Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
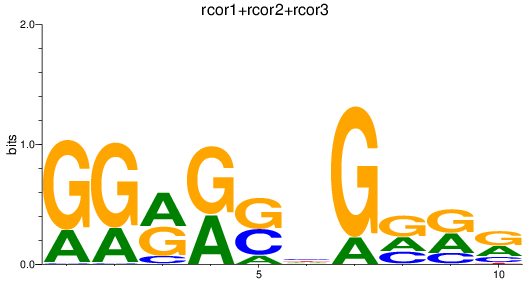
Results for rcor1+rcor2+rcor3
Z-value: 0.83
Transcription factors associated with rcor1+rcor2+rcor3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
rcor3
|
ENSDARG00000004502 | REST corepressor 3 |
|
rcor2
|
ENSDARG00000008278 | REST corepressor 2 |
|
rcor1
|
ENSDARG00000031434 | REST corepressor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| rcor2 | dr11_v1_chr7_-_24995631_24995640 | 0.68 | 1.5e-03 | Click! |
| rcor3 | dr11_v1_chr22_+_38276024_38276024 | 0.65 | 2.5e-03 | Click! |
| rcor1 | dr11_v1_chr17_-_29271359_29271359 | 0.52 | 2.3e-02 | Click! |
Activity profile of rcor1+rcor2+rcor3 motif
Sorted Z-values of rcor1+rcor2+rcor3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_56095275 | 0.86 |
ENSDART00000154701
|
si:ch211-178n15.1
|
si:ch211-178n15.1 |
| chr3_+_41917499 | 0.80 |
ENSDART00000028673
|
lfng
|
LFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr3_-_62527675 | 0.78 |
ENSDART00000155048
ENSDART00000064500 |
sox9b
|
SRY (sex determining region Y)-box 9b |
| chr1_-_59571758 | 0.71 |
ENSDART00000193546
ENSDART00000167087 |
wfikkn1
|
WAP, follistatin/kazal, immunoglobulin, kunitz and netrin domain containing 1 |
| chr7_+_21752168 | 0.68 |
ENSDART00000173641
|
kdm6ba
|
lysine (K)-specific demethylase 6B, a |
| chr8_-_36554675 | 0.68 |
ENSDART00000132804
ENSDART00000078746 |
ccdc157
|
coiled-coil domain containing 157 |
| chr11_+_14937904 | 0.68 |
ENSDART00000185103
|
CR550302.3
|
|
| chr17_+_1514711 | 0.67 |
ENSDART00000165641
|
akt1
|
v-akt murine thymoma viral oncogene homolog 1 |
| chr22_+_569565 | 0.66 |
ENSDART00000037069
|
usp49
|
ubiquitin specific peptidase 49 |
| chr5_+_22098591 | 0.64 |
ENSDART00000143676
|
zc3h12b
|
zinc finger CCCH-type containing 12B |
| chr2_+_11685742 | 0.62 |
ENSDART00000138562
|
greb1l
|
growth regulation by estrogen in breast cancer-like |
| chr17_+_41992054 | 0.59 |
ENSDART00000182878
ENSDART00000111537 |
kiz
|
kizuna centrosomal protein |
| chr22_+_10215558 | 0.59 |
ENSDART00000063274
|
kctd6a
|
potassium channel tetramerization domain containing 6a |
| chr2_+_30032303 | 0.58 |
ENSDART00000151841
|
rbm33b
|
RNA binding motif protein 33b |
| chr19_+_48060464 | 0.57 |
ENSDART00000123163
|
zgc:85936
|
zgc:85936 |
| chr25_-_37258653 | 0.57 |
ENSDART00000131076
|
rfwd3
|
ring finger and WD repeat domain 3 |
| chr16_+_29043813 | 0.56 |
ENSDART00000122681
|
nes
|
nestin |
| chr4_-_19693978 | 0.56 |
ENSDART00000100974
ENSDART00000040405 |
snd1
|
staphylococcal nuclease and tudor domain containing 1 |
| chr12_+_8003872 | 0.55 |
ENSDART00000180300
ENSDART00000126661 |
rhobtb1
|
Rho-related BTB domain containing 1 |
| chr3_-_38777553 | 0.55 |
ENSDART00000193045
|
znf281a
|
zinc finger protein 281a |
| chr7_+_51103416 | 0.53 |
ENSDART00000174236
|
col4a5
|
collagen, type IV, alpha 5 (Alport syndrome) |
| chr4_-_64703 | 0.53 |
ENSDART00000167851
|
CU856344.1
|
|
| chr6_+_31684 | 0.53 |
ENSDART00000188853
ENSDART00000184553 |
CZQB01141835.2
|
|
| chr19_+_7559901 | 0.53 |
ENSDART00000141189
|
pbxip1a
|
pre-B-cell leukemia homeobox interacting protein 1a |
| chr8_+_29749017 | 0.52 |
ENSDART00000185144
|
mapk4
|
mitogen-activated protein kinase 4 |
| chr21_-_30787414 | 0.52 |
ENSDART00000012091
|
rrm1
|
ribonucleotide reductase M1 polypeptide |
| chr10_-_36618674 | 0.52 |
ENSDART00000135302
|
rsf1b.1
|
remodeling and spacing factor 1b, tandem duplicate 1 |
| chr20_-_32981053 | 0.51 |
ENSDART00000138708
|
nbas
|
neuroblastoma amplified sequence |
| chr22_-_11724563 | 0.51 |
ENSDART00000190796
ENSDART00000184744 |
krt222
|
keratin 222 |
| chr1_+_58990121 | 0.51 |
ENSDART00000171654
|
CABZ01083166.1
|
|
| chr19_+_14352332 | 0.51 |
ENSDART00000164386
|
arid1ab
|
AT rich interactive domain 1Ab (SWI-like) |
| chr17_-_10059557 | 0.50 |
ENSDART00000092209
ENSDART00000161243 |
baz1a
|
bromodomain adjacent to zinc finger domain, 1A |
| chr13_-_51922290 | 0.50 |
ENSDART00000168648
|
srfb
|
serum response factor b |
| chr22_+_22438783 | 0.49 |
ENSDART00000147825
|
kif14
|
kinesin family member 14 |
| chr8_+_45381298 | 0.49 |
ENSDART00000149451
ENSDART00000110364 |
tti2
|
TELO2 interacting protein 2 |
| chr19_-_2231146 | 0.49 |
ENSDART00000181909
ENSDART00000043595 |
twist1a
|
twist family bHLH transcription factor 1a |
| chr18_+_19772874 | 0.49 |
ENSDART00000043455
|
smad3b
|
SMAD family member 3b |
| chr12_-_49168398 | 0.48 |
ENSDART00000186608
|
FO704624.1
|
|
| chr7_+_36898622 | 0.48 |
ENSDART00000190773
|
tox3
|
TOX high mobility group box family member 3 |
| chr24_-_26518972 | 0.48 |
ENSDART00000097792
|
tnikb
|
TRAF2 and NCK interacting kinase b |
| chr14_-_1949277 | 0.47 |
ENSDART00000159435
|
pcdh2g5
|
protocadherin 2 gamma 5 |
| chr7_+_16835218 | 0.46 |
ENSDART00000173580
|
nav2a
|
neuron navigator 2a |
| chr17_-_23412705 | 0.45 |
ENSDART00000126995
|
si:ch211-149k12.3
|
si:ch211-149k12.3 |
| chr17_+_26783828 | 0.45 |
ENSDART00000154921
|
larp1b
|
La ribonucleoprotein domain family, member 1B |
| chr7_+_39720317 | 0.44 |
ENSDART00000164996
|
si:dkey-58i8.5
|
si:dkey-58i8.5 |
| chr17_+_28624321 | 0.44 |
ENSDART00000122260
|
heatr5a
|
HEAT repeat containing 5a |
| chr3_+_60957512 | 0.44 |
ENSDART00000044096
|
helz
|
helicase with zinc finger |
| chr1_+_14454663 | 0.44 |
ENSDART00000005067
|
rbpja
|
recombination signal binding protein for immunoglobulin kappa J region a |
| chr13_+_15941850 | 0.43 |
ENSDART00000016294
|
fignl1
|
fidgetin-like 1 |
| chr5_+_59234421 | 0.43 |
ENSDART00000045518
|
prkrip1
|
PRKR interacting protein 1 |
| chr14_-_2004291 | 0.42 |
ENSDART00000114039
|
pcdh2g5
|
protocadherin 2 gamma 5 |
| chr25_-_27729046 | 0.42 |
ENSDART00000131437
|
zgc:153935
|
zgc:153935 |
| chr22_-_10539180 | 0.42 |
ENSDART00000131217
|
ippk
|
inositol 1,3,4,5,6-pentakisphosphate 2-kinase |
| chr13_-_40316367 | 0.41 |
ENSDART00000009343
|
pyroxd2
|
pyridine nucleotide-disulphide oxidoreductase domain 2 |
| chr12_+_14149686 | 0.41 |
ENSDART00000123741
|
kbtbd2
|
kelch repeat and BTB (POZ) domain containing 2 |
| chr3_+_14157090 | 0.41 |
ENSDART00000156766
|
si:ch211-108d22.2
|
si:ch211-108d22.2 |
| chr19_+_49721 | 0.41 |
ENSDART00000160489
|
col14a1b
|
collagen, type XIV, alpha 1b |
| chr17_-_25630822 | 0.40 |
ENSDART00000126201
ENSDART00000105503 ENSDART00000151878 |
rab3gap2
|
RAB3 GTPase activating protein subunit 2 (non-catalytic) |
| chr20_-_1560724 | 0.40 |
ENSDART00000179193
ENSDART00000092508 ENSDART00000133369 |
ptprk
|
protein tyrosine phosphatase, receptor type, K |
| chr24_-_24060460 | 0.39 |
ENSDART00000142813
|
abcc13
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 13 |
| chr8_+_54137350 | 0.39 |
ENSDART00000164153
|
brpf1
|
bromodomain and PHD finger containing, 1 |
| chr6_+_58543336 | 0.39 |
ENSDART00000157018
|
stmn3
|
stathmin-like 3 |
| chr6_-_60104628 | 0.38 |
ENSDART00000057463
ENSDART00000169188 |
pmepa1
|
prostate transmembrane protein, androgen induced 1 |
| chr3_-_15667713 | 0.38 |
ENSDART00000026658
|
zgc:66474
|
zgc:66474 |
| chr14_+_14806851 | 0.38 |
ENSDART00000169235
|
fhdc2
|
FH2 domain containing 2 |
| chr23_-_6722101 | 0.37 |
ENSDART00000157828
|
baz2a
|
bromodomain adjacent to zinc finger domain, 2A |
| chr21_-_13972745 | 0.37 |
ENSDART00000143874
|
akna
|
AT-hook transcription factor |
| chr19_+_43256986 | 0.37 |
ENSDART00000182336
|
DGKD
|
diacylglycerol kinase delta |
| chr17_-_22048233 | 0.37 |
ENSDART00000155203
|
ttbk1b
|
tau tubulin kinase 1b |
| chr18_+_62932 | 0.36 |
ENSDART00000052638
|
slc27a2a
|
solute carrier family 27 (fatty acid transporter), member 2a |
| chr4_+_14971239 | 0.36 |
ENSDART00000005985
|
smo
|
smoothened, frizzled class receptor |
| chr24_+_24170914 | 0.35 |
ENSDART00000127842
|
si:dkey-226l10.6
|
si:dkey-226l10.6 |
| chr19_-_2029777 | 0.34 |
ENSDART00000128639
|
CABZ01071939.1
|
|
| chr6_-_10788065 | 0.34 |
ENSDART00000190968
|
wipf1b
|
WAS/WASL interacting protein family, member 1b |
| chr19_-_31765615 | 0.34 |
ENSDART00000103636
|
si:dkeyp-120h9.1
|
si:dkeyp-120h9.1 |
| chr5_+_24087035 | 0.34 |
ENSDART00000183644
|
tp53
|
tumor protein p53 |
| chr25_+_2993855 | 0.34 |
ENSDART00000163009
|
neo1b
|
neogenin 1b |
| chr3_-_24205339 | 0.34 |
ENSDART00000157135
|
si:dkey-110g7.8
|
si:dkey-110g7.8 |
| chr2_-_43123949 | 0.33 |
ENSDART00000075347
ENSDART00000132346 |
lrrc6
|
leucine rich repeat containing 6 |
| chr6_+_269204 | 0.33 |
ENSDART00000191678
|
atf4a
|
activating transcription factor 4a |
| chr19_+_14059349 | 0.33 |
ENSDART00000166230
|
tpbga
|
trophoblast glycoprotein a |
| chr23_-_45504991 | 0.33 |
ENSDART00000148761
|
col24a1
|
collagen type XXIV alpha 1 |
| chr1_+_45056371 | 0.33 |
ENSDART00000073689
ENSDART00000167309 |
btr01
|
bloodthirsty-related gene family, member 1 |
| chr15_-_434503 | 0.33 |
ENSDART00000122286
|
CABZ01056629.1
|
|
| chr14_+_23518110 | 0.32 |
ENSDART00000112930
|
si:ch211-221f10.2
|
si:ch211-221f10.2 |
| chr22_+_30009926 | 0.32 |
ENSDART00000142529
|
si:dkey-286j15.1
|
si:dkey-286j15.1 |
| chr1_+_21725821 | 0.32 |
ENSDART00000132602
|
pax5
|
paired box 5 |
| chr10_-_641609 | 0.32 |
ENSDART00000041236
|
rfx3
|
regulatory factor X, 3 (influences HLA class II expression) |
| chr15_-_33807758 | 0.32 |
ENSDART00000158445
|
pds5b
|
PDS5 cohesin associated factor B |
| chr1_-_39943596 | 0.31 |
ENSDART00000149730
|
stox2a
|
storkhead box 2a |
| chr11_+_807153 | 0.31 |
ENSDART00000173289
|
vgll4b
|
vestigial-like family member 4b |
| chr7_+_51103232 | 0.31 |
ENSDART00000073827
|
col4a5
|
collagen, type IV, alpha 5 (Alport syndrome) |
| chr22_+_17531214 | 0.31 |
ENSDART00000161174
ENSDART00000088356 |
hnrnpm
|
heterogeneous nuclear ribonucleoprotein M |
| chr12_+_16452575 | 0.31 |
ENSDART00000058665
|
kif20bb
|
kinesin family member 20Bb |
| chr22_+_17531020 | 0.30 |
ENSDART00000137689
|
hnrnpm
|
heterogeneous nuclear ribonucleoprotein M |
| chr14_+_18785727 | 0.30 |
ENSDART00000184452
|
si:ch211-111e20.1
|
si:ch211-111e20.1 |
| chr7_+_13491452 | 0.30 |
ENSDART00000053535
|
arih1l
|
ariadne homolog, ubiquitin-conjugating enzyme E2 binding protein, 1 like |
| chr9_-_32668928 | 0.30 |
ENSDART00000135798
|
gzm3.3
|
granzyme 3, tandem duplicate 3 |
| chr10_-_35177257 | 0.30 |
ENSDART00000077426
|
pom121
|
POM121 transmembrane nucleoporin |
| chr2_-_44282796 | 0.30 |
ENSDART00000163040
ENSDART00000166923 ENSDART00000056372 ENSDART00000109251 ENSDART00000132682 |
mpz
|
myelin protein zero |
| chr11_-_44999858 | 0.30 |
ENSDART00000167759
ENSDART00000126845 |
ldb1b
|
LIM-domain binding 1b |
| chr25_+_36311333 | 0.30 |
ENSDART00000190174
|
hist1h2a2
|
histone cluster 1 H2A family member 2 |
| chr17_+_38573471 | 0.30 |
ENSDART00000040627
|
sptb
|
spectrin, beta, erythrocytic |
| chr11_+_12811906 | 0.30 |
ENSDART00000123445
|
rtel1
|
regulator of telomere elongation helicase 1 |
| chr23_+_2717950 | 0.29 |
ENSDART00000137641
|
ncoa6
|
nuclear receptor coactivator 6 |
| chr6_+_59991076 | 0.29 |
ENSDART00000163575
|
CABZ01100888.1
|
|
| chr23_+_43770149 | 0.29 |
ENSDART00000024313
|
rnf150b
|
ring finger protein 150b |
| chr1_+_55080797 | 0.28 |
ENSDART00000077390
|
lgalsla
|
lectin, galactoside-binding-like a |
| chr1_-_45584407 | 0.28 |
ENSDART00000149155
|
atf7ip
|
activating transcription factor 7 interacting protein |
| chr8_+_36554816 | 0.28 |
ENSDART00000126687
|
sf3a1
|
splicing factor 3a, subunit 1 |
| chr3_-_27065477 | 0.27 |
ENSDART00000185660
|
atf7ip2
|
activating transcription factor 7 interacting protein 2 |
| chr7_-_56831621 | 0.27 |
ENSDART00000182912
|
senp8
|
SUMO/sentrin peptidase family member, NEDD8 specific |
| chr20_+_305035 | 0.27 |
ENSDART00000104807
|
si:dkey-119m7.4
|
si:dkey-119m7.4 |
| chr2_+_1989941 | 0.26 |
ENSDART00000190814
|
ssx2ipa
|
synovial sarcoma, X breakpoint 2 interacting protein a |
| chr22_-_876506 | 0.26 |
ENSDART00000137522
|
cept1b
|
choline/ethanolamine phosphotransferase 1b |
| chr5_-_32445835 | 0.26 |
ENSDART00000170919
|
ncs1a
|
neuronal calcium sensor 1a |
| chr11_+_31324335 | 0.26 |
ENSDART00000088093
|
sipa1l2
|
signal-induced proliferation-associated 1 like 2 |
| chr5_-_2672492 | 0.26 |
ENSDART00000192337
|
CABZ01072548.1
|
|
| chr19_-_15855427 | 0.26 |
ENSDART00000133059
|
cited4a
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 4a |
| chr12_+_16452912 | 0.25 |
ENSDART00000192467
|
kif20bb
|
kinesin family member 20Bb |
| chr2_+_50094873 | 0.25 |
ENSDART00000132307
|
zcchc2
|
zinc finger, CCHC domain containing 2 |
| chr22_-_968484 | 0.25 |
ENSDART00000105895
|
cacna1sa
|
calcium channel, voltage-dependent, L type, alpha 1S subunit, a |
| chr15_-_25198107 | 0.25 |
ENSDART00000159793
|
mnta
|
MAX network transcriptional repressor a |
| chr7_-_29223614 | 0.25 |
ENSDART00000173598
|
herc1
|
HECT and RLD domain containing E3 ubiquitin protein ligase family member 1 |
| chr2_+_45374163 | 0.24 |
ENSDART00000193650
|
camsap2b
|
calmodulin regulated spectrin-associated protein family, member 2b |
| chr12_+_18744610 | 0.24 |
ENSDART00000153456
|
mkl1b
|
megakaryoblastic leukemia (translocation) 1b |
| chr1_+_6243906 | 0.24 |
ENSDART00000127329
|
gtf3c3
|
general transcription factor IIIC, polypeptide 3 |
| chr14_+_39156 | 0.24 |
ENSDART00000082184
|
tmem107
|
transmembrane protein 107 |
| chr2_+_49572059 | 0.24 |
ENSDART00000108861
|
sema4e
|
semaphorin 4e |
| chr7_-_25697285 | 0.24 |
ENSDART00000082620
|
dysf
|
dysferlin, limb girdle muscular dystrophy 2B (autosomal recessive) |
| chr13_-_10431476 | 0.24 |
ENSDART00000133968
|
camkmt
|
calmodulin-lysine N-methyltransferase |
| chr1_-_44928987 | 0.24 |
ENSDART00000134635
|
si:dkey-9i23.15
|
si:dkey-9i23.15 |
| chr20_+_54333774 | 0.23 |
ENSDART00000144633
|
cipcb
|
CLOCK-interacting pacemaker b |
| chr9_+_21535885 | 0.23 |
ENSDART00000141408
|
arhgef7a
|
Rho guanine nucleotide exchange factor (GEF) 7a |
| chr9_-_30371247 | 0.23 |
ENSDART00000079068
|
asb11
|
ankyrin repeat and SOCS box containing 11 |
| chr2_-_7185460 | 0.23 |
ENSDART00000092078
|
rc3h1b
|
ring finger and CCCH-type domains 1b |
| chr10_-_7666810 | 0.23 |
ENSDART00000191479
|
pcyox1
|
prenylcysteine oxidase 1 |
| chr8_-_18613948 | 0.23 |
ENSDART00000089172
|
cpox
|
coproporphyrinogen oxidase |
| chr8_+_26868105 | 0.23 |
ENSDART00000005337
|
rimkla
|
ribosomal modification protein rimK-like family member A |
| chr12_+_13348918 | 0.23 |
ENSDART00000181373
|
rnasen
|
ribonuclease type III, nuclear |
| chr4_+_63928858 | 0.23 |
ENSDART00000161958
|
si:dkey-179k24.2
|
si:dkey-179k24.2 |
| chr21_-_15200556 | 0.23 |
ENSDART00000141809
|
sfswap
|
splicing factor SWAP |
| chr24_-_26715174 | 0.23 |
ENSDART00000079726
|
pld1b
|
phospholipase D1b |
| chr14_-_36325558 | 0.23 |
ENSDART00000172783
|
egf
|
epidermal growth factor |
| chr5_-_32505276 | 0.22 |
ENSDART00000034705
ENSDART00000187597 |
ntmt1
|
N-terminal Xaa-Pro-Lys N-methyltransferase 1 |
| chr24_+_28525507 | 0.22 |
ENSDART00000191121
|
arhgap29a
|
Rho GTPase activating protein 29a |
| chr20_-_46694573 | 0.22 |
ENSDART00000182557
|
AL929435.2
|
|
| chr3_+_16229911 | 0.22 |
ENSDART00000121728
|
rpl19
|
ribosomal protein L19 |
| chr5_+_21225017 | 0.22 |
ENSDART00000141573
|
si:dkey-13n15.11
|
si:dkey-13n15.11 |
| chr13_-_4367083 | 0.22 |
ENSDART00000124716
|
si:dkeyp-121d4.3
|
si:dkeyp-121d4.3 |
| chr1_-_40102836 | 0.22 |
ENSDART00000147317
|
cntf
|
ciliary neurotrophic factor |
| chr11_+_13424116 | 0.21 |
ENSDART00000125563
|
homer3b
|
homer scaffolding protein 3b |
| chr21_+_39941559 | 0.21 |
ENSDART00000189718
ENSDART00000160875 ENSDART00000135235 |
slc47a1
|
solute carrier family 47 (multidrug and toxin extrusion), member 1 |
| chr13_+_31070181 | 0.21 |
ENSDART00000110560
ENSDART00000146088 |
si:ch211-223a10.1
|
si:ch211-223a10.1 |
| chr14_-_23801389 | 0.21 |
ENSDART00000054264
|
nr3c1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| chr14_-_237130 | 0.21 |
ENSDART00000164988
|
bod1l1
|
biorientation of chromosomes in cell division 1-like 1 |
| chr18_-_17074155 | 0.21 |
ENSDART00000143304
|
tango6
|
transport and golgi organization 6 homolog (Drosophila) |
| chr7_-_41554047 | 0.21 |
ENSDART00000174144
|
plxdc2
|
plexin domain containing 2 |
| chr6_-_43387920 | 0.21 |
ENSDART00000154434
|
frmd4ba
|
FERM domain containing 4Ba |
| chr19_-_3298352 | 0.21 |
ENSDART00000147182
|
si:ch211-133n4.7
|
si:ch211-133n4.7 |
| chr1_+_16600690 | 0.20 |
ENSDART00000162164
|
mtus1b
|
microtubule associated tumor suppressor 1b |
| chr6_-_31233696 | 0.20 |
ENSDART00000079173
|
lepr
|
leptin receptor |
| chr19_+_4892281 | 0.20 |
ENSDART00000150969
|
cdk12
|
cyclin-dependent kinase 12 |
| chr19_+_6938289 | 0.20 |
ENSDART00000139122
ENSDART00000178832 |
flot1b
|
flotillin 1b |
| chr2_+_4383061 | 0.20 |
ENSDART00000163986
|
wacb
|
WW domain containing adaptor with coiled-coil b |
| chr21_+_25643880 | 0.19 |
ENSDART00000192392
ENSDART00000145091 |
tmem151a
|
transmembrane protein 151A |
| chr5_+_53482597 | 0.19 |
ENSDART00000180333
|
BX323994.1
|
|
| chr13_-_280652 | 0.19 |
ENSDART00000193627
|
slc30a6
|
solute carrier family 30 (zinc transporter), member 6 |
| chr9_-_27717006 | 0.19 |
ENSDART00000146860
|
gtf2e1
|
general transcription factor IIE, polypeptide 1, alpha |
| chr9_-_54344405 | 0.19 |
ENSDART00000182939
|
CT998556.1
|
|
| chr11_-_25734417 | 0.19 |
ENSDART00000103570
|
brpf3a
|
bromodomain and PHD finger containing, 3a |
| chr6_-_52156427 | 0.19 |
ENSDART00000082821
|
rims4
|
regulating synaptic membrane exocytosis 4 |
| chr5_-_32505109 | 0.19 |
ENSDART00000188219
|
ntmt1
|
N-terminal Xaa-Pro-Lys N-methyltransferase 1 |
| chr12_-_33893381 | 0.19 |
ENSDART00000153280
|
sema4gb
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4Gb |
| chr1_-_19502322 | 0.19 |
ENSDART00000181888
ENSDART00000044030 |
kitb
|
v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog b |
| chr23_+_2825940 | 0.18 |
ENSDART00000135781
|
plcg1
|
phospholipase C, gamma 1 |
| chr18_-_17087138 | 0.18 |
ENSDART00000135597
|
zc3h18
|
zinc finger CCCH-type containing 18 |
| chr11_-_42315748 | 0.18 |
ENSDART00000169853
|
slmapa
|
sarcolemma associated protein a |
| chr12_-_37449396 | 0.18 |
ENSDART00000152951
|
cdc42ep4b
|
CDC42 effector protein (Rho GTPase binding) 4b |
| chr17_+_49081828 | 0.18 |
ENSDART00000156492
|
tiam2a
|
T cell lymphoma invasion and metastasis 2a |
| chr25_+_336503 | 0.18 |
ENSDART00000160395
|
CU929262.1
|
|
| chr19_-_82504 | 0.17 |
ENSDART00000027864
ENSDART00000160560 |
hnrnpr
|
heterogeneous nuclear ribonucleoprotein R |
| chr12_+_44610912 | 0.17 |
ENSDART00000179030
ENSDART00000178008 |
FAM196A (1 of many)
|
family with sequence similarity 196 member A |
| chr6_-_48347724 | 0.17 |
ENSDART00000046973
|
capza1a
|
capping protein (actin filament) muscle Z-line, alpha 1a |
| chr11_+_45343245 | 0.17 |
ENSDART00000160904
|
si:ch73-100l22.3
|
si:ch73-100l22.3 |
| chr10_-_40826657 | 0.17 |
ENSDART00000076304
|
pcna
|
proliferating cell nuclear antigen |
| chr10_+_34377697 | 0.17 |
ENSDART00000189441
|
stard13a
|
StAR-related lipid transfer (START) domain containing 13a |
| chr15_+_1397811 | 0.17 |
ENSDART00000102125
|
schip1
|
schwannomin interacting protein 1 |
| chr2_-_2642476 | 0.17 |
ENSDART00000124032
|
serbp1b
|
SERPINE1 mRNA binding protein 1b |
| chr24_-_25166416 | 0.17 |
ENSDART00000111552
ENSDART00000169495 |
phldb2b
|
pleckstrin homology-like domain, family B, member 2b |
| chr10_-_42237304 | 0.17 |
ENSDART00000140341
|
tcf7l1a
|
transcription factor 7 like 1a |
| chr8_+_13106760 | 0.17 |
ENSDART00000029308
|
itgb4
|
integrin, beta 4 |
| chr21_-_11646878 | 0.17 |
ENSDART00000162426
ENSDART00000135937 ENSDART00000131448 ENSDART00000148097 ENSDART00000133443 |
cast
|
calpastatin |
| chr14_+_16287968 | 0.16 |
ENSDART00000106593
|
prpf19
|
pre-mRNA processing factor 19 |
| chr22_-_26524596 | 0.16 |
ENSDART00000087623
|
zgc:194330
|
zgc:194330 |
| chr18_-_44356656 | 0.16 |
ENSDART00000193084
|
prdm10
|
PR domain containing 10 |
| chr4_+_33537372 | 0.16 |
ENSDART00000181450
|
si:dkey-84h14.2
|
si:dkey-84h14.2 |
| chr3_+_28502419 | 0.16 |
ENSDART00000151081
|
sept12
|
septin 12 |
| chr24_+_18689980 | 0.16 |
ENSDART00000160941
ENSDART00000180303 ENSDART00000190435 |
cspp1a
|
centrosome and spindle pole associated protein 1a |
| chr24_-_14711597 | 0.16 |
ENSDART00000131830
|
jph1a
|
junctophilin 1a |
Network of associatons between targets according to the STRING database.
First level regulatory network of rcor1+rcor2+rcor3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0061032 | visceral serous pericardium development(GO:0061032) |
| 0.2 | 0.8 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.2 | 0.6 | GO:0010610 | regulation of mRNA stability involved in response to stress(GO:0010610) regulation of mRNA stability involved in response to oxidative stress(GO:2000815) |
| 0.1 | 0.4 | GO:0072314 | glomerular visceral epithelial cell fate commitment(GO:0072149) glomerular epithelial cell fate commitment(GO:0072314) |
| 0.1 | 0.6 | GO:1901881 | positive regulation of protein depolymerization(GO:1901881) |
| 0.1 | 0.4 | GO:0006480 | N-terminal protein amino acid methylation(GO:0006480) |
| 0.1 | 0.8 | GO:0055016 | hypochord development(GO:0055016) |
| 0.1 | 0.5 | GO:2000622 | negative regulation of mRNA catabolic process(GO:1902373) regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.1 | 0.5 | GO:0060829 | negative regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060829) |
| 0.1 | 0.4 | GO:0060844 | arterial endothelial cell fate commitment(GO:0060844) |
| 0.1 | 0.3 | GO:1901216 | regulation of transposition, RNA-mediated(GO:0010525) negative regulation of transposition, RNA-mediated(GO:0010526) transposition, RNA-mediated(GO:0032197) positive regulation of neuron apoptotic process(GO:0043525) positive regulation of neuron death(GO:1901216) |
| 0.1 | 0.4 | GO:0052746 | inositol phosphorylation(GO:0052746) |
| 0.1 | 0.2 | GO:0035477 | regulation of angioblast cell migration involved in selective angioblast sprouting(GO:0035477) |
| 0.1 | 0.5 | GO:2001106 | regulation of guanyl-nucleotide exchange factor activity(GO:1905097) regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.1 | 0.7 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.1 | 0.3 | GO:0006521 | regulation of cellular amino acid metabolic process(GO:0006521) regulation of cellular amine metabolic process(GO:0033238) |
| 0.1 | 0.2 | GO:1901890 | positive regulation of cell junction assembly(GO:1901890) |
| 0.1 | 0.2 | GO:0001112 | DNA-templated transcriptional open complex formation(GO:0001112) transcriptional open complex formation at RNA polymerase II promoter(GO:0001113) protein-DNA complex remodeling(GO:0001120) |
| 0.1 | 0.2 | GO:0042117 | plasma membrane repair(GO:0001778) monocyte activation(GO:0042117) |
| 0.1 | 0.2 | GO:0032530 | regulation of microvillus organization(GO:0032530) regulation of microvillus assembly(GO:0032534) |
| 0.1 | 0.3 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) collagen-activated signaling pathway(GO:0038065) |
| 0.1 | 0.4 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.1 | 0.2 | GO:0071387 | cellular response to cortisol stimulus(GO:0071387) response to dexamethasone(GO:0071548) |
| 0.1 | 0.5 | GO:0032205 | negative regulation of telomere maintenance(GO:0032205) |
| 0.0 | 0.1 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.0 | 0.2 | GO:0010693 | regulation of alkaline phosphatase activity(GO:0010692) negative regulation of alkaline phosphatase activity(GO:0010693) |
| 0.0 | 0.2 | GO:0010332 | response to gamma radiation(GO:0010332) cellular response to gamma radiation(GO:0071480) |
| 0.0 | 0.5 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.0 | 0.2 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.2 | GO:0030329 | prenylated protein catabolic process(GO:0030327) prenylcysteine catabolic process(GO:0030328) prenylcysteine metabolic process(GO:0030329) |
| 0.0 | 0.2 | GO:0038093 | Fc receptor signaling pathway(GO:0038093) |
| 0.0 | 0.2 | GO:1903723 | negative regulation of centriole elongation(GO:1903723) |
| 0.0 | 0.2 | GO:0006272 | leading strand elongation(GO:0006272) |
| 0.0 | 0.6 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.0 | 0.2 | GO:0043518 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) |
| 0.0 | 0.1 | GO:0097384 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) cellular lipid biosynthetic process(GO:0097384) ether biosynthetic process(GO:1901503) |
| 0.0 | 0.2 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.3 | GO:0060118 | reflex(GO:0060004) vestibular reflex(GO:0060005) vestibular receptor cell differentiation(GO:0060114) vestibular receptor cell development(GO:0060118) |
| 0.0 | 0.1 | GO:1904983 | transmembrane glycine transport from cytosol to mitochondrion(GO:1904983) |
| 0.0 | 0.1 | GO:0072156 | distal tubule morphogenesis(GO:0072156) |
| 0.0 | 0.2 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.0 | 0.2 | GO:0035332 | positive regulation of hippo signaling(GO:0035332) |
| 0.0 | 0.1 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.0 | 0.3 | GO:0070309 | intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress(GO:0070059) lens fiber cell development(GO:0070307) lens fiber cell morphogenesis(GO:0070309) |
| 0.0 | 0.1 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.0 | 0.2 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.0 | 0.2 | GO:0097340 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.0 | 0.1 | GO:0071034 | cerebellar Purkinje cell layer structural organization(GO:0021693) cerebellar cortex structural organization(GO:0021698) CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.0 | 0.5 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.0 | 0.3 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.3 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.0 | 0.3 | GO:0031937 | methylation-dependent chromatin silencing(GO:0006346) positive regulation of chromatin silencing(GO:0031937) regulation of methylation-dependent chromatin silencing(GO:0090308) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.0 | 0.1 | GO:0048755 | branching morphogenesis of a nerve(GO:0048755) |
| 0.0 | 0.2 | GO:0031111 | negative regulation of microtubule depolymerization(GO:0007026) negative regulation of microtubule polymerization or depolymerization(GO:0031111) |
| 0.0 | 0.1 | GO:0010591 | regulation of lamellipodium assembly(GO:0010591) |
| 0.0 | 0.5 | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.0 | 0.1 | GO:0070973 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.2 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.0 | 0.1 | GO:0010801 | regulation of peptidyl-threonine phosphorylation(GO:0010799) negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.0 | 0.2 | GO:0031272 | pseudopodium organization(GO:0031268) pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.6 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.0 | 0.4 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.0 | 0.8 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.0 | 0.5 | GO:0043652 | engulfment of apoptotic cell(GO:0043652) |
| 0.0 | 0.1 | GO:0006212 | uracil catabolic process(GO:0006212) beta-alanine metabolic process(GO:0019482) beta-alanine biosynthetic process(GO:0019483) uracil metabolic process(GO:0019860) 'de novo' UMP biosynthetic process(GO:0044205) |
| 0.0 | 0.1 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.0 | 0.2 | GO:0046501 | protoporphyrinogen IX biosynthetic process(GO:0006782) protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.0 | 0.3 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.1 | GO:0036060 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.0 | 0.7 | GO:0043491 | protein kinase B signaling(GO:0043491) |
| 0.0 | 0.3 | GO:0048752 | semicircular canal morphogenesis(GO:0048752) |
| 0.0 | 0.5 | GO:0060037 | pharyngeal system development(GO:0060037) |
| 0.0 | 0.3 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 | 0.3 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 | 0.2 | GO:0070328 | acylglycerol homeostasis(GO:0055090) triglyceride homeostasis(GO:0070328) |
| 0.0 | 1.0 | GO:0031047 | gene silencing by RNA(GO:0031047) |
| 0.0 | 0.1 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0031213 | RSF complex(GO:0031213) |
| 0.1 | 0.5 | GO:0070209 | ASTRA complex(GO:0070209) |
| 0.1 | 0.3 | GO:0005592 | collagen type XI trimer(GO:0005592) |
| 0.1 | 0.6 | GO:0031332 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.1 | 0.5 | GO:0035060 | brahma complex(GO:0035060) |
| 0.1 | 0.3 | GO:0008091 | spectrin(GO:0008091) |
| 0.1 | 0.3 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 0.2 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.0 | 0.6 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.0 | 0.2 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.2 | GO:0005673 | transcription factor TFIIE complex(GO:0005673) |
| 0.0 | 0.7 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.2 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.0 | 0.2 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.0 | 0.6 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.3 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.0 | 0.5 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.2 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.0 | 0.2 | GO:0019908 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) nuclear cyclin-dependent protein kinase holoenzyme complex(GO:0019908) |
| 0.0 | 0.6 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.3 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.2 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.2 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.2 | GO:0035101 | FACT complex(GO:0035101) |
| 0.0 | 0.4 | GO:0043186 | P granule(GO:0043186) |
| 0.0 | 0.3 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 0.8 | GO:0030173 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 0.2 | GO:0070187 | telosome(GO:0070187) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.1 | 0.4 | GO:0071885 | N-terminal protein N-methyltransferase activity(GO:0071885) |
| 0.1 | 0.6 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.1 | 0.2 | GO:0072590 | N-acetyl-L-aspartate-L-glutamate ligase activity(GO:0072590) citrate-L-glutamate ligase activity(GO:0072591) |
| 0.1 | 0.4 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.1 | 0.3 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.1 | 0.5 | GO:0061731 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.1 | 0.7 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.1 | 0.3 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.1 | 0.4 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.1 | 0.7 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 0.3 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) ethanolaminephosphotransferase activity(GO:0004307) |
| 0.0 | 0.3 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.2 | GO:0001735 | prenylcysteine oxidase activity(GO:0001735) |
| 0.0 | 0.2 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.0 | 0.3 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.6 | GO:0043994 | H3 histone acetyltransferase activity(GO:0010484) histone acetyltransferase activity (H3-K23 specific)(GO:0043994) |
| 0.0 | 0.4 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.2 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 0.2 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.0 | 0.3 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.2 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.0 | 0.5 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.4 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.0 | 0.2 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 0.1 | GO:0001047 | core promoter sequence-specific DNA binding(GO:0001046) core promoter binding(GO:0001047) |
| 0.0 | 0.2 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.0 | 0.2 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.1 | GO:0016635 | oxidoreductase activity, acting on the CH-CH group of donors, quinone or related compound as acceptor(GO:0016635) |
| 0.0 | 0.2 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.1 | GO:0072518 | Rho-dependent protein serine/threonine kinase activity(GO:0072518) |
| 0.0 | 0.1 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.4 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) kinase inhibitor activity(GO:0019210) |
| 0.0 | 1.0 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.0 | 0.1 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.3 | GO:0042910 | xenobiotic transporter activity(GO:0042910) |
| 0.0 | 0.2 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 0.5 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.1 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.0 | 0.3 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.3 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.2 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.0 | 0.1 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.7 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.5 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 0.2 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.0 | 0.1 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.0 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 1.2 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.3 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.2 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 0.4 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.2 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 1.0 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.2 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.2 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.0 | 0.8 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.3 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.5 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 1.2 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.4 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.3 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.1 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.2 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.3 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.3 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.4 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.2 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.2 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.0 | 0.1 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.2 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.0 | 0.2 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.3 | REACTOME RNA POL II PRE TRANSCRIPTION EVENTS | Genes involved in RNA Polymerase II Pre-transcription Events |
| 0.0 | 0.2 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |