Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
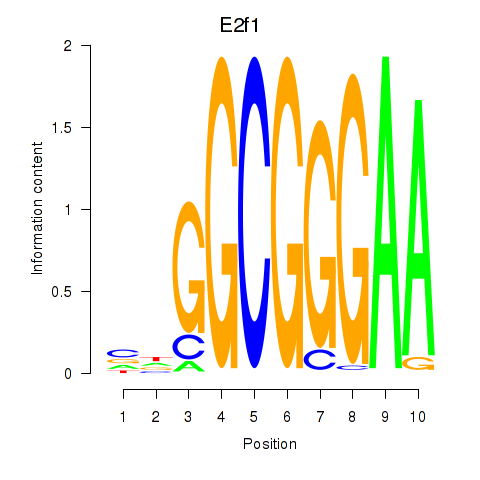
Results for E2f1
Z-value: 1.00
Transcription factors associated with E2f1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
E2f1
|
ENSRNOG00000016708 | E2F transcription factor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| E2f1 | rn6_v1_chr3_-_150062311_150062311 | -0.37 | 5.4e-01 | Click! |
Activity profile of E2f1 motif
Sorted Z-values of E2f1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_33885495 | 1.43 |
ENSRNOT00000086633
|
Sdc1
|
syndecan 1 |
| chr7_-_12221330 | 1.02 |
ENSRNOT00000042204
|
Plk5
|
polo-like kinase 5 |
| chr2_+_155555840 | 0.48 |
ENSRNOT00000080951
|
AABR07010944.1
|
|
| chr1_-_184188911 | 0.47 |
ENSRNOT00000055124
ENSRNOT00000014948 |
Calca
|
calcitonin-related polypeptide alpha |
| chr2_+_27107318 | 0.44 |
ENSRNOT00000019553
|
Arhgef26
|
Rho guanine nucleotide exchange factor 26 |
| chr18_-_63394690 | 0.44 |
ENSRNOT00000090078
ENSRNOT00000029431 |
Cep76
|
centrosomal protein 76 |
| chr11_+_89511191 | 0.43 |
ENSRNOT00000002510
|
Mcm4
|
minichromosome maintenance complex component 4 |
| chr16_+_7303578 | 0.39 |
ENSRNOT00000025656
|
Sema3g
|
semaphorin 3G |
| chr16_-_73827488 | 0.38 |
ENSRNOT00000064070
|
Ank1
|
ankyrin 1 |
| chr10_-_56558487 | 0.38 |
ENSRNOT00000023256
|
Slc2a4
|
solute carrier family 2 member 4 |
| chr10_-_40953651 | 0.34 |
ENSRNOT00000063889
|
Glra1
|
glycine receptor, alpha 1 |
| chr10_+_72909550 | 0.33 |
ENSRNOT00000004540
|
Ppm1d
|
protein phosphatase, Mg2+/Mn2+ dependent, 1D |
| chr10_-_18131562 | 0.33 |
ENSRNOT00000006676
|
Tlx3
|
T-cell leukemia, homeobox 3 |
| chr19_+_14523554 | 0.30 |
ENSRNOT00000084271
ENSRNOT00000064731 |
Mcm5
|
minichromosome maintenance complex component 5 |
| chr6_-_99783047 | 0.27 |
ENSRNOT00000009028
|
Sptb
|
spectrin, beta, erythrocytic |
| chr20_+_4967194 | 0.27 |
ENSRNOT00000070846
|
Lsm2
|
LSM2 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr1_-_31777070 | 0.26 |
ENSRNOT00000020711
|
Slc9a3
|
solute carrier family 9 member A3 |
| chr7_+_2435553 | 0.26 |
ENSRNOT00000065593
|
Prim1
|
primase (DNA) subunit 1 |
| chr4_+_145264462 | 0.26 |
ENSRNOT00000011246
|
Brpf1
|
bromodomain and PHD finger containing, 1 |
| chr10_+_36098051 | 0.25 |
ENSRNOT00000083971
|
Adamts2
|
ADAM metallopeptidase with thrombospondin type 1 motif, 2 |
| chr3_+_37599728 | 0.25 |
ENSRNOT00000090575
|
Rif1
|
replication timing regulatory factor 1 |
| chr10_-_40953467 | 0.25 |
ENSRNOT00000092189
|
Glra1
|
glycine receptor, alpha 1 |
| chr3_-_60795951 | 0.23 |
ENSRNOT00000002174
|
Atf2
|
activating transcription factor 2 |
| chr10_-_39373437 | 0.23 |
ENSRNOT00000058907
|
Slc22a5
|
solute carrier family 22 member 5 |
| chr17_+_88215834 | 0.23 |
ENSRNOT00000034098
|
Gpr158
|
G protein-coupled receptor 158 |
| chr2_+_241029693 | 0.22 |
ENSRNOT00000033413
|
Slc39a8
|
solute carrier family 39 member 8 |
| chr14_+_87312203 | 0.22 |
ENSRNOT00000088032
|
Adcy1
|
adenylate cyclase 1 |
| chr6_+_144291974 | 0.21 |
ENSRNOT00000035255
|
Ncapg2
|
non-SMC condensin II complex, subunit G2 |
| chr7_-_58219790 | 0.21 |
ENSRNOT00000067907
|
Tbc1d15
|
TBC1 domain family, member 15 |
| chr7_-_93826665 | 0.21 |
ENSRNOT00000011344
|
Tnfrsf11b
|
TNF receptor superfamily member 11B |
| chr6_+_104340206 | 0.20 |
ENSRNOT00000058301
|
Plekhd1
|
pleckstrin homology and coiled-coil domain containing D1 |
| chr13_-_25262469 | 0.20 |
ENSRNOT00000019921
|
Rnf152
|
ring finger protein 152 |
| chr6_-_111222858 | 0.20 |
ENSRNOT00000074707
|
Tmed8
|
transmembrane p24 trafficking protein 8 |
| chr10_+_14373679 | 0.20 |
ENSRNOT00000022063
|
Ift140
|
intraflagellar transport 140 |
| chr5_-_109621170 | 0.20 |
ENSRNOT00000093007
|
Elavl2
|
ELAV like RNA binding protein 2 |
| chr5_-_104980641 | 0.19 |
ENSRNOT00000071192
|
Haus6
|
HAUS augmin-like complex, subunit 6 |
| chr12_+_28381982 | 0.19 |
ENSRNOT00000076101
|
Wbscr17
|
Williams-Beuren syndrome chromosome region 17 |
| chr4_+_57019941 | 0.19 |
ENSRNOT00000011356
|
Smo
|
smoothened, frizzled class receptor |
| chr11_-_31847490 | 0.19 |
ENSRNOT00000061304
|
Donson
|
downstream neighbor of SON |
| chr1_+_214454090 | 0.19 |
ENSRNOT00000049448
|
Tspan4
|
tetraspanin 4 |
| chr4_+_153217782 | 0.19 |
ENSRNOT00000015499
|
Cecr2
|
CECR2, histone acetyl-lysine reader |
| chrX_+_158835811 | 0.19 |
ENSRNOT00000071888
ENSRNOT00000080110 |
Ints6l
|
integrator complex subunit 6 like |
| chr5_+_154522119 | 0.18 |
ENSRNOT00000072618
|
E2f2
|
E2F transcription factor 2 |
| chr13_-_45068077 | 0.18 |
ENSRNOT00000004969
|
Mcm6
|
minichromosome maintenance complex component 6 |
| chr10_-_72235888 | 0.18 |
ENSRNOT00000039481
|
Znhit3
|
zinc finger, HIT-type containing 3 |
| chr4_+_57823411 | 0.18 |
ENSRNOT00000030462
ENSRNOT00000088235 |
Ssmem1
|
serine-rich single-pass membrane protein 1 |
| chr10_-_83128297 | 0.18 |
ENSRNOT00000082160
|
Kat7
|
lysine acetyltransferase 7 |
| chr5_+_24410863 | 0.17 |
ENSRNOT00000010591
|
Tp53inp1
|
tumor protein p53 inducible nuclear protein 1 |
| chr20_-_8574082 | 0.17 |
ENSRNOT00000048845
|
Mdga1
|
MAM domain containing glycosylphosphatidylinositol anchor 1 |
| chrX_+_15098904 | 0.17 |
ENSRNOT00000007367
ENSRNOT00000087033 |
Rbm3
|
RNA binding motif (RNP1, RRM) protein 3 |
| chr14_+_33580541 | 0.17 |
ENSRNOT00000002905
|
Ppat
|
phosphoribosyl pyrophosphate amidotransferase |
| chr9_-_9912276 | 0.17 |
ENSRNOT00000071515
|
Tnfsf9
|
tumor necrosis factor superfamily member 9 |
| chr3_-_159802952 | 0.17 |
ENSRNOT00000011610
|
Oser1
|
oxidative stress responsive serine-rich 1 |
| chr16_-_74072541 | 0.17 |
ENSRNOT00000089722
|
AABR07026361.2
|
|
| chr1_+_228337767 | 0.17 |
ENSRNOT00000066247
|
Patl1
|
PAT1 homolog 1, processing body mRNA decay factor |
| chr1_-_281756159 | 0.17 |
ENSRNOT00000013170
|
Prlhr
|
prolactin releasing hormone receptor |
| chr5_-_169658875 | 0.17 |
ENSRNOT00000015840
|
Kcnab2
|
potassium voltage-gated channel subfamily A regulatory beta subunit 2 |
| chr10_-_19652898 | 0.17 |
ENSRNOT00000009648
|
Spdl1
|
spindle apparatus coiled-coil protein 1 |
| chr1_+_145770135 | 0.17 |
ENSRNOT00000015320
|
Stard5
|
StAR-related lipid transfer domain containing 5 |
| chr10_+_27973681 | 0.17 |
ENSRNOT00000004970
|
Gabrb2
|
gamma-aminobutyric acid type A receptor beta 2 subunit |
| chr17_-_1085885 | 0.17 |
ENSRNOT00000026287
|
Ptch1
|
patched 1 |
| chr8_-_132753145 | 0.17 |
ENSRNOT00000007467
ENSRNOT00000008172 |
RGD1566368
|
similar to Solute carrier family 6 (neurotransmitter transporter), member 20 |
| chr17_-_24182425 | 0.17 |
ENSRNOT00000024206
|
Mcur1
|
mitochondrial calcium uniporter regulator 1 |
| chr3_-_5975734 | 0.17 |
ENSRNOT00000081376
|
Vav2
|
vav guanine nucleotide exchange factor 2 |
| chr10_-_82887301 | 0.16 |
ENSRNOT00000035894
|
Itga3
|
integrin subunit alpha 3 |
| chr4_-_57823283 | 0.16 |
ENSRNOT00000032772
ENSRNOT00000091255 |
Tmem209
|
transmembrane protein 209 |
| chr7_+_53630621 | 0.16 |
ENSRNOT00000067011
ENSRNOT00000080598 |
Csrp2
|
cysteine and glycine-rich protein 2 |
| chr18_-_16497886 | 0.16 |
ENSRNOT00000021624
|
Rprd1a
|
regulation of nuclear pre-mRNA domain containing 1A |
| chr16_+_27399467 | 0.16 |
ENSRNOT00000065642
|
Tll1
|
tolloid-like 1 |
| chr7_+_78092037 | 0.16 |
ENSRNOT00000050753
|
Rims2
|
regulating synaptic membrane exocytosis 2 |
| chr4_-_148803988 | 0.16 |
ENSRNOT00000018101
|
Rassf4
|
Ras association domain family member 4 |
| chr10_+_47282208 | 0.15 |
ENSRNOT00000057953
|
Kcnj12
|
potassium voltage-gated channel subfamily J member 12 |
| chr9_-_10757954 | 0.15 |
ENSRNOT00000075265
|
Uhrf1
|
ubiquitin-like with PHD and ring finger domains 1 |
| chr5_-_152762165 | 0.15 |
ENSRNOT00000022921
|
Selenon
|
selenoprotein N |
| chr3_-_7422738 | 0.15 |
ENSRNOT00000088339
|
Gtf3c4
|
general transcription factor IIIC subunit 4 |
| chr10_-_53037816 | 0.15 |
ENSRNOT00000057509
|
Shisa6
|
shisa family member 6 |
| chr5_+_114940053 | 0.15 |
ENSRNOT00000012396
|
Hook1
|
hook microtubule-tethering protein 1 |
| chr1_+_263186235 | 0.15 |
ENSRNOT00000021876
|
Cnnm1
|
cyclin and CBS domain divalent metal cation transport mediator 1 |
| chr7_-_47586137 | 0.15 |
ENSRNOT00000006086
|
Tmtc2
|
transmembrane and tetratricopeptide repeat containing 2 |
| chr11_-_83867203 | 0.15 |
ENSRNOT00000002394
|
Chrd
|
chordin |
| chr1_+_101541266 | 0.15 |
ENSRNOT00000028436
|
Hsd17b14
|
hydroxysteroid (17-beta) dehydrogenase 14 |
| chr1_+_211423022 | 0.15 |
ENSRNOT00000029587
|
Dpysl4
|
dihydropyrimidinase-like 4 |
| chr1_+_221710670 | 0.15 |
ENSRNOT00000064798
|
Map4k2
|
mitogen activated protein kinase kinase kinase kinase 2 |
| chr20_+_12944786 | 0.15 |
ENSRNOT00000048218
|
Pcnt
|
pericentrin |
| chr7_+_125649688 | 0.15 |
ENSRNOT00000017906
|
Phf21b
|
PHD finger protein 21B |
| chrX_-_75566481 | 0.14 |
ENSRNOT00000003714
|
Zdhhc15
|
zinc finger, DHHC-type containing 15 |
| chr10_-_64202380 | 0.14 |
ENSRNOT00000008982
|
Rflnb
|
refilin B |
| chr4_-_68597586 | 0.14 |
ENSRNOT00000015921
|
RGD1563986
|
similar to RIKEN cDNA E330009J07 gene |
| chr5_-_135663284 | 0.14 |
ENSRNOT00000087237
ENSRNOT00000066869 |
Toe1
|
target of EGR1, member 1 (nuclear) |
| chr15_-_24056073 | 0.14 |
ENSRNOT00000015100
|
Wdhd1
|
WD repeat and HMG-box DNA binding protein 1 |
| chr5_-_129352886 | 0.14 |
ENSRNOT00000012088
|
Cdkn2c
|
cyclin-dependent kinase inhibitor 2C |
| chr1_+_241594565 | 0.14 |
ENSRNOT00000020123
|
Apba1
|
amyloid beta precursor protein binding family A member 1 |
| chrX_+_63343724 | 0.14 |
ENSRNOT00000076530
ENSRNOT00000008558 |
Klhl15
|
kelch-like family member 15 |
| chrX_+_104882704 | 0.14 |
ENSRNOT00000079572
ENSRNOT00000074330 ENSRNOT00000082983 |
Cstf2
|
cleavage stimulation factor subunit 2 |
| chr7_-_123088819 | 0.14 |
ENSRNOT00000056077
|
AC096601.1
|
|
| chr1_+_15412603 | 0.14 |
ENSRNOT00000051496
ENSRNOT00000067070 |
Map3k5
|
mitogen-activated protein kinase kinase kinase 5 |
| chr13_+_97838361 | 0.14 |
ENSRNOT00000003641
|
Cnst
|
consortin, connexin sorting protein |
| chr4_-_152380023 | 0.14 |
ENSRNOT00000012397
|
Erc1
|
ELKS/RAB6-interacting/CAST family member 1 |
| chr1_-_182140570 | 0.14 |
ENSRNOT00000060098
|
Ythdc2
|
YTH domain containing 2 |
| chr10_+_104582955 | 0.14 |
ENSRNOT00000009733
|
Unk
|
unkempt family zinc finger |
| chr2_+_58462949 | 0.13 |
ENSRNOT00000080618
|
Nadk2
|
NAD kinase 2, mitochondrial |
| chr1_+_167539036 | 0.13 |
ENSRNOT00000093112
|
Rrm1
|
ribonucleotide reductase catalytic subunit M1 |
| chr1_+_101884019 | 0.13 |
ENSRNOT00000028650
|
Tmem143
|
transmembrane protein 143 |
| chr6_+_99402360 | 0.13 |
ENSRNOT00000078498
|
Zbtb1
|
zinc finger and BTB domain containing 1 |
| chr5_-_133709712 | 0.13 |
ENSRNOT00000010201
|
Foxd2
|
forkhead box D2 |
| chr9_+_10013854 | 0.13 |
ENSRNOT00000077653
ENSRNOT00000072033 |
Khsrp
|
KH-type splicing regulatory protein |
| chr10_-_57638485 | 0.13 |
ENSRNOT00000009468
|
Dhx33
|
DEAH-box helicase 33 |
| chr14_+_82356916 | 0.13 |
ENSRNOT00000040229
|
Slbp
|
stem-loop binding protein |
| chr1_+_198214797 | 0.13 |
ENSRNOT00000068543
|
Tbx6
|
T-box 6 |
| chrX_+_31140960 | 0.13 |
ENSRNOT00000084395
ENSRNOT00000004494 |
Mospd2
|
motile sperm domain containing 2 |
| chr17_-_10640548 | 0.13 |
ENSRNOT00000064274
|
Simc1
|
SUMO-interacting motifs containing 1 |
| chr1_+_85386470 | 0.13 |
ENSRNOT00000093332
ENSRNOT00000044326 |
Plekhg2
|
pleckstrin homology and RhoGEF domain containing G2 |
| chr8_+_114866768 | 0.13 |
ENSRNOT00000076731
|
Wdr82
|
WD repeat domain 82 |
| chr10_+_11206226 | 0.13 |
ENSRNOT00000006979
|
Tfap4
|
transcription factor AP-4 |
| chr12_+_38345456 | 0.12 |
ENSRNOT00000001685
|
Clip1
|
CAP-GLY domain containing linker protein 1 |
| chr4_-_82258765 | 0.12 |
ENSRNOT00000008523
|
Hoxa5
|
homeo box A5 |
| chrX_+_157353730 | 0.12 |
ENSRNOT00000080691
|
Haus7
|
HAUS augmin-like complex, subunit 7 |
| chr14_-_82287706 | 0.12 |
ENSRNOT00000080695
|
Fgfr3
|
fibroblast growth factor receptor 3 |
| chr16_+_20555395 | 0.12 |
ENSRNOT00000026652
|
Gdf15
|
growth differentiation factor 15 |
| chr11_+_87522971 | 0.12 |
ENSRNOT00000043545
|
Smpd4
|
sphingomyelin phosphodiesterase 4 |
| chr10_-_14373334 | 0.12 |
ENSRNOT00000022050
|
Cramp1
|
cramped chromatin regulator homolog 1 |
| chr16_+_48863418 | 0.12 |
ENSRNOT00000029868
|
Primpol
|
primase and DNA directed polymerase |
| chr8_-_48619592 | 0.12 |
ENSRNOT00000012534
|
Abcg4
|
ATP binding cassette subfamily G member 4 |
| chr2_+_127686925 | 0.12 |
ENSRNOT00000086653
ENSRNOT00000016946 |
Plk4
|
polo-like kinase 4 |
| chr3_+_176865156 | 0.12 |
ENSRNOT00000019084
|
Zgpat
|
zinc finger CCCH-type and G-patch domain containing |
| chr3_-_163477715 | 0.12 |
ENSRNOT00000009517
|
Prex1
|
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 1 |
| chr3_+_124896618 | 0.12 |
ENSRNOT00000028888
|
Cds2
|
CDP-diacylglycerol synthase 2 |
| chr6_-_122897997 | 0.12 |
ENSRNOT00000057601
|
Eml5
|
echinoderm microtubule associated protein like 5 |
| chr19_-_19832812 | 0.12 |
ENSRNOT00000084657
|
Papd5
|
poly(A) RNA polymerase D5, non-canonical |
| chr8_-_79119720 | 0.12 |
ENSRNOT00000086562
|
Tex9
|
testis expressed 9 |
| chr5_+_135663371 | 0.12 |
ENSRNOT00000024375
|
Mutyh
|
mutY DNA glycosylase |
| chr12_-_44279002 | 0.12 |
ENSRNOT00000064900
|
Fbxo21
|
F-box protein 21 |
| chr15_+_59216705 | 0.12 |
ENSRNOT00000030097
|
Ccdc122
|
coiled-coil domain containing 122 |
| chr8_-_13909061 | 0.12 |
ENSRNOT00000078372
ENSRNOT00000083011 |
Cep295
|
centrosomal protein 295 |
| chr2_+_184600721 | 0.12 |
ENSRNOT00000075823
|
Gatb
|
glutamyl-tRNA amidotransferase subunit B |
| chr2_-_252691886 | 0.12 |
ENSRNOT00000068739
|
Prkacb
|
protein kinase cAMP-activated catalytic subunit beta |
| chr1_+_201981357 | 0.12 |
ENSRNOT00000027999
|
Acadsb
|
acyl-CoA dehydrogenase, short/branched chain |
| chr19_-_914880 | 0.12 |
ENSRNOT00000017127
|
Cklf
|
chemokine-like factor |
| chr9_-_65387169 | 0.12 |
ENSRNOT00000051843
|
Orc2
|
origin recognition complex, subunit 2 |
| chr14_+_71533063 | 0.12 |
ENSRNOT00000004231
|
Prom1
|
prominin 1 |
| chr6_+_126636491 | 0.12 |
ENSRNOT00000089601
|
Ubr7
|
ubiquitin protein ligase E3 component n-recognin 7 (putative) |
| chr5_-_150609352 | 0.12 |
ENSRNOT00000074374
|
Phactr4
|
phosphatase and actin regulator 4 |
| chr16_+_37177443 | 0.12 |
ENSRNOT00000014083
|
Cep44
|
centrosomal protein 44 |
| chr6_+_99433550 | 0.11 |
ENSRNOT00000079359
ENSRNOT00000008504 |
Hspa2
|
heat shock protein family A member 2 |
| chr3_+_154507035 | 0.11 |
ENSRNOT00000017265
|
Rprd1b
|
regulation of nuclear pre-mRNA domain containing 1B |
| chr7_-_123088279 | 0.11 |
ENSRNOT00000071998
|
Tob2
|
transducer of ERBB2, 2 |
| chr1_+_126749508 | 0.11 |
ENSRNOT00000015845
|
Pcsk6
|
proprotein convertase subtilisin/kexin type 6 |
| chr16_-_71319449 | 0.11 |
ENSRNOT00000029284
|
Fgfr1
|
Fibroblast growth factor receptor 1 |
| chr6_+_86651196 | 0.11 |
ENSRNOT00000034756
|
RGD1307621
|
hypothetical LOC314168 |
| chr20_+_13940877 | 0.11 |
ENSRNOT00000093587
|
Cabin1
|
calcineurin binding protein 1 |
| chr1_-_190370499 | 0.11 |
ENSRNOT00000084389
|
AABR07005618.1
|
|
| chr13_-_48284990 | 0.11 |
ENSRNOT00000086928
|
Srgap2
|
SLIT-ROBO Rho GTPase activating protein 2 |
| chr1_-_188713270 | 0.11 |
ENSRNOT00000082192
ENSRNOT00000065892 |
Gprc5b
|
G protein-coupled receptor, class C, group 5, member B |
| chr7_-_12673659 | 0.11 |
ENSRNOT00000091650
ENSRNOT00000041277 ENSRNOT00000044865 |
Ptbp1
|
polypyrimidine tract binding protein 1 |
| chr7_+_37812831 | 0.11 |
ENSRNOT00000005910
|
Btg1
|
BTG anti-proliferation factor 1 |
| chr3_-_12007570 | 0.11 |
ENSRNOT00000060186
|
Lrsam1
|
leucine rich repeat and sterile alpha motif containing 1 |
| chr5_+_139783951 | 0.11 |
ENSRNOT00000081333
|
Rims3
|
regulating synaptic membrane exocytosis 3 |
| chr5_-_60559533 | 0.11 |
ENSRNOT00000092899
|
Zbtb5
|
zinc finger and BTB domain containing 5 |
| chr1_+_280103695 | 0.11 |
ENSRNOT00000065289
|
Eno4
|
enolase family member 4 |
| chr9_+_43049587 | 0.11 |
ENSRNOT00000021434
ENSRNOT00000079234 |
Cnnm4
|
cyclin and CBS domain divalent metal cation transport mediator 4 |
| chr5_+_47853818 | 0.11 |
ENSRNOT00000009228
ENSRNOT00000079656 |
Casp8ap2
|
caspase 8 associated protein 2 |
| chr8_+_108958046 | 0.11 |
ENSRNOT00000079618
ENSRNOT00000066917 |
Stag1
|
stromal antigen 1 |
| chr1_-_163129641 | 0.11 |
ENSRNOT00000083055
|
Capn5
|
calpain 5 |
| chr11_-_61294022 | 0.11 |
ENSRNOT00000064385
|
Spice1
|
spindle and centriole associated protein 1 |
| chr1_+_173607101 | 0.11 |
ENSRNOT00000074636
|
Tub
|
tubby bipartite transcription factor |
| chr13_-_73704480 | 0.11 |
ENSRNOT00000005296
|
Tor1aip1
|
torsin 1A interacting protein 1 |
| chr5_+_122647281 | 0.11 |
ENSRNOT00000066041
|
Mier1
|
mesoderm induction early response 1, transcriptional regulator |
| chr17_+_36334147 | 0.10 |
ENSRNOT00000050261
|
E2f3
|
E2F transcription factor 3 |
| chrX_-_63291371 | 0.10 |
ENSRNOT00000082421
|
Eif2s3
|
eukaryotic translation initiation factor 2 subunit gamma |
| chr10_-_31041626 | 0.10 |
ENSRNOT00000007267
|
Lsm11
|
LSM11, U7 small nuclear RNA associated |
| chr1_-_219682287 | 0.10 |
ENSRNOT00000026092
|
Rhod
|
ras homolog family member D |
| chr1_+_264670841 | 0.10 |
ENSRNOT00000034814
ENSRNOT00000082412 |
Slf2
|
SMC5-SMC6 complex localization factor 2 |
| chr10_+_17421075 | 0.10 |
ENSRNOT00000047011
|
Stk10
|
serine/threonine kinase 10 |
| chr5_+_79382096 | 0.10 |
ENSRNOT00000085461
|
Tmem268
|
transmembrane protein 268 |
| chr18_-_65768781 | 0.10 |
ENSRNOT00000016189
|
Poli
|
DNA polymerase iota |
| chr4_+_95884743 | 0.10 |
ENSRNOT00000008585
|
Smarcad1
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1` |
| chr2_-_116413873 | 0.10 |
ENSRNOT00000010344
|
Mynn
|
myoneurin |
| chr12_-_30033357 | 0.10 |
ENSRNOT00000001198
|
Kctd7
|
potassium channel tetramerization domain containing 7 |
| chr7_+_140742418 | 0.10 |
ENSRNOT00000089211
|
Prph
|
peripherin |
| chr3_-_3691972 | 0.10 |
ENSRNOT00000061735
|
Qsox2
|
quiescin sulfhydryl oxidase 2 |
| chr10_-_84881190 | 0.10 |
ENSRNOT00000073746
|
Pnpo
|
pyridoxamine 5'-phosphate oxidase |
| chr5_+_152721940 | 0.10 |
ENSRNOT00000039322
|
Aunip
|
aurora kinase A and ninein interacting protein |
| chr11_-_27080701 | 0.10 |
ENSRNOT00000002180
|
Ltn1
|
listerin E3 ubiquitin protein ligase 1 |
| chr7_+_97984862 | 0.10 |
ENSRNOT00000008508
|
Atad2
|
ATPase family, AAA domain containing 2 |
| chr10_-_15228235 | 0.10 |
ENSRNOT00000027121
|
Wdr90
|
WD repeat domain 90 |
| chr7_+_2643288 | 0.10 |
ENSRNOT00000047241
|
Timeless
|
timeless circadian clock |
| chr3_+_170354141 | 0.10 |
ENSRNOT00000005901
|
Fam210b
|
family with sequence similarity 210, member B |
| chr10_-_14324170 | 0.10 |
ENSRNOT00000035513
ENSRNOT00000090587 |
Hn1l
|
hematological and neurological expressed 1-like |
| chr2_-_219741886 | 0.10 |
ENSRNOT00000085122
|
Slc35a3
|
solute carrier family 35 member A3 |
| chr1_-_43884267 | 0.10 |
ENSRNOT00000024418
|
Cnksr3
|
Cnksr family member 3 |
| chr8_-_78655856 | 0.10 |
ENSRNOT00000081185
|
Tcf12
|
transcription factor 12 |
| chr4_-_159482869 | 0.10 |
ENSRNOT00000088333
|
Rad51ap1
|
RAD51 associated protein 1 |
| chr4_-_116278615 | 0.10 |
ENSRNOT00000020505
|
Cyp26b1
|
cytochrome P450, family 26, subfamily b, polypeptide 1 |
| chr5_+_147185474 | 0.10 |
ENSRNOT00000000134
|
Ak2
|
adenylate kinase 2 |
| chr9_-_10757720 | 0.10 |
ENSRNOT00000083848
|
Uhrf1
|
ubiquitin-like with PHD and ring finger domains 1 |
| chr9_+_61823531 | 0.10 |
ENSRNOT00000070844
|
Mars2
|
methionyl-tRNA synthetase 2, mitochondrial |
| chr2_-_124149621 | 0.10 |
ENSRNOT00000023448
ENSRNOT00000084825 ENSRNOT00000023437 |
Nudt6
|
nudix hydrolase 6 |
| chr10_+_64360390 | 0.10 |
ENSRNOT00000010168
|
Fam57a
|
family with sequence similarity 57, member A |
| chr5_-_160619650 | 0.10 |
ENSRNOT00000089981
|
Tmem51
|
transmembrane protein 51 |
| chr3_+_108944141 | 0.10 |
ENSRNOT00000034950
|
Fam98b
|
family with sequence similarity 98, member B |
Network of associatons between targets according to the STRING database.
First level regulatory network of E2f1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.4 | GO:1903553 | positive regulation of extracellular exosome assembly(GO:1903553) |
| 0.2 | 0.5 | GO:0001983 | baroreceptor response to increased systemic arterial blood pressure(GO:0001983) |
| 0.1 | 0.7 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.1 | 0.6 | GO:2000344 | positive regulation of acrosome reaction(GO:2000344) |
| 0.1 | 0.3 | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 0.1 | 0.2 | GO:0032915 | positive regulation of transforming growth factor beta2 production(GO:0032915) |
| 0.1 | 0.3 | GO:0042891 | antibiotic transport(GO:0042891) |
| 0.1 | 0.2 | GO:0036446 | myofibroblast differentiation(GO:0036446) regulation of myofibroblast differentiation(GO:1904760) |
| 0.1 | 0.2 | GO:0042489 | negative regulation of odontogenesis of dentin-containing tooth(GO:0042489) |
| 0.1 | 0.2 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.1 | 0.3 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.1 | 0.2 | GO:0072720 | response to dithiothreitol(GO:0072720) |
| 0.1 | 0.2 | GO:0045065 | cytotoxic T cell differentiation(GO:0045065) |
| 0.1 | 0.2 | GO:0021997 | epidermal cell fate specification(GO:0009957) response to chlorate(GO:0010157) neural plate axis specification(GO:0021997) |
| 0.1 | 1.0 | GO:0002347 | response to tumor cell(GO:0002347) |
| 0.1 | 0.2 | GO:0014873 | response to muscle activity involved in regulation of muscle adaptation(GO:0014873) |
| 0.1 | 0.4 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.1 | 0.2 | GO:0016598 | protein arginylation(GO:0016598) |
| 0.0 | 0.1 | GO:1900158 | regulation of chondrocyte development(GO:0061181) negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 0.0 | 0.1 | GO:0048372 | ventricular zone neuroblast division(GO:0021847) fibroblast growth factor receptor signaling pathway involved in orbitofrontal cortex development(GO:0035607) lateral mesodermal cell fate commitment(GO:0048372) lateral mesodermal cell fate specification(GO:0048377) regulation of lateral mesodermal cell fate specification(GO:0048378) |
| 0.0 | 0.1 | GO:1990166 | protein localization to site of double-strand break(GO:1990166) |
| 0.0 | 0.2 | GO:0007228 | positive regulation of hh target transcription factor activity(GO:0007228) |
| 0.0 | 0.1 | GO:0046601 | positive regulation of centriole replication(GO:0046601) |
| 0.0 | 0.1 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.0 | 0.6 | GO:0046599 | regulation of centriole replication(GO:0046599) |
| 0.0 | 0.2 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.1 | GO:0060764 | cell-cell signaling involved in mammary gland development(GO:0060764) |
| 0.0 | 0.3 | GO:0010216 | maintenance of DNA methylation(GO:0010216) |
| 0.0 | 0.1 | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
| 0.0 | 0.2 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.1 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.0 | 0.1 | GO:0003363 | lamellipodium assembly involved in ameboidal cell migration(GO:0003363) extension of a leading process involved in cell motility in cerebral cortex radial glia guided migration(GO:0021816) |
| 0.0 | 0.4 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.0 | 0.1 | GO:0042998 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) |
| 0.0 | 0.1 | GO:0042822 | pyridoxal phosphate metabolic process(GO:0042822) |
| 0.0 | 0.1 | GO:0032066 | nucleolus to nucleoplasm transport(GO:0032066) |
| 0.0 | 0.1 | GO:1904976 | cellular response to bleomycin(GO:1904976) |
| 0.0 | 0.1 | GO:1902809 | regulation of skeletal muscle fiber differentiation(GO:1902809) |
| 0.0 | 0.1 | GO:1904882 | telomerase catalytic core complex assembly(GO:1904868) regulation of telomerase catalytic core complex assembly(GO:1904882) positive regulation of telomerase catalytic core complex assembly(GO:1904884) |
| 0.0 | 0.1 | GO:0006431 | methionyl-tRNA aminoacylation(GO:0006431) |
| 0.0 | 0.4 | GO:1990403 | embryonic brain development(GO:1990403) |
| 0.0 | 0.2 | GO:0070574 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.0 | 0.1 | GO:0002572 | pro-T cell differentiation(GO:0002572) |
| 0.0 | 0.2 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.0 | 0.2 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.0 | 0.1 | GO:0030327 | prenylated protein catabolic process(GO:0030327) |
| 0.0 | 0.1 | GO:0042148 | strand invasion(GO:0042148) |
| 0.0 | 0.2 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.0 | 0.2 | GO:0090080 | positive regulation of MAPKKK cascade by fibroblast growth factor receptor signaling pathway(GO:0090080) |
| 0.0 | 0.1 | GO:0010825 | positive regulation of centrosome duplication(GO:0010825) |
| 0.0 | 0.1 | GO:2000049 | positive regulation of cell-cell adhesion mediated by cadherin(GO:2000049) |
| 0.0 | 0.1 | GO:0072139 | glomerular parietal epithelial cell differentiation(GO:0072139) |
| 0.0 | 0.1 | GO:0060821 | inactivation of X chromosome by DNA methylation(GO:0060821) |
| 0.0 | 0.1 | GO:1905245 | regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902959) positive regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902961) regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) regulation of aspartic-type peptidase activity(GO:1905245) positive regulation of aspartic-type peptidase activity(GO:1905247) |
| 0.0 | 0.1 | GO:0014043 | negative regulation of neuron maturation(GO:0014043) |
| 0.0 | 0.3 | GO:0098719 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.1 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 0.0 | 0.1 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.0 | 0.1 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 0.0 | 0.1 | GO:0015781 | pyrimidine nucleotide-sugar transport(GO:0015781) |
| 0.0 | 0.1 | GO:0071630 | nucleus-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071630) |
| 0.0 | 0.1 | GO:0090172 | microtubule cytoskeleton organization involved in homologous chromosome segregation(GO:0090172) |
| 0.0 | 0.1 | GO:1905168 | positive regulation of double-strand break repair via homologous recombination(GO:1905168) replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.0 | 0.1 | GO:0009992 | cellular water homeostasis(GO:0009992) |
| 0.0 | 0.1 | GO:0006172 | ADP biosynthetic process(GO:0006172) |
| 0.0 | 0.1 | GO:1901896 | positive regulation of calcium-transporting ATPase activity(GO:1901896) |
| 0.0 | 0.1 | GO:0035986 | senescence-associated heterochromatin focus assembly(GO:0035986) |
| 0.0 | 0.4 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.1 | GO:0051088 | PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 0.0 | 0.1 | GO:0070827 | chromatin maintenance(GO:0070827) |
| 0.0 | 0.1 | GO:0014826 | vein smooth muscle contraction(GO:0014826) |
| 0.0 | 0.1 | GO:0019065 | receptor-mediated endocytosis of virus by host cell(GO:0019065) endocytosis involved in viral entry into host cell(GO:0075509) |
| 0.0 | 0.1 | GO:0007354 | zygotic determination of anterior/posterior axis, embryo(GO:0007354) |
| 0.0 | 0.1 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.0 | 0.1 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.0 | 0.2 | GO:0001842 | neural fold formation(GO:0001842) |
| 0.0 | 0.1 | GO:0036337 | Fas signaling pathway(GO:0036337) |
| 0.0 | 0.1 | GO:0060014 | granulosa cell differentiation(GO:0060014) |
| 0.0 | 0.1 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.0 | 0.2 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.0 | 0.6 | GO:0006779 | porphyrin-containing compound biosynthetic process(GO:0006779) |
| 0.0 | 0.1 | GO:0006713 | glucocorticoid catabolic process(GO:0006713) |
| 0.0 | 0.1 | GO:1904354 | negative regulation of telomere capping(GO:1904354) |
| 0.0 | 0.1 | GO:1903999 | negative regulation of eating behavior(GO:1903999) |
| 0.0 | 0.1 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.0 | 0.1 | GO:0090625 | mRNA cleavage involved in gene silencing by siRNA(GO:0090625) |
| 0.0 | 0.2 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 0.1 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 | 0.1 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.0 | 0.1 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.0 | 0.2 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.0 | 0.1 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.0 | 0.3 | GO:0071044 | histone mRNA catabolic process(GO:0071044) |
| 0.0 | 0.1 | GO:0043570 | maintenance of DNA repeat elements(GO:0043570) |
| 0.0 | 0.1 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 0.5 | GO:0044381 | glucose import in response to insulin stimulus(GO:0044381) |
| 0.0 | 0.1 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 0.1 | GO:2000327 | positive regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000327) |
| 0.0 | 0.1 | GO:0031053 | primary miRNA processing(GO:0031053) |
| 0.0 | 0.1 | GO:0046351 | disaccharide biosynthetic process(GO:0046351) |
| 0.0 | 0.2 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.1 | GO:1901994 | negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 0.0 | 0.1 | GO:0072697 | protein localization to cell cortex(GO:0072697) |
| 0.0 | 0.2 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.0 | 0.0 | GO:0010045 | response to nickel cation(GO:0010045) |
| 0.0 | 0.2 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.0 | 0.1 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.0 | 0.0 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.0 | 0.1 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.0 | 0.0 | GO:0071930 | negative regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071930) |
| 0.0 | 0.1 | GO:0035188 | blastocyst hatching(GO:0001835) hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.0 | 0.1 | GO:1903215 | negative regulation of protein targeting to mitochondrion(GO:1903215) |
| 0.0 | 0.1 | GO:0019074 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.0 | 0.1 | GO:0044829 | positive regulation by host of viral genome replication(GO:0044829) |
| 0.0 | 0.2 | GO:0035376 | sterol import(GO:0035376) cholesterol import(GO:0070508) |
| 0.0 | 0.3 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 0.0 | 0.3 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.0 | 0.1 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.0 | 0.1 | GO:0016188 | synaptic vesicle maturation(GO:0016188) |
| 0.0 | 0.1 | GO:2000628 | regulation of miRNA metabolic process(GO:2000628) |
| 0.0 | 0.1 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.0 | 0.1 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.0 | 0.1 | GO:0002326 | B cell lineage commitment(GO:0002326) |
| 0.0 | 0.1 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 0.0 | 0.0 | GO:0015864 | pyrimidine nucleoside transport(GO:0015864) |
| 0.0 | 0.1 | GO:0031937 | positive regulation of chromatin silencing(GO:0031937) |
| 0.0 | 0.1 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.0 | 0.2 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.0 | 0.1 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.1 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.0 | 0.1 | GO:0090071 | negative regulation of ribosome biogenesis(GO:0090071) |
| 0.0 | 0.1 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.0 | 0.2 | GO:0043923 | positive regulation by host of viral transcription(GO:0043923) |
| 0.0 | 0.0 | GO:1903944 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.0 | 0.1 | GO:0046061 | dATP catabolic process(GO:0046061) |
| 0.0 | 0.1 | GO:0070417 | cellular response to cold(GO:0070417) |
| 0.0 | 0.1 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.0 | 0.1 | GO:0044691 | tooth eruption(GO:0044691) |
| 0.0 | 0.2 | GO:0098814 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.0 | 0.1 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.0 | 0.1 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.0 | 0.1 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.1 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.0 | 0.1 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.2 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.0 | 0.2 | GO:0090503 | RNA phosphodiester bond hydrolysis, exonucleolytic(GO:0090503) |
| 0.0 | 0.0 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.0 | 0.1 | GO:0071494 | cellular response to UV-C(GO:0071494) |
| 0.0 | 0.0 | GO:0045054 | constitutive secretory pathway(GO:0045054) |
| 0.0 | 0.1 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.0 | 0.1 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 0.1 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.0 | 0.0 | GO:0061744 | motor behavior(GO:0061744) |
| 0.0 | 0.1 | GO:0060907 | positive regulation of macrophage cytokine production(GO:0060907) |
| 0.0 | 0.1 | GO:0008616 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.0 | 0.1 | GO:0032439 | endosome localization(GO:0032439) |
| 0.0 | 0.0 | GO:0048341 | apoptotic process involved in outflow tract morphogenesis(GO:0003275) paraxial mesoderm formation(GO:0048341) negative regulation of lymphangiogenesis(GO:1901491) regulation of apoptotic process involved in outflow tract morphogenesis(GO:1902256) |
| 0.0 | 0.1 | GO:0072423 | response to cell cycle checkpoint signaling(GO:0072396) response to DNA integrity checkpoint signaling(GO:0072402) response to DNA damage checkpoint signaling(GO:0072423) |
| 0.0 | 0.4 | GO:0030517 | negative regulation of axon extension(GO:0030517) |
| 0.0 | 0.0 | GO:1905071 | proepicardium development(GO:0003342) septum transversum development(GO:0003343) occluding junction disassembly(GO:1905071) regulation of occluding junction disassembly(GO:1905073) positive regulation of occluding junction disassembly(GO:1905075) |
| 0.0 | 0.1 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.1 | GO:0000492 | small nucleolar ribonucleoprotein complex assembly(GO:0000491) box C/D snoRNP assembly(GO:0000492) |
| 0.0 | 0.0 | GO:0019676 | ammonia assimilation cycle(GO:0019676) |
| 0.0 | 0.1 | GO:0006353 | DNA-templated transcription, termination(GO:0006353) |
| 0.0 | 0.1 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 0.0 | 0.1 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.0 | 0.1 | GO:2000781 | positive regulation of double-strand break repair(GO:2000781) |
| 0.0 | 0.1 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.0 | 0.0 | GO:0015960 | diadenosine polyphosphate biosynthetic process(GO:0015960) diadenosine tetraphosphate metabolic process(GO:0015965) diadenosine tetraphosphate biosynthetic process(GO:0015966) |
| 0.0 | 0.1 | GO:0050434 | positive regulation of viral transcription(GO:0050434) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | GO:0042555 | MCM complex(GO:0042555) |
| 0.1 | 0.7 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.1 | 0.3 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.1 | 0.2 | GO:0034667 | integrin alpha3-beta1 complex(GO:0034667) |
| 0.0 | 0.2 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.1 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
| 0.0 | 0.3 | GO:0000796 | condensin complex(GO:0000796) |
| 0.0 | 0.3 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.1 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.0 | 0.2 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.4 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.0 | 0.4 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.2 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 0.3 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.0 | 0.2 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.0 | 0.2 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 0.1 | GO:0044299 | C-fiber(GO:0044299) |
| 0.0 | 0.1 | GO:0098536 | deuterosome(GO:0098536) |
| 0.0 | 0.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.1 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.0 | 0.2 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.0 | GO:0097124 | cyclin A2-CDK2 complex(GO:0097124) |
| 0.0 | 0.1 | GO:0071012 | catalytic step 1 spliceosome(GO:0071012) |
| 0.0 | 0.3 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.1 | GO:0071914 | prominosome(GO:0071914) |
| 0.0 | 0.3 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.1 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.0 | 0.2 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.1 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 0.1 | GO:0035363 | histone locus body(GO:0035363) |
| 0.0 | 0.1 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.1 | GO:0030690 | Noc1p-Noc2p complex(GO:0030690) |
| 0.0 | 0.1 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.0 | 0.1 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.0 | 0.2 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 0.6 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.1 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 0.0 | 0.1 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 0.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.2 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.0 | GO:0032301 | MutSalpha complex(GO:0032301) |
| 0.0 | 0.8 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.1 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 0.0 | 0.1 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.1 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.1 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.0 | GO:1990590 | ATF1-ATF4 transcription factor complex(GO:1990590) |
| 0.0 | 0.1 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 0.0 | 0.1 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 0.2 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.1 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 0.1 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) |
| 0.0 | 0.2 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.1 | GO:0030893 | meiotic cohesin complex(GO:0030893) |
| 0.0 | 0.1 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.0 | 0.1 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.0 | 0.3 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.0 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.0 | 0.2 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.1 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 0.0 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.0 | 0.0 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.0 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 0.0 | 0.1 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.0 | 0.0 | GO:0031308 | intrinsic component of nuclear outer membrane(GO:0031308) integral component of nuclear outer membrane(GO:0031309) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0030977 | taurine binding(GO:0030977) |
| 0.1 | 0.3 | GO:0042895 | antibiotic transporter activity(GO:0042895) |
| 0.1 | 0.4 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 0.3 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 0.1 | 0.4 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.1 | 0.2 | GO:0005119 | smoothened binding(GO:0005119) |
| 0.0 | 0.1 | GO:0047045 | testosterone 17-beta-dehydrogenase (NADP+) activity(GO:0047045) |
| 0.0 | 0.1 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.0 | 0.6 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.1 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 0.0 | 0.1 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.0 | 0.1 | GO:0050567 | glutaminyl-tRNA synthase (glutamine-hydrolyzing) activity(GO:0050567) |
| 0.0 | 0.2 | GO:0000700 | mismatch base pair DNA N-glycosylase activity(GO:0000700) |
| 0.0 | 0.1 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.0 | 0.2 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.0 | 0.1 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.0 | 0.1 | GO:0071209 | U7 snRNA binding(GO:0071209) |
| 0.0 | 0.3 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.3 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.3 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.1 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.1 | GO:0004825 | methionine-tRNA ligase activity(GO:0004825) |
| 0.0 | 0.2 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.2 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.0 | 0.2 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.0 | 0.9 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 0.1 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.1 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 0.0 | 0.2 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.1 | GO:0032142 | single guanine insertion binding(GO:0032142) |
| 0.0 | 0.1 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.1 | GO:0003945 | N-acetyllactosamine synthase activity(GO:0003945) |
| 0.0 | 0.1 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 1.4 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.1 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.0 | 0.1 | GO:0015292 | uniporter activity(GO:0015292) |
| 0.0 | 0.1 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 0.0 | 0.1 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.1 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.2 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.2 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.0 | 0.1 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.1 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.0 | 0.1 | GO:0001225 | RNA polymerase II transcription coactivator binding(GO:0001225) |
| 0.0 | 0.1 | GO:0055105 | ubiquitin-protein transferase inhibitor activity(GO:0055105) |
| 0.0 | 0.1 | GO:0032551 | UTP binding(GO:0002134) pyrimidine ribonucleoside binding(GO:0032551) |
| 0.0 | 0.1 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 0.1 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.1 | GO:0071253 | connexin binding(GO:0071253) |
| 0.0 | 0.1 | GO:0031750 | D3 dopamine receptor binding(GO:0031750) |
| 0.0 | 0.1 | GO:0032296 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.0 | 0.0 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.0 | 0.1 | GO:0016670 | oxidoreductase activity, acting on a sulfur group of donors, oxygen as acceptor(GO:0016670) |
| 0.0 | 0.1 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.0 | 0.1 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.0 | 0.3 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 0.1 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.0 | 0.0 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.0 | 0.1 | GO:0016717 | oxidoreductase activity, acting on paired donors, with oxidation of a pair of donors resulting in the reduction of molecular oxygen to two molecules of water(GO:0016717) |
| 0.0 | 0.2 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 0.0 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.0 | 0.0 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.0 | 0.0 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.0 | 0.0 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.0 | 0.2 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 0.0 | 0.2 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.1 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.0 | 0.0 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.2 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.0 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.1 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.1 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.0 | 0.1 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.0 | 0.1 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.1 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 0.1 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.0 | 0.3 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.1 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.1 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.1 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.0 | 0.1 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.1 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.1 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.0 | 1.0 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.3 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.1 | GO:0032554 | purine deoxyribonucleotide binding(GO:0032554) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.5 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.4 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.3 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.0 | 0.6 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 0.3 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.0 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.0 | 0.3 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.2 | PID IL1 PATHWAY | IL1-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.4 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 0.1 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.1 | 0.7 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.8 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.0 | 0.4 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.5 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 0.0 | 0.2 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 0.1 | REACTOME LAGGING STRAND SYNTHESIS | Genes involved in Lagging Strand Synthesis |
| 0.0 | 0.7 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.3 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.3 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.0 | 0.2 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.2 | REACTOME RESOLUTION OF AP SITES VIA THE MULTIPLE NUCLEOTIDE PATCH REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the multiple-nucleotide patch replacement pathway |
| 0.0 | 0.2 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.2 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.0 | 0.1 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 0.1 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.2 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.1 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.2 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |