Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
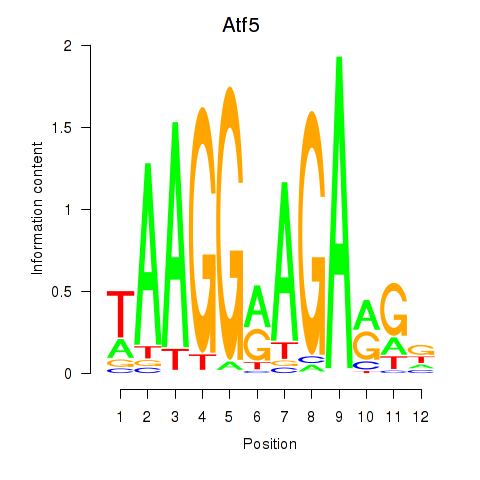
Results for Atf5
Z-value: 0.39
Transcription factors associated with Atf5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Atf5
|
ENSRNOG00000020060 | activating transcription factor 5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Atf5 | rn6_v1_chr1_-_100810522_100810522 | -0.10 | 8.7e-01 | Click! |
Activity profile of Atf5 motif
Sorted Z-values of Atf5 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_21345805 | 0.39 |
ENSRNOT00000081296
ENSRNOT00000007802 |
Car8
|
carbonic anhydrase 8 |
| chr10_-_88551056 | 0.28 |
ENSRNOT00000032737
|
Zfp385c
|
zinc finger protein 385C |
| chr5_-_150167077 | 0.21 |
ENSRNOT00000088493
ENSRNOT00000013761 ENSRNOT00000067349 ENSRNOT00000051097 ENSRNOT00000067189 |
Epb41
|
erythrocyte membrane protein band 4.1 |
| chr1_-_255815733 | 0.10 |
ENSRNOT00000047387
|
Cpeb3
|
cytoplasmic polyadenylation element binding protein 3 |
| chr7_+_79638046 | 0.09 |
ENSRNOT00000029419
|
Zfpm2
|
zinc finger protein, multitype 2 |
| chr10_+_84131495 | 0.09 |
ENSRNOT00000009986
|
Hoxb8
|
homeo box B8 |
| chr8_+_81766041 | 0.08 |
ENSRNOT00000032280
|
Onecut1
|
one cut homeobox 1 |
| chr16_+_27399467 | 0.08 |
ENSRNOT00000065642
|
Tll1
|
tolloid-like 1 |
| chrX_-_29648359 | 0.07 |
ENSRNOT00000086721
ENSRNOT00000006777 |
Gpm6b
|
glycoprotein m6b |
| chr3_-_11417546 | 0.07 |
ENSRNOT00000018776
|
Lcn2
|
lipocalin 2 |
| chr1_-_7480825 | 0.07 |
ENSRNOT00000048754
|
Phactr2
|
phosphatase and actin regulator 2 |
| chr11_-_31180616 | 0.07 |
ENSRNOT00000065535
ENSRNOT00000052033 ENSRNOT00000002812 |
Synj1
|
synaptojanin 1 |
| chr5_+_79055521 | 0.06 |
ENSRNOT00000010333
|
Col27a1
|
collagen type XXVII alpha 1 chain |
| chr7_-_142348232 | 0.06 |
ENSRNOT00000078848
|
Galnt6
|
polypeptide N-acetylgalactosaminyltransferase 6 |
| chr3_-_80601410 | 0.06 |
ENSRNOT00000022841
|
Atg13
|
autophagy related 13 |
| chr20_-_14193690 | 0.06 |
ENSRNOT00000058237
|
Upb1
|
beta-ureidopropionase 1 |
| chr4_+_60549197 | 0.05 |
ENSRNOT00000071249
ENSRNOT00000075621 |
Exoc4
|
exocyst complex component 4 |
| chr7_+_80750725 | 0.05 |
ENSRNOT00000079962
|
Oxr1
|
oxidation resistance 1 |
| chr3_+_116899878 | 0.05 |
ENSRNOT00000090802
ENSRNOT00000066101 |
Sema6d
|
semaphorin 6D |
| chr8_-_131899023 | 0.05 |
ENSRNOT00000034690
|
Zfp445
|
zinc finger protein 445 |
| chr9_-_45206427 | 0.05 |
ENSRNOT00000042227
|
Aff3
|
AF4/FMR2 family, member 3 |
| chr8_+_52751854 | 0.04 |
ENSRNOT00000072518
|
Nxpe1
|
neurexophilin and PC-esterase domain family, member 1 |
| chr9_-_60672246 | 0.04 |
ENSRNOT00000017761
|
Hecw2
|
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 2 |
| chr6_-_100011226 | 0.04 |
ENSRNOT00000083286
ENSRNOT00000041442 |
Max
|
MYC associated factor X |
| chr9_+_121802673 | 0.04 |
ENSRNOT00000086534
|
Yes1
|
YES proto-oncogene 1, Src family tyrosine kinase |
| chr14_-_87701884 | 0.04 |
ENSRNOT00000079338
|
Mospd1
|
motile sperm domain containing 1 |
| chr20_-_5020150 | 0.04 |
ENSRNOT00000001146
|
Sapcd1
|
suppressor APC domain containing 1 |
| chr19_+_9668186 | 0.04 |
ENSRNOT00000016563
|
Cnot1
|
CCR4-NOT transcription complex, subunit 1 |
| chr7_-_118332577 | 0.04 |
ENSRNOT00000090370
|
Rbfox2
|
RNA binding protein, fox-1 homolog 2 |
| chr9_-_28972835 | 0.03 |
ENSRNOT00000086967
ENSRNOT00000079684 |
Rims1
|
regulating synaptic membrane exocytosis 1 |
| chr6_-_102472926 | 0.03 |
ENSRNOT00000079351
|
Zfyve26
|
zinc finger FYVE-type containing 26 |
| chr16_-_7007287 | 0.03 |
ENSRNOT00000041216
|
Itih3
|
inter-alpha trypsin inhibitor, heavy chain 3 |
| chr1_-_57815038 | 0.02 |
ENSRNOT00000075401
|
Rgmb
|
repulsive guidance molecule family member B |
| chr1_+_245237736 | 0.02 |
ENSRNOT00000035814
|
Vldlr
|
very low density lipoprotein receptor |
| chr2_+_188141350 | 0.02 |
ENSRNOT00000078913
|
Gon4l
|
gon-4 like |
| chr5_+_129052043 | 0.02 |
ENSRNOT00000013459
|
LOC108348114
|
tetratricopeptide repeat protein 39A |
| chr9_+_49479023 | 0.02 |
ENSRNOT00000050922
ENSRNOT00000077111 |
Pou3f3
|
POU class 3 homeobox 3 |
| chr1_+_192379543 | 0.02 |
ENSRNOT00000078705
|
Prkcb
|
protein kinase C, beta |
| chrX_-_159841072 | 0.02 |
ENSRNOT00000001158
|
Arhgef6
|
Rac/Cdc42 guanine nucleotide exchange factor 6 |
| chr13_+_43818010 | 0.02 |
ENSRNOT00000005532
|
LOC100909664
|
centrosomal protein of 170 kDa-like |
| chrX_+_5549419 | 0.02 |
ENSRNOT00000004676
|
Fundc1
|
FUN14 domain containing 1 |
| chr15_-_51834030 | 0.02 |
ENSRNOT00000024895
|
Ccar2
|
cell cycle and apoptosis regulator 2 |
| chr5_-_138462864 | 0.02 |
ENSRNOT00000011325
|
Ccdc30
|
coiled-coil domain containing 30 |
| chr8_+_117679278 | 0.02 |
ENSRNOT00000042114
|
Uqcrc1
|
ubiquinol-cytochrome c reductase core protein I |
| chr18_-_86279680 | 0.02 |
ENSRNOT00000006169
|
LOC689166
|
hypothetical protein LOC689166 |
| chr3_+_172510874 | 0.01 |
ENSRNOT00000073634
|
Nelfcd
|
negative elongation factor complex member C/D |
| chr13_-_94859390 | 0.01 |
ENSRNOT00000005467
ENSRNOT00000079763 |
AABR07021840.1
|
|
| chr1_+_106998623 | 0.01 |
ENSRNOT00000022383
|
Slc17a6
|
solute carrier family 17 member 6 |
| chr7_-_121943121 | 0.01 |
ENSRNOT00000046460
|
Rpl26-ps1
|
ribosomal protein L26, pseudogene 1 |
| chr5_-_173209768 | 0.01 |
ENSRNOT00000045053
|
Atad3a
|
ATPase family, AAA domain containing 3A |
| chr9_-_16643182 | 0.01 |
ENSRNOT00000024266
|
LOC108348250
|
cullin-7 |
| chr9_-_63637677 | 0.01 |
ENSRNOT00000049259
|
Satb2
|
SATB homeobox 2 |
| chr6_+_27768943 | 0.01 |
ENSRNOT00000015820
|
Kif3c
|
kinesin family member 3C |
| chr13_-_89291371 | 0.01 |
ENSRNOT00000004181
|
Fcrlb
|
Fc receptor-like B |
| chr4_-_129569470 | 0.01 |
ENSRNOT00000008893
|
Uba3
|
ubiquitin-like modifier activating enzyme 3 |
| chr19_+_34139997 | 0.01 |
ENSRNOT00000017867
|
Arhgap10
|
Rho GTPase activating protein 10 |
| chr1_+_200037749 | 0.01 |
ENSRNOT00000027630
|
Inpp5f
|
inositol polyphosphate-5-phosphatase F |
| chr1_+_199495298 | 0.00 |
ENSRNOT00000086003
ENSRNOT00000026748 |
Itgad
|
integrin subunit alpha D |
| chr5_+_151211342 | 0.00 |
ENSRNOT00000067939
|
Ahdc1
|
AT hook, DNA binding motif, containing 1 |
| chr20_+_1889652 | 0.00 |
ENSRNOT00000068508
ENSRNOT00000000996 |
Olr1746
|
olfactory receptor 1746 |
| chr9_-_71788281 | 0.00 |
ENSRNOT00000020078
|
Crygc
|
crystallin, gamma C |
| chr19_+_41631715 | 0.00 |
ENSRNOT00000022749
|
Chst4
|
carbohydrate sulfotransferase 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Atf5
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0045900 | negative regulation of translational elongation(GO:0045900) |
| 0.0 | 0.1 | GO:0015891 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 0.0 | 0.1 | GO:2000195 | negative regulation of female gonad development(GO:2000195) |
| 0.0 | 0.1 | GO:0019483 | beta-alanine biosynthetic process(GO:0019483) |
| 0.0 | 0.1 | GO:0051612 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.0 | 0.1 | GO:0003431 | growth plate cartilage chondrocyte development(GO:0003431) |
| 0.0 | 0.0 | GO:0031632 | positive regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031632) |
| 0.0 | 0.1 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.0 | 0.4 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.1 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |