400323 | 0.00 |
ENSRNOT00000024364
|
Ubap2l
|
ubiquitin associated protein 2-like |
| chrX_+_15155230 | 0.00 |
ENSRNOT00000073289
ENSRNOT00000051439 |
Was
|
Wiskott-Aldrich syndrome |
| chrX_-_1704033 | 0.00 |
ENSRNOT00000051956
|
Usp11
|
ubiquitin specific peptidase 11 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Etv6
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:1990790 | response to glial cell derived neurotrophic factor(GO:1990790) cellular response to glial cell derived neurotrophic factor(GO:1990792) |
| 0.1 | 0.2 | GO:0060084 | synaptic transmission involved in micturition(GO:0060084) |
| 0.0 | 0.1 | GO:0036451 | cap mRNA methylation(GO:0036451) |
| 0.0 | 0.2 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 0.0 | 0.3 | GO:2000812 | regulation of barbed-end actin filament capping(GO:2000812) |
| 0.0 | 0.1 | GO:0007228 | positive regulation of hh target transcription factor activity(GO:0007228) |
| 0.0 | 0.2 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.0 | 0.1 | GO:0002084 | protein depalmitoylation(GO:0002084) |
| 0.0 | 0.1 | GO:0042998 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) |
| 0.0 | 0.1 | GO:0019086 | late viral transcription(GO:0019086) |
| 0.0 | 0.2 | GO:1904717 | regulation of AMPA glutamate receptor clustering(GO:1904717) |
| 0.0 | 0.1 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.0 | 0.1 | GO:0010847 | regulation of chromatin assembly(GO:0010847) |
| 0.0 | 0.1 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.0 | 0.2 | GO:0035878 | nail development(GO:0035878) |
| 0.0 | 0.2 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 | 0.1 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 0.0 | 0.1 | GO:0090309 | DNA methylation on cytosine within a CG sequence(GO:0010424) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.0 | 0.2 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.0 | 0.1 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.0 | 0.0 | GO:0034499 | late endosome to Golgi transport(GO:0034499) |
| 0.0 | 0.1 | GO:0034184 | positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) |
| 0.0 | 0.1 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.0 | 0.1 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.0 | 0.1 | GO:0071502 | cellular response to temperature stimulus(GO:0071502) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.0 | 0.1 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.0 | 0.2 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 0.1 | GO:0035101 | FACT complex(GO:0035101) |
| 0.0 | 0.1 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.0 | 0.1 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.0 | 0.1 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.1 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.1 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.0 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.1 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.1 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.0 | GO:0031515 | tRNA (m1A) methyltransferase complex(GO:0031515) |
| 0.0 | 0.2 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.0 | GO:1990415 | Pex17p-Pex14p docking complex(GO:1990415) peroxisomal importomer complex(GO:1990429) |
| 0.0 | 0.0 | GO:0072487 | MSL complex(GO:0072487) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.0 | 0.1 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.0 | 0.1 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.0 | 0.2 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.2 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 0.2 | GO:0071253 | connexin binding(GO:0071253) |
| 0.0 | 0.1 | GO:0004040 | amidase activity(GO:0004040) |
| 0.0 | 0.1 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.2 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.0 | 0.2 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 0.1 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.1 | GO:1990955 | G-rich single-stranded DNA binding(GO:1990955) |
| 0.0 | 0.2 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.0 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.0 | 0.1 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.0 | 0.1 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.0 | GO:0000829 | inositol heptakisphosphate kinase activity(GO:0000829) |
| 0.0 | 0.2 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 0.0 | GO:0016426 | tRNA (adenine) methyltransferase activity(GO:0016426) |
| 0.0 | 0.1 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.0 | 0.2 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.2 | REACTOME HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Highly calcium permeable postsynaptic nicotinic acetylcholine receptors |