Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
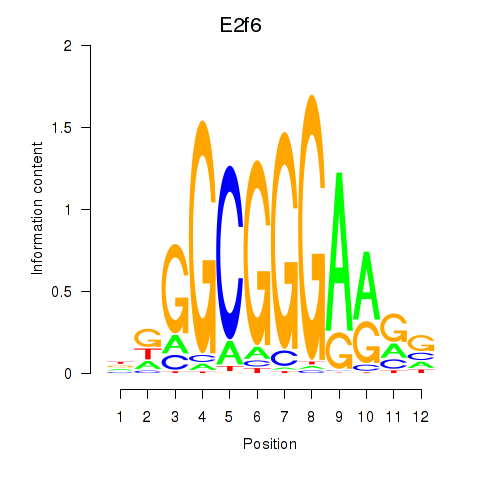
Results for E2f6
Z-value: 0.95
Transcription factors associated with E2f6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
E2f6
|
ENSRNOG00000004449 | E2F transcription factor 6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| E2f6 | rn6_v1_chr6_+_42092467_42092467 | 0.66 | 2.3e-01 | Click! |
Activity profile of E2f6 motif
Sorted Z-values of E2f6 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_109621170 | 0.56 |
ENSRNOT00000093007
|
Elavl2
|
ELAV like RNA binding protein 2 |
| chr14_+_19866408 | 0.50 |
ENSRNOT00000060465
|
Adamts3
|
ADAM metallopeptidase with thrombospondin type 1, motif 3 |
| chr6_+_9483594 | 0.49 |
ENSRNOT00000089272
|
AABR07062799.2
|
|
| chr3_+_110442637 | 0.43 |
ENSRNOT00000010471
|
Pak6
|
p21 (RAC1) activated kinase 6 |
| chr7_-_63407241 | 0.41 |
ENSRNOT00000024679
|
Tbc1d30
|
TBC1 domain family, member 30 |
| chr20_-_5485837 | 0.41 |
ENSRNOT00000092272
ENSRNOT00000000559 ENSRNOT00000092597 |
Daxx
|
death-domain associated protein |
| chr7_+_70364813 | 0.40 |
ENSRNOT00000084012
ENSRNOT00000031230 |
Agap2
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 |
| chr1_-_260992291 | 0.40 |
ENSRNOT00000035415
ENSRNOT00000034758 |
Slit1
|
slit guidance ligand 1 |
| chr1_-_220265772 | 0.39 |
ENSRNOT00000027119
|
Npas4
|
neuronal PAS domain protein 4 |
| chr6_+_119519714 | 0.39 |
ENSRNOT00000004955
|
Flrt2
|
fibronectin leucine rich transmembrane protein 2 |
| chr13_+_70379346 | 0.38 |
ENSRNOT00000038183
|
Nmnat2
|
nicotinamide nucleotide adenylyltransferase 2 |
| chr1_-_165967069 | 0.35 |
ENSRNOT00000089359
|
Arhgef17
|
Rho guanine nucleotide exchange factor (GEF) 17 |
| chr10_+_70689863 | 0.35 |
ENSRNOT00000091122
ENSRNOT00000086987 ENSRNOT00000082030 |
Taf15
|
TATA-box binding protein associated factor 15 |
| chr10_-_85574889 | 0.35 |
ENSRNOT00000072274
|
LOC691153
|
hypothetical protein LOC691153 |
| chr7_-_126465723 | 0.34 |
ENSRNOT00000021196
|
Wnt7b
|
wingless-type MMTV integration site family, member 7B |
| chr15_+_51756978 | 0.33 |
ENSRNOT00000024067
|
Egr3
|
early growth response 3 |
| chr9_+_54212767 | 0.33 |
ENSRNOT00000078073
ENSRNOT00000090026 |
Gls
|
glutaminase |
| chr18_-_55916220 | 0.32 |
ENSRNOT00000025934
|
Synpo
|
synaptopodin |
| chr9_+_10013854 | 0.30 |
ENSRNOT00000077653
ENSRNOT00000072033 |
Khsrp
|
KH-type splicing regulatory protein |
| chr1_-_166911694 | 0.30 |
ENSRNOT00000066915
|
Inppl1
|
inositol polyphosphate phosphatase-like 1 |
| chr7_+_25919867 | 0.29 |
ENSRNOT00000009625
ENSRNOT00000090153 |
Ric8b
|
RIC8 guanine nucleotide exchange factor B |
| chr18_-_57245666 | 0.29 |
ENSRNOT00000080365
|
Ablim3
|
actin binding LIM protein family, member 3 |
| chr10_+_11786121 | 0.28 |
ENSRNOT00000033919
|
Slx4
|
SLX4 structure-specific endonuclease subunit |
| chr12_-_30033357 | 0.28 |
ENSRNOT00000001198
|
Kctd7
|
potassium channel tetramerization domain containing 7 |
| chr16_-_69132584 | 0.28 |
ENSRNOT00000017776
|
Adgra2
|
adhesion G protein-coupled receptor A2 |
| chr5_+_63781801 | 0.28 |
ENSRNOT00000008302
|
Nr4a3
|
nuclear receptor subfamily 4, group A, member 3 |
| chr8_+_114867062 | 0.27 |
ENSRNOT00000074771
|
Wdr82
|
WD repeat domain 82 |
| chr7_+_99954492 | 0.27 |
ENSRNOT00000005885
|
Trib1
|
tribbles pseudokinase 1 |
| chr20_+_5535432 | 0.26 |
ENSRNOT00000040859
|
Syngap1
|
synaptic Ras GTPase activating protein 1 |
| chr3_-_60795951 | 0.26 |
ENSRNOT00000002174
|
Atf2
|
activating transcription factor 2 |
| chr10_+_104582955 | 0.25 |
ENSRNOT00000009733
|
Unk
|
unkempt family zinc finger |
| chr1_+_100297152 | 0.25 |
ENSRNOT00000026100
|
Shank1
|
SH3 and multiple ankyrin repeat domains 1 |
| chr3_-_58181971 | 0.25 |
ENSRNOT00000002076
|
Dlx2
|
distal-less homeobox 2 |
| chr17_+_88215834 | 0.24 |
ENSRNOT00000034098
|
Gpr158
|
G protein-coupled receptor 158 |
| chr10_-_90030423 | 0.23 |
ENSRNOT00000092150
|
Mpp2
|
membrane palmitoylated protein 2 |
| chr12_-_19599374 | 0.23 |
ENSRNOT00000001849
|
Gpc2
|
glypican 2 |
| chr5_+_172364421 | 0.23 |
ENSRNOT00000018769
|
Hes5
|
hes family bHLH transcription factor 5 |
| chr9_+_10471742 | 0.23 |
ENSRNOT00000072276
|
Safb2
|
scaffold attachment factor B2 |
| chr7_+_143707237 | 0.22 |
ENSRNOT00000074212
|
Tns2
|
tensin 2 |
| chr1_+_221792221 | 0.22 |
ENSRNOT00000054828
|
Nrxn2
|
neurexin 2 |
| chr12_+_27155587 | 0.22 |
ENSRNOT00000044800
|
AABR07035916.1
|
|
| chr20_-_3419831 | 0.21 |
ENSRNOT00000046798
|
Mdc1
|
mediator of DNA damage checkpoint 1 |
| chr10_+_61685645 | 0.21 |
ENSRNOT00000003933
|
Mnt
|
MAX network transcriptional repressor |
| chr8_+_78858570 | 0.21 |
ENSRNOT00000089335
|
Zfp280d
|
zinc finger protein 280D |
| chr1_-_164590562 | 0.21 |
ENSRNOT00000024157
|
Tpbgl
|
trophoblast glycoprotein-like |
| chr10_-_57638485 | 0.21 |
ENSRNOT00000009468
|
Dhx33
|
DEAH-box helicase 33 |
| chr7_+_123381077 | 0.21 |
ENSRNOT00000082603
ENSRNOT00000056041 |
Srebf2
|
sterol regulatory element binding transcription factor 2 |
| chr5_-_169658875 | 0.21 |
ENSRNOT00000015840
|
Kcnab2
|
potassium voltage-gated channel subfamily A regulatory beta subunit 2 |
| chr15_+_34167945 | 0.21 |
ENSRNOT00000032252
|
Carmil3
|
capping protein regulator and myosin 1 linker 3 |
| chr4_+_157523110 | 0.21 |
ENSRNOT00000081640
|
Zfp384
|
zinc finger protein 384 |
| chr2_+_226900619 | 0.20 |
ENSRNOT00000019638
|
Pde5a
|
phosphodiesterase 5A |
| chr17_+_6909728 | 0.20 |
ENSRNOT00000061231
|
LOC681410
|
hypothetical protein LOC681410 |
| chr5_+_128923934 | 0.20 |
ENSRNOT00000064145
|
Eps15
|
epidermal growth factor receptor pathway substrate 15 |
| chr10_-_90030639 | 0.20 |
ENSRNOT00000090456
|
Mpp2
|
membrane palmitoylated protein 2 |
| chr16_+_71058022 | 0.20 |
ENSRNOT00000066901
|
Bag4
|
BCL2-associated athanogene 4 |
| chr1_-_222628596 | 0.20 |
ENSRNOT00000083157
ENSRNOT00000034477 |
RGD1308106
|
LOC361719 |
| chr3_+_58164931 | 0.20 |
ENSRNOT00000002078
|
Dlx1
|
distal-less homeobox 1 |
| chr1_-_64147251 | 0.19 |
ENSRNOT00000088502
|
Cnot3
|
CCR4-NOT transcription complex, subunit 3 |
| chrX_+_39711201 | 0.19 |
ENSRNOT00000080512
ENSRNOT00000009802 |
Cnksr2
|
connector enhancer of kinase suppressor of Ras 2 |
| chr1_+_144831523 | 0.19 |
ENSRNOT00000039748
|
Mex3b
|
mex-3 RNA binding family member B |
| chr7_-_138039630 | 0.19 |
ENSRNOT00000008138
|
Slc38a1
|
solute carrier family 38, member 1 |
| chr13_+_110677810 | 0.19 |
ENSRNOT00000006340
|
Slc30a1
|
solute carrier family 30 member 1 |
| chr2_-_211352540 | 0.19 |
ENSRNOT00000023728
|
LOC108349010
|
protein kish-B |
| chr1_+_165382690 | 0.19 |
ENSRNOT00000023802
|
C2cd3
|
C2 calcium-dependent domain containing 3 |
| chr1_+_15412603 | 0.19 |
ENSRNOT00000051496
ENSRNOT00000067070 |
Map3k5
|
mitogen-activated protein kinase kinase kinase 5 |
| chr4_+_160020472 | 0.18 |
ENSRNOT00000078802
|
Parp11
|
poly (ADP-ribose) polymerase family, member 11 |
| chr10_-_85049331 | 0.18 |
ENSRNOT00000012538
|
Tbx21
|
T-box 21 |
| chr8_-_115358046 | 0.18 |
ENSRNOT00000017607
|
Grm2
|
glutamate metabotropic receptor 2 |
| chr17_-_10208360 | 0.18 |
ENSRNOT00000087397
|
Unc5a
|
unc-5 netrin receptor A |
| chr1_+_43475589 | 0.18 |
ENSRNOT00000092034
ENSRNOT00000024682 |
Oprm1
|
opioid receptor, mu 1 |
| chr6_+_99402360 | 0.17 |
ENSRNOT00000078498
|
Zbtb1
|
zinc finger and BTB domain containing 1 |
| chr10_+_62674561 | 0.17 |
ENSRNOT00000019946
ENSRNOT00000056110 |
Ankrd13b
|
ankyrin repeat domain 13B |
| chr3_+_124896618 | 0.17 |
ENSRNOT00000028888
|
Cds2
|
CDP-diacylglycerol synthase 2 |
| chr2_-_207300854 | 0.17 |
ENSRNOT00000018061
|
Mov10
|
Mov10 RISC complex RNA helicase |
| chr3_-_7422738 | 0.17 |
ENSRNOT00000088339
|
Gtf3c4
|
general transcription factor IIIC subunit 4 |
| chr9_+_49479023 | 0.17 |
ENSRNOT00000050922
ENSRNOT00000077111 |
Pou3f3
|
POU class 3 homeobox 3 |
| chr1_+_198932870 | 0.16 |
ENSRNOT00000055003
|
Fbrs
|
fibrosin |
| chr3_-_141411170 | 0.16 |
ENSRNOT00000017364
|
Nkx2-2
|
NK2 homeobox 2 |
| chr10_-_56444847 | 0.16 |
ENSRNOT00000056872
ENSRNOT00000092662 |
Nlgn2
|
neuroligin 2 |
| chr13_+_82231030 | 0.16 |
ENSRNOT00000003551
|
Scyl3
|
SCY1 like pseudokinase 3 |
| chr13_+_97838361 | 0.16 |
ENSRNOT00000003641
|
Cnst
|
consortin, connexin sorting protein |
| chr4_-_183293999 | 0.16 |
ENSRNOT00000079695
|
Ipo8
|
importin 8 |
| chr1_+_167539036 | 0.16 |
ENSRNOT00000093112
|
Rrm1
|
ribonucleotide reductase catalytic subunit M1 |
| chr18_+_51785111 | 0.16 |
ENSRNOT00000019351
|
Lmnb1
|
lamin B1 |
| chr8_+_71514281 | 0.16 |
ENSRNOT00000022256
|
Ns5atp9
|
NS5A (hepatitis C virus) transactivated protein 9 |
| chr7_+_80713321 | 0.16 |
ENSRNOT00000091355
ENSRNOT00000078354 |
Oxr1
|
oxidation resistance 1 |
| chr16_-_61091169 | 0.16 |
ENSRNOT00000016328
|
Dusp4
|
dual specificity phosphatase 4 |
| chr1_-_101046208 | 0.15 |
ENSRNOT00000091711
|
Prr12
|
proline rich 12 |
| chr5_-_166133491 | 0.15 |
ENSRNOT00000087739
ENSRNOT00000089099 |
Kif1b
|
kinesin family member 1B |
| chr12_-_9331195 | 0.15 |
ENSRNOT00000044134
|
Pan3
|
PAN3 poly(A) specific ribonuclease subunit |
| chr6_+_80159364 | 0.15 |
ENSRNOT00000005439
|
Pnn
|
pinin, desmosome associated protein |
| chr1_-_220644636 | 0.15 |
ENSRNOT00000027632
|
Pacs1
|
phosphofurin acidic cluster sorting protein 1 |
| chr7_-_31784192 | 0.15 |
ENSRNOT00000010869
|
Apaf1
|
apoptotic peptidase activating factor 1 |
| chr10_+_83655460 | 0.14 |
ENSRNOT00000008011
|
Gngt2
|
G protein subunit gamma transducin 2 |
| chr11_-_71695000 | 0.14 |
ENSRNOT00000073550
|
Ubxn7
|
UBX domain protein 7 |
| chr18_-_56115593 | 0.14 |
ENSRNOT00000045041
|
Tcof1
|
treacle ribosome biogenesis factor 1 |
| chrX_+_31140960 | 0.14 |
ENSRNOT00000084395
ENSRNOT00000004494 |
Mospd2
|
motile sperm domain containing 2 |
| chr7_-_138039984 | 0.14 |
ENSRNOT00000089806
|
Slc38a1
|
solute carrier family 38, member 1 |
| chr1_+_222746023 | 0.14 |
ENSRNOT00000028787
|
Atl3
|
atlastin GTPase 3 |
| chr8_-_78655856 | 0.14 |
ENSRNOT00000081185
|
Tcf12
|
transcription factor 12 |
| chr7_-_123445613 | 0.14 |
ENSRNOT00000070937
|
Shisa8
|
shisa family member 8 |
| chr20_-_4561062 | 0.14 |
ENSRNOT00000065044
ENSRNOT00000092698 ENSRNOT00000060607 |
Cfb
C2
|
complement factor B complement C2 |
| chr11_-_30428073 | 0.14 |
ENSRNOT00000047741
|
Scaf4
|
SR-related CTD-associated factor 4 |
| chr8_-_123371257 | 0.14 |
ENSRNOT00000017243
|
Stt3b
|
STT3B, catalytic subunit of the oligosaccharyltransferase complex |
| chr14_-_82287108 | 0.13 |
ENSRNOT00000023144
|
Fgfr3
|
fibroblast growth factor receptor 3 |
| chr10_-_13892997 | 0.13 |
ENSRNOT00000004192
|
Traf7
|
TNF receptor associated factor 7 |
| chr18_+_56180089 | 0.13 |
ENSRNOT00000030966
|
Arsi
|
arylsulfatase family, member I |
| chr6_-_100011226 | 0.13 |
ENSRNOT00000083286
ENSRNOT00000041442 |
Max
|
MYC associated factor X |
| chr19_-_37303788 | 0.13 |
ENSRNOT00000084734
|
Fhod1
|
formin homology 2 domain containing 1 |
| chr10_-_82197520 | 0.13 |
ENSRNOT00000092024
|
Cacna1g
|
calcium voltage-gated channel subunit alpha1 G |
| chr20_-_29199224 | 0.13 |
ENSRNOT00000071477
|
Mcu
|
mitochondrial calcium uniporter |
| chr10_-_15228235 | 0.13 |
ENSRNOT00000027121
|
Wdr90
|
WD repeat domain 90 |
| chr15_-_24056073 | 0.13 |
ENSRNOT00000015100
|
Wdhd1
|
WD repeat and HMG-box DNA binding protein 1 |
| chr11_-_70322690 | 0.13 |
ENSRNOT00000002443
|
Heg1
|
heart development protein with EGF-like domains 1 |
| chr4_+_157523320 | 0.13 |
ENSRNOT00000023192
|
Zfp384
|
zinc finger protein 384 |
| chr10_+_90550147 | 0.13 |
ENSRNOT00000032944
|
Fzd2
|
frizzled class receptor 2 |
| chr7_+_73588163 | 0.13 |
ENSRNOT00000015707
|
Kcns2
|
potassium voltage-gated channel, modifier subfamily S, member 2 |
| chr6_+_110624856 | 0.13 |
ENSRNOT00000014017
|
Vash1
|
vasohibin 1 |
| chr5_+_173660921 | 0.13 |
ENSRNOT00000066561
|
Noc2l
|
NOC2-like nucleolar associated transcriptional repressor |
| chr8_-_55408464 | 0.12 |
ENSRNOT00000066893
|
Sik2
|
salt-inducible kinase 2 |
| chr9_+_66568074 | 0.12 |
ENSRNOT00000035238
|
Bmpr2
|
bone morphogenetic protein receptor type 2 |
| chr6_-_122897997 | 0.12 |
ENSRNOT00000057601
|
Eml5
|
echinoderm microtubule associated protein like 5 |
| chr13_-_103264906 | 0.12 |
ENSRNOT00000090786
ENSRNOT00000085211 ENSRNOT00000057395 |
Iars2
|
isoleucyl-tRNA synthetase 2, mitochondrial |
| chrX_+_10964067 | 0.12 |
ENSRNOT00000093181
ENSRNOT00000066480 |
Med14
|
mediator complex subunit 14 |
| chr7_-_63045728 | 0.12 |
ENSRNOT00000039532
|
Lemd3
|
LEM domain containing 3 |
| chr2_+_113984646 | 0.12 |
ENSRNOT00000016799
|
Tnik
|
TRAF2 and NCK interacting kinase |
| chr8_-_62424303 | 0.12 |
ENSRNOT00000091223
|
Csk
|
c-src tyrosine kinase |
| chr18_-_31071371 | 0.12 |
ENSRNOT00000060432
|
Diaph1
|
diaphanous-related formin 1 |
| chr12_-_24537313 | 0.12 |
ENSRNOT00000001975
|
Baz1b
|
bromodomain adjacent to zinc finger domain, 1B |
| chr1_+_225015616 | 0.12 |
ENSRNOT00000026416
|
Hnrnpul2
|
heterogeneous nuclear ribonucleoprotein U-like 2 |
| chr14_+_33580541 | 0.12 |
ENSRNOT00000002905
|
Ppat
|
phosphoribosyl pyrophosphate amidotransferase |
| chr14_+_87312203 | 0.12 |
ENSRNOT00000088032
|
Adcy1
|
adenylate cyclase 1 |
| chr17_-_30714410 | 0.12 |
ENSRNOT00000022902
ENSRNOT00000081993 |
Prpf4b
|
pre-mRNA processing factor 4B |
| chr14_-_82287706 | 0.12 |
ENSRNOT00000080695
|
Fgfr3
|
fibroblast growth factor receptor 3 |
| chr2_+_58462949 | 0.11 |
ENSRNOT00000080618
|
Nadk2
|
NAD kinase 2, mitochondrial |
| chr6_+_111049559 | 0.11 |
ENSRNOT00000015571
|
Tmem63c
|
transmembrane protein 63c |
| chr15_+_34520142 | 0.11 |
ENSRNOT00000074659
|
Nynrin
|
NYN domain and retroviral integrase containing |
| chr12_-_46493203 | 0.11 |
ENSRNOT00000057036
|
Cit
|
citron rho-interacting serine/threonine kinase |
| chr6_-_99783047 | 0.11 |
ENSRNOT00000009028
|
Sptb
|
spectrin, beta, erythrocytic |
| chr11_-_86890390 | 0.11 |
ENSRNOT00000046183
|
Trmt2a
|
tRNA methyltransferase 2 homolog A |
| chrX_-_157331204 | 0.11 |
ENSRNOT00000085201
|
Bgn
|
biglycan |
| chr17_-_10766253 | 0.11 |
ENSRNOT00000000117
|
Cplx2
|
complexin 2 |
| chr5_+_22380334 | 0.11 |
ENSRNOT00000009744
|
Clvs1
|
clavesin 1 |
| chr1_-_219450451 | 0.11 |
ENSRNOT00000025317
|
Rad9a
|
RAD9 checkpoint clamp component A |
| chr8_+_77107536 | 0.11 |
ENSRNOT00000083255
|
Adam10
|
ADAM metallopeptidase domain 10 |
| chr1_+_265829108 | 0.11 |
ENSRNOT00000025791
|
Nolc1
|
nucleolar and coiled-body phosphoprotein 1 |
| chr20_+_3823596 | 0.11 |
ENSRNOT00000087670
|
Rxrb
|
retinoid X receptor beta |
| chr10_+_55927223 | 0.11 |
ENSRNOT00000011523
|
Kcnab3
|
potassium voltage-gated channel subfamily A regulatory beta subunit 3 |
| chr9_+_81783349 | 0.11 |
ENSRNOT00000021548
|
Cnot9
|
CCR4-NOT transcription complex subunit 9 |
| chr1_+_154377447 | 0.11 |
ENSRNOT00000084268
ENSRNOT00000092086 ENSRNOT00000091470 ENSRNOT00000025415 |
Picalm
|
phosphatidylinositol binding clathrin assembly protein |
| chr3_+_148722955 | 0.11 |
ENSRNOT00000013730
|
Pofut1
|
protein O-fucosyltransferase 1 |
| chr1_+_264670841 | 0.11 |
ENSRNOT00000034814
ENSRNOT00000082412 |
Slf2
|
SMC5-SMC6 complex localization factor 2 |
| chr2_-_30340103 | 0.11 |
ENSRNOT00000024023
ENSRNOT00000067722 |
Bdp1
|
B double prime 1, subunit of RNA polymerase III transcription initiation factor IIIB |
| chr16_-_47874647 | 0.10 |
ENSRNOT00000035323
|
Rwdd4
|
RWD domain containing 4 |
| chr5_-_144479306 | 0.10 |
ENSRNOT00000087697
|
Ago1
|
argonaute 1, RISC catalytic component |
| chr4_+_157513414 | 0.10 |
ENSRNOT00000078769
|
Pianp
|
PILR alpha associated neural protein |
| chr5_-_169167831 | 0.10 |
ENSRNOT00000012407
|
Phf13
|
PHD finger protein 13 |
| chr13_-_74077783 | 0.10 |
ENSRNOT00000005677
|
Soat1
|
sterol O-acyltransferase 1 |
| chr7_+_80750725 | 0.10 |
ENSRNOT00000079962
|
Oxr1
|
oxidation resistance 1 |
| chr4_+_133286114 | 0.10 |
ENSRNOT00000084158
|
Ppp4r2
|
protein phosphatase 4, regulatory subunit 2 |
| chr10_-_4910305 | 0.10 |
ENSRNOT00000033122
|
Rmi2
|
RecQ mediated genome instability 2 |
| chr5_+_135351816 | 0.10 |
ENSRNOT00000082602
|
Ipp
|
intracisternal A particle-promoted polypeptide |
| chr2_-_186515135 | 0.10 |
ENSRNOT00000077375
|
Kirrel
|
kin of IRRE like (Drosophila) |
| chr9_+_82674202 | 0.10 |
ENSRNOT00000027208
|
Tmem198
|
transmembrane protein 198 |
| chr1_+_221710670 | 0.10 |
ENSRNOT00000064798
|
Map4k2
|
mitogen activated protein kinase kinase kinase kinase 2 |
| chr8_+_62925357 | 0.10 |
ENSRNOT00000011074
ENSRNOT00000090588 |
Stra6
|
stimulated by retinoic acid 6 |
| chr13_+_44475970 | 0.10 |
ENSRNOT00000024602
ENSRNOT00000091645 |
Ccnt2
|
cyclin T2 |
| chr10_+_67325347 | 0.10 |
ENSRNOT00000079002
ENSRNOT00000085914 |
Suz12
|
suppressor of zeste 12 homolog (Drosophila) |
| chrX_+_62366453 | 0.10 |
ENSRNOT00000089828
|
Arx
|
aristaless related homeobox |
| chr1_-_131454689 | 0.10 |
ENSRNOT00000014152
|
Nr2f2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr13_+_44812567 | 0.10 |
ENSRNOT00000005372
|
R3hdm1
|
R3H domain containing 1 |
| chr10_-_82197848 | 0.10 |
ENSRNOT00000081307
|
Cacna1g
|
calcium voltage-gated channel subunit alpha1 G |
| chr8_-_22821223 | 0.10 |
ENSRNOT00000014135
|
Kank2
|
KN motif and ankyrin repeat domains 2 |
| chr3_+_100770975 | 0.10 |
ENSRNOT00000089233
|
Bdnf
|
brain-derived neurotrophic factor |
| chr7_-_143852119 | 0.10 |
ENSRNOT00000016801
|
Rarg
|
retinoic acid receptor, gamma |
| chr7_+_120125633 | 0.10 |
ENSRNOT00000012480
|
Sh3bp1
|
SH3-domain binding protein 1 |
| chr20_-_46666830 | 0.10 |
ENSRNOT00000000331
|
Cep57l1
|
centrosomal protein 57-like 1 |
| chr7_-_2986935 | 0.09 |
ENSRNOT00000081125
ENSRNOT00000006578 |
Pa2g4
|
proliferation-associated 2G4 |
| chr5_+_169519212 | 0.09 |
ENSRNOT00000024732
|
Chd5
|
chromodomain helicase DNA binding protein 5 |
| chr13_+_110257571 | 0.09 |
ENSRNOT00000005715
|
Ints7
|
integrator complex subunit 7 |
| chr10_-_76039964 | 0.09 |
ENSRNOT00000003164
|
Msi2
|
musashi RNA-binding protein 2 |
| chr15_-_45927804 | 0.09 |
ENSRNOT00000086271
|
Ints6
|
integrator complex subunit 6 |
| chr1_+_199225100 | 0.09 |
ENSRNOT00000088606
|
Setd1a
|
SET domain containing 1A |
| chr2_-_197971456 | 0.09 |
ENSRNOT00000063866
ENSRNOT00000085971 |
Prpf3
|
pre-mRNA processing factor 3 |
| chr2_+_195996521 | 0.09 |
ENSRNOT00000082849
|
Pogz
|
pogo transposable element with ZNF domain |
| chr2_+_198823366 | 0.09 |
ENSRNOT00000083769
ENSRNOT00000028814 |
Pias3
|
protein inhibitor of activated STAT, 3 |
| chr7_+_31784438 | 0.09 |
ENSRNOT00000010914
ENSRNOT00000010929 |
Ikbip
|
IKBKB interacting protein |
| chr19_+_24044103 | 0.09 |
ENSRNOT00000004621
|
Rnf150
|
ring finger protein 150 |
| chr1_-_71135764 | 0.09 |
ENSRNOT00000064949
|
Smim17
|
small integral membrane protein 17 |
| chr11_-_27080701 | 0.09 |
ENSRNOT00000002180
|
Ltn1
|
listerin E3 ubiquitin protein ligase 1 |
| chr1_+_277355619 | 0.09 |
ENSRNOT00000022788
|
Nhlrc2
|
NHL repeat containing 2 |
| chr8_-_47393503 | 0.09 |
ENSRNOT00000059868
|
Arhgef12
|
Rho guanine nucleotide exchange factor 12 |
| chr9_-_66033871 | 0.09 |
ENSRNOT00000035209
|
Als2
|
ALS2, alsin Rho guanine nucleotide exchange factor |
| chr12_-_38691075 | 0.09 |
ENSRNOT00000084192
|
Bcl7a
|
BCL tumor suppressor 7A |
| chr7_+_40217269 | 0.09 |
ENSRNOT00000082090
|
Cep290
|
centrosomal protein 290 |
| chr3_-_177089199 | 0.09 |
ENSRNOT00000020873
|
Zfp512b
|
zinc finger protein 512B |
| chr1_-_266074181 | 0.09 |
ENSRNOT00000026378
|
Psd
|
pleckstrin and Sec7 domain containing |
| chr10_-_14788617 | 0.09 |
ENSRNOT00000043626
|
Cacna1h
|
calcium voltage-gated channel subunit alpha1 H |
| chr6_+_28235695 | 0.09 |
ENSRNOT00000047210
|
Dnmt3a
|
DNA methyltransferase 3 alpha |
| chr9_+_88357556 | 0.09 |
ENSRNOT00000020669
|
Col4a3
|
collagen type IV alpha 3 chain |
Network of associatons between targets according to the STRING database.
First level regulatory network of E2f6
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.1 | 0.4 | GO:0021898 | regulation of transcription from RNA polymerase II promoter involved in forebrain neuron fate commitment(GO:0021882) commitment of multipotent stem cells to neuronal lineage in forebrain(GO:0021898) |
| 0.1 | 0.4 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.1 | 0.4 | GO:0071454 | cellular response to anoxia(GO:0071454) |
| 0.1 | 0.4 | GO:0019355 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.1 | 0.3 | GO:0072054 | renal outer medulla development(GO:0072054) |
| 0.1 | 0.4 | GO:0021834 | chemorepulsion involved in embryonic olfactory bulb interneuron precursor migration(GO:0021834) |
| 0.1 | 0.3 | GO:0090210 | regulation of establishment of blood-brain barrier(GO:0090210) |
| 0.1 | 0.6 | GO:0071386 | cellular response to corticosterone stimulus(GO:0071386) |
| 0.1 | 0.3 | GO:0032915 | positive regulation of transforming growth factor beta2 production(GO:0032915) |
| 0.1 | 0.3 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 0.1 | 0.2 | GO:2000978 | auditory receptor cell fate determination(GO:0042668) negative regulation of pro-B cell differentiation(GO:2000974) negative regulation of forebrain neuron differentiation(GO:2000978) |
| 0.1 | 0.3 | GO:0072429 | meiotic DNA double-strand break processing(GO:0000706) response to intra-S DNA damage checkpoint signaling(GO:0072429) positive regulation of t-circle formation(GO:1904431) |
| 0.1 | 0.3 | GO:1900625 | regulation of mast cell cytokine production(GO:0032763) regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.1 | 0.1 | GO:0003430 | growth plate cartilage chondrocyte growth(GO:0003430) |
| 0.1 | 0.2 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.1 | 0.3 | GO:0045658 | regulation of neutrophil differentiation(GO:0045658) |
| 0.1 | 0.4 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.1 | 0.2 | GO:0032066 | nucleolus to nucleoplasm transport(GO:0032066) |
| 0.1 | 0.2 | GO:0071579 | regulation of zinc ion transport(GO:0071579) |
| 0.1 | 0.3 | GO:0050893 | sensory processing(GO:0050893) |
| 0.1 | 0.2 | GO:0021997 | neural plate axis specification(GO:0021997) |
| 0.1 | 0.2 | GO:0031635 | adenylate cyclase-inhibiting opioid receptor signaling pathway(GO:0031635) |
| 0.1 | 0.2 | GO:0021530 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.1 | 0.3 | GO:0035279 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.1 | 0.2 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.1 | 0.3 | GO:0072697 | protein localization to cell cortex(GO:0072697) |
| 0.0 | 0.3 | GO:0006868 | glutamine transport(GO:0006868) |
| 0.0 | 0.1 | GO:0090271 | positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.0 | 0.1 | GO:0007199 | G-protein coupled receptor signaling pathway coupled to cGMP nucleotide second messenger(GO:0007199) |
| 0.0 | 0.1 | GO:0003249 | cell proliferation involved in heart valve morphogenesis(GO:0003249) regulation of cell proliferation involved in heart valve morphogenesis(GO:0003250) regulation of lung blood pressure(GO:0014916) negative regulation of cell proliferation involved in heart morphogenesis(GO:2000137) |
| 0.0 | 0.2 | GO:0072021 | ascending thin limb development(GO:0072021) metanephric ascending thin limb development(GO:0072218) |
| 0.0 | 0.2 | GO:0055118 | negative regulation of cardiac muscle contraction(GO:0055118) positive regulation of oocyte development(GO:0060282) |
| 0.0 | 0.2 | GO:0042998 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) |
| 0.0 | 0.1 | GO:0019065 | receptor-mediated endocytosis of virus by host cell(GO:0019065) endocytosis involved in viral entry into host cell(GO:0075509) |
| 0.0 | 0.3 | GO:2000628 | regulation of miRNA metabolic process(GO:2000628) |
| 0.0 | 0.2 | GO:0010045 | response to nickel cation(GO:0010045) |
| 0.0 | 0.0 | GO:0072034 | renal vesicle induction(GO:0072034) |
| 0.0 | 0.1 | GO:0051088 | PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 0.0 | 0.1 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.0 | 0.1 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.0 | 0.1 | GO:1990166 | protein localization to site of double-strand break(GO:1990166) |
| 0.0 | 0.2 | GO:1903215 | negative regulation of mRNA modification(GO:0090367) negative regulation of protein targeting to mitochondrion(GO:1903215) |
| 0.0 | 0.0 | GO:0061443 | endocardial cushion cell differentiation(GO:0061443) |
| 0.0 | 0.1 | GO:0046865 | isoprenoid transport(GO:0046864) terpenoid transport(GO:0046865) |
| 0.0 | 0.1 | GO:0019086 | late viral transcription(GO:0019086) |
| 0.0 | 0.1 | GO:0060849 | lymphatic endothelial cell fate commitment(GO:0060838) regulation of transcription involved in lymphatic endothelial cell fate commitment(GO:0060849) |
| 0.0 | 0.1 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.0 | 0.4 | GO:0046643 | regulation of gamma-delta T cell differentiation(GO:0045586) regulation of gamma-delta T cell activation(GO:0046643) |
| 0.0 | 0.1 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 0.0 | 0.4 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.0 | 0.2 | GO:0071864 | positive regulation of cell proliferation in bone marrow(GO:0071864) positive regulation of MAPKKK cascade by fibroblast growth factor receptor signaling pathway(GO:0090080) |
| 0.0 | 0.1 | GO:0006428 | isoleucyl-tRNA aminoacylation(GO:0006428) |
| 0.0 | 0.2 | GO:0019520 | aldonic acid metabolic process(GO:0019520) D-gluconate metabolic process(GO:0019521) |
| 0.0 | 0.2 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.0 | 0.1 | GO:0048210 | Golgi vesicle fusion to target membrane(GO:0048210) |
| 0.0 | 0.1 | GO:1900368 | regulation of RNA interference(GO:1900368) |
| 0.0 | 0.2 | GO:0090116 | C-5 methylation of cytosine(GO:0090116) |
| 0.0 | 0.3 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.1 | GO:0044725 | chromatin reprogramming in the zygote(GO:0044725) |
| 0.0 | 0.3 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.1 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.0 | 0.2 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 | 0.1 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.0 | 0.2 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.1 | GO:0007228 | positive regulation of hh target transcription factor activity(GO:0007228) |
| 0.0 | 0.3 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.0 | 0.1 | GO:0097116 | gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 0.0 | 0.3 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.0 | 0.1 | GO:0018992 | germ-line sex determination(GO:0018992) |
| 0.0 | 0.1 | GO:2000327 | positive regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000327) |
| 0.0 | 0.1 | GO:0070827 | chromatin maintenance(GO:0070827) |
| 0.0 | 0.0 | GO:0032899 | regulation of neurotrophin production(GO:0032899) |
| 0.0 | 0.2 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.0 | 0.1 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.0 | 0.1 | GO:0042984 | amyloid precursor protein biosynthetic process(GO:0042983) regulation of amyloid precursor protein biosynthetic process(GO:0042984) |
| 0.0 | 0.1 | GO:0042997 | negative regulation of low-density lipoprotein particle clearance(GO:0010989) negative regulation of Golgi to plasma membrane protein transport(GO:0042997) |
| 0.0 | 0.1 | GO:0043686 | co-translational protein modification(GO:0043686) |
| 0.0 | 0.1 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
| 0.0 | 0.2 | GO:0032825 | positive regulation of natural killer cell differentiation(GO:0032825) |
| 0.0 | 0.1 | GO:0002125 | maternal aggressive behavior(GO:0002125) |
| 0.0 | 0.1 | GO:0044330 | cell wall macromolecule metabolic process(GO:0044036) canonical Wnt signaling pathway involved in positive regulation of wound healing(GO:0044330) cell wall organization or biogenesis(GO:0071554) metanephric cap development(GO:0072185) metanephric cap morphogenesis(GO:0072186) metanephric cap mesenchymal cell proliferation involved in metanephros development(GO:0090094) response to putrescine(GO:1904585) cellular response to putrescine(GO:1904586) |
| 0.0 | 0.2 | GO:0033601 | positive regulation of mammary gland epithelial cell proliferation(GO:0033601) |
| 0.0 | 0.1 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.1 | GO:0035093 | spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.0 | 0.1 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) |
| 0.0 | 0.1 | GO:1903943 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.0 | 0.1 | GO:1902659 | regulation of glucose mediated signaling pathway(GO:1902659) |
| 0.0 | 0.1 | GO:0070172 | positive regulation of tooth mineralization(GO:0070172) |
| 0.0 | 0.1 | GO:1904721 | negative regulation of mRNA cleavage(GO:0031438) negative regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904721) |
| 0.0 | 0.2 | GO:0048304 | positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 0.0 | 0.1 | GO:0045716 | positive regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045716) |
| 0.0 | 0.1 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.0 | 0.2 | GO:0006655 | phosphatidylglycerol biosynthetic process(GO:0006655) |
| 0.0 | 0.1 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.0 | 0.1 | GO:0014826 | vein smooth muscle contraction(GO:0014826) |
| 0.0 | 0.2 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.0 | 0.1 | GO:1904937 | pancreatic A cell development(GO:0003322) forebrain-midbrain boundary formation(GO:0021905) somatic motor neuron fate commitment(GO:0021917) regulation of transcription from RNA polymerase II promoter involved in somatic motor neuron fate commitment(GO:0021918) sensory neuron migration(GO:1904937) |
| 0.0 | 0.2 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.1 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.1 | GO:0044828 | negative regulation by host of viral genome replication(GO:0044828) |
| 0.0 | 0.1 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.0 | 0.3 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.0 | 0.1 | GO:0060355 | positive regulation of cell adhesion molecule production(GO:0060355) |
| 0.0 | 0.1 | GO:0000019 | regulation of mitotic recombination(GO:0000019) |
| 0.0 | 0.0 | GO:0060082 | eye blink reflex(GO:0060082) |
| 0.0 | 0.1 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 0.1 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.0 | 0.1 | GO:0034696 | response to prostaglandin F(GO:0034696) |
| 0.0 | 0.1 | GO:0015851 | purine nucleobase transport(GO:0006863) nucleobase transport(GO:0015851) |
| 0.0 | 0.1 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 0.0 | 0.0 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.0 | 0.1 | GO:0061193 | taste bud development(GO:0061193) |
| 0.0 | 0.1 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.2 | GO:1900745 | positive regulation of p38MAPK cascade(GO:1900745) |
| 0.0 | 0.0 | GO:0060447 | bud outgrowth involved in lung branching(GO:0060447) |
| 0.0 | 0.0 | GO:0021730 | trigeminal sensory nucleus development(GO:0021730) principal sensory nucleus of trigeminal nerve development(GO:0021740) |
| 0.0 | 0.1 | GO:1902036 | regulation of hematopoietic stem cell differentiation(GO:1902036) |
| 0.0 | 0.1 | GO:0048715 | negative regulation of oligodendrocyte differentiation(GO:0048715) |
| 0.0 | 0.0 | GO:0006116 | NADH oxidation(GO:0006116) |
| 0.0 | 0.0 | GO:1905247 | regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902959) positive regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902961) regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) regulation of aspartic-type peptidase activity(GO:1905245) positive regulation of aspartic-type peptidase activity(GO:1905247) |
| 0.0 | 0.1 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.1 | GO:0051036 | regulation of endosome size(GO:0051036) |
| 0.0 | 0.1 | GO:2000344 | cortisol metabolic process(GO:0034650) cortisol biosynthetic process(GO:0034651) positive regulation of acrosome reaction(GO:2000344) |
| 0.0 | 0.1 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.0 | 0.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.0 | GO:0071298 | cellular response to L-ascorbic acid(GO:0071298) |
| 0.0 | 0.1 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.0 | 0.0 | GO:0032100 | positive regulation of response to food(GO:0032097) positive regulation of appetite(GO:0032100) |
| 0.0 | 0.1 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.0 | 0.1 | GO:0072103 | glomerulus vasculature morphogenesis(GO:0072103) glomerular capillary formation(GO:0072104) |
| 0.0 | 0.0 | GO:2000523 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.0 | 0.0 | GO:1903699 | tarsal gland development(GO:1903699) |
| 0.0 | 0.1 | GO:0035330 | regulation of hippo signaling(GO:0035330) |
| 0.0 | 0.1 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.0 | 0.1 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.0 | 0.1 | GO:0009758 | carbohydrate utilization(GO:0009758) |
| 0.0 | 0.0 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.0 | 0.2 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.0 | 0.1 | GO:0002118 | aggressive behavior(GO:0002118) |
| 0.0 | 0.2 | GO:0045943 | positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.0 | 0.1 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 0.1 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.0 | 0.2 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
| 0.0 | 0.0 | GO:0071529 | cementum mineralization(GO:0071529) |
| 0.0 | 0.1 | GO:0031915 | positive regulation of synaptic plasticity(GO:0031915) |
| 0.0 | 0.1 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0097444 | spine apparatus(GO:0097444) |
| 0.1 | 0.3 | GO:0033557 | Slx1-Slx4 complex(GO:0033557) |
| 0.1 | 0.2 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.1 | 0.3 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.2 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.0 | 0.1 | GO:0030690 | Noc1p-Noc2p complex(GO:0030690) |
| 0.0 | 0.1 | GO:0002945 | cyclin K-CDK12 complex(GO:0002944) cyclin K-CDK13 complex(GO:0002945) |
| 0.0 | 0.5 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.3 | GO:0090576 | RNA polymerase III transcription factor complex(GO:0090576) |
| 0.0 | 0.1 | GO:0043293 | apoptosome(GO:0043293) |
| 0.0 | 0.4 | GO:0001741 | XY body(GO:0001741) |
| 0.0 | 0.1 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.0 | 0.4 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.1 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.1 | GO:0031074 | nucleocytoplasmic shuttling complex(GO:0031074) |
| 0.0 | 0.1 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.0 | 0.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.1 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.2 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.1 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) |
| 0.0 | 0.2 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.0 | 0.2 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 0.3 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.1 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 0.3 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.1 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 0.0 | 0.0 | GO:0000811 | GINS complex(GO:0000811) |
| 0.0 | 0.2 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.1 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.0 | 0.1 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 0.1 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.0 | 0.4 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.2 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.1 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.0 | 0.0 | GO:0005592 | collagen type XI trimer(GO:0005592) |
| 0.0 | 0.1 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.2 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.1 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.0 | 0.3 | GO:0042629 | mast cell granule(GO:0042629) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0000309 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.1 | 0.3 | GO:1902379 | chemoattractant activity involved in axon guidance(GO:1902379) |
| 0.1 | 0.3 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.1 | 0.3 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.1 | 0.3 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.1 | 0.2 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 0.1 | 0.4 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.1 | 0.2 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.1 | 0.2 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.1 | 0.3 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.0 | 0.2 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.0 | 0.1 | GO:0015292 | uniporter activity(GO:0015292) |
| 0.0 | 0.2 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 0.0 | 0.3 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 0.0 | 0.3 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) ankyrin repeat binding(GO:0071532) |
| 0.0 | 0.2 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.2 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.3 | GO:0004445 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) |
| 0.0 | 0.4 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.4 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.1 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.0 | 0.2 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.1 | GO:0004822 | isoleucine-tRNA ligase activity(GO:0004822) |
| 0.0 | 0.4 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.1 | GO:0001156 | transcription factor activity, core RNA polymerase III binding(GO:0000995) TFIIIC-class transcription factor binding(GO:0001156) |
| 0.0 | 0.2 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 0.1 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 0.0 | 0.1 | GO:0051916 | granulocyte colony-stimulating factor binding(GO:0051916) |
| 0.0 | 0.1 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.0 | 0.1 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.1 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.0 | 0.2 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.0 | 0.2 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.0 | 0.2 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 0.1 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.0 | 0.2 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.1 | GO:0051880 | G-quadruplex DNA binding(GO:0051880) |
| 0.0 | 0.1 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 0.0 | 0.1 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.1 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.0 | 0.4 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.1 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.0 | 0.2 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.0 | 0.2 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.1 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.1 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.0 | 0.1 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.3 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.1 | GO:0001640 | adenylate cyclase inhibiting G-protein coupled glutamate receptor activity(GO:0001640) |
| 0.0 | 0.1 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.2 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) |
| 0.0 | 0.3 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.0 | 0.3 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.1 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 0.0 | 0.1 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.2 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.1 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) |
| 0.0 | 0.0 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.0 | 0.1 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.0 | 0.2 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.5 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 0.1 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 0.5 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 0.1 | PID ALK2 PATHWAY | ALK2 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.1 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.2 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.3 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.7 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.0 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.4 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.3 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.0 | 0.3 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.0 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 0.1 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.2 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.3 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 0.1 | REACTOME PRE NOTCH EXPRESSION AND PROCESSING | Genes involved in Pre-NOTCH Expression and Processing |
| 0.0 | 0.1 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.1 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |