Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
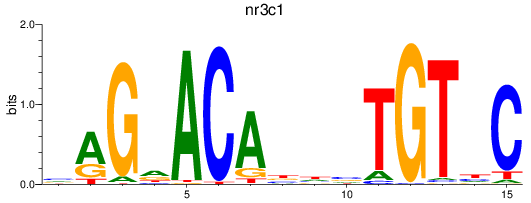
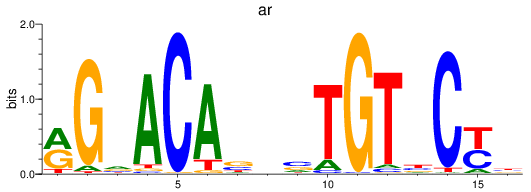
Results for nr3c1_ar
Z-value: 2.35
Transcription factors associated with nr3c1_ar
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nr3c1
|
ENSDARG00000025032 | nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
|
nr3c1
|
ENSDARG00000112480 | nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
|
nr3c1
|
ENSDARG00000116957 | nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
|
ar
|
ENSDARG00000067976 | androgen receptor |
|
ar
|
ENSDARG00000114287 | androgen receptor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nr3c1 | dr11_v1_chr14_-_23801389_23801389 | -0.85 | 4.0e-06 | Click! |
| ar | dr11_v1_chr5_+_35561607_35561607 | 0.51 | 2.4e-02 | Click! |
Activity profile of nr3c1_ar motif
Sorted Z-values of nr3c1_ar motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_41096058 | 17.94 |
ENSDART00000028373
|
fkbp5
|
FK506 binding protein 5 |
| chr6_+_41099787 | 12.86 |
ENSDART00000186884
|
fkbp5
|
FK506 binding protein 5 |
| chr12_+_13256415 | 12.41 |
ENSDART00000144542
|
atp2a1l
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 1, like |
| chr3_-_31079186 | 10.83 |
ENSDART00000145636
ENSDART00000140569 |
ELOB (1 of many)
elob
|
elongin B elongin B |
| chr17_-_37395460 | 10.20 |
ENSDART00000148160
ENSDART00000075975 |
crip1
|
cysteine-rich protein 1 |
| chr16_+_50089417 | 7.98 |
ENSDART00000153675
|
nr1d2a
|
nuclear receptor subfamily 1, group D, member 2a |
| chr24_-_38097305 | 7.78 |
ENSDART00000124321
|
crp2
|
C-reactive protein 2 |
| chr6_-_46875310 | 7.66 |
ENSDART00000154442
|
igfn1.3
|
immunoglobulin-like and fibronectin type III domain containing 1, tandem duplicate 3 |
| chr5_-_24238733 | 5.94 |
ENSDART00000138170
|
plscr3a
|
phospholipid scramblase 3a |
| chr15_-_9031996 | 5.91 |
ENSDART00000124998
|
rtn2a
|
reticulon 2a |
| chr16_+_10777116 | 5.64 |
ENSDART00000190902
|
atp1a3b
|
ATPase Na+/K+ transporting subunit alpha 3b |
| chr17_-_6730247 | 5.54 |
ENSDART00000031091
|
vsnl1b
|
visinin-like 1b |
| chr2_+_52232630 | 5.49 |
ENSDART00000006216
|
plpp2a
|
phospholipid phosphatase 2a |
| chr12_-_4683325 | 5.37 |
ENSDART00000152771
|
si:ch211-255p10.3
|
si:ch211-255p10.3 |
| chr10_+_29431529 | 5.37 |
ENSDART00000158154
|
dlg2
|
discs, large homolog 2 (Drosophila) |
| chr2_-_22688651 | 5.37 |
ENSDART00000013863
|
agxtb
|
alanine-glyoxylate aminotransferase b |
| chr5_+_9382301 | 5.33 |
ENSDART00000124017
|
ugt2a7
|
UDP glucuronosyltransferase 2 family, polypeptide A7 |
| chr16_+_10776688 | 5.27 |
ENSDART00000161969
ENSDART00000172657 |
atp1a3b
|
ATPase Na+/K+ transporting subunit alpha 3b |
| chr14_-_46228094 | 5.11 |
ENSDART00000172788
|
si:ch211-113d11.5
|
si:ch211-113d11.5 |
| chr10_-_24343507 | 5.11 |
ENSDART00000002974
|
pitpnab
|
phosphatidylinositol transfer protein, alpha b |
| chr18_-_24996634 | 4.97 |
ENSDART00000170210
|
si:ch211-196l7.4
|
si:ch211-196l7.4 |
| chr7_+_48288762 | 4.96 |
ENSDART00000083569
|
oaz2b
|
ornithine decarboxylase antizyme 2b |
| chr22_-_968484 | 4.89 |
ENSDART00000105895
|
cacna1sa
|
calcium channel, voltage-dependent, L type, alpha 1S subunit, a |
| chr20_-_791788 | 4.76 |
ENSDART00000134128
|
impg1a
|
interphotoreceptor matrix proteoglycan 1a |
| chr21_+_6290566 | 4.53 |
ENSDART00000161647
|
fnbp1b
|
formin binding protein 1b |
| chr2_-_155270 | 4.44 |
ENSDART00000131177
|
adcy1b
|
adenylate cyclase 1b |
| chr5_+_9360394 | 4.34 |
ENSDART00000124642
|
FP236810.2
|
|
| chr14_-_2355833 | 4.31 |
ENSDART00000157677
|
PCDHGC3 (1 of many)
|
si:ch73-233f7.6 |
| chr9_-_9977827 | 4.29 |
ENSDART00000187315
ENSDART00000010246 |
ugt1ab
|
UDP glucuronosyltransferase 1 family a, b |
| chr19_+_19412692 | 4.19 |
ENSDART00000113580
|
wu:fc38h03
|
wu:fc38h03 |
| chr7_+_58699718 | 4.09 |
ENSDART00000049264
|
sdr16c5b
|
short chain dehydrogenase/reductase family 16C, member 5b |
| chr9_-_9982696 | 4.08 |
ENSDART00000192548
ENSDART00000125852 |
ugt1ab
|
UDP glucuronosyltransferase 1 family a, b |
| chr1_-_26782573 | 3.89 |
ENSDART00000090611
|
sh3gl2a
|
SH3 domain containing GRB2 like 2a, endophilin A1 |
| chr14_+_32838110 | 3.65 |
ENSDART00000158077
|
arr3b
|
arrestin 3b, retinal (X-arrestin) |
| chr8_+_53452681 | 3.54 |
ENSDART00000166705
|
cacna1db
|
calcium channel, voltage-dependent, L type, alpha 1D subunit, b |
| chr11_+_6152643 | 3.48 |
ENSDART00000012789
|
slc1a6
|
solute carrier family 1 (high affinity aspartate/glutamate transporter), member 6 |
| chr2_+_38556195 | 3.45 |
ENSDART00000138769
|
cdh24b
|
cadherin 24, type 2b |
| chr3_-_5067585 | 3.43 |
ENSDART00000169609
|
tefb
|
thyrotrophic embryonic factor b |
| chr16_+_46430627 | 3.37 |
ENSDART00000127681
|
rpz6
|
rapunzel 6 |
| chr21_+_25054420 | 3.35 |
ENSDART00000065132
|
zgc:171740
|
zgc:171740 |
| chr11_+_34522554 | 3.32 |
ENSDART00000109833
|
zmat3
|
zinc finger, matrin-type 3 |
| chr7_-_32833153 | 3.15 |
ENSDART00000099871
ENSDART00000099872 |
slc17a6b
|
solute carrier family 17 (vesicular glutamate transporter), member 6b |
| chr3_-_32337653 | 3.13 |
ENSDART00000156918
ENSDART00000156551 |
si:dkey-16p21.8
|
si:dkey-16p21.8 |
| chr12_-_46959990 | 3.12 |
ENSDART00000084557
|
lhpp
|
phospholysine phosphohistidine inorganic pyrophosphate phosphatase |
| chr14_-_2036604 | 3.12 |
ENSDART00000192446
|
BX005294.2
|
|
| chr5_-_5669879 | 3.07 |
ENSDART00000191963
|
CABZ01075628.1
|
|
| chr15_-_24883956 | 2.99 |
ENSDART00000113199
|
aipl1
|
aryl hydrocarbon receptor interacting protein-like 1 |
| chr4_-_2014406 | 2.96 |
ENSDART00000180463
|
si:dkey-97m3.1
|
si:dkey-97m3.1 |
| chr9_+_22677503 | 2.95 |
ENSDART00000131429
ENSDART00000080005 ENSDART00000101756 ENSDART00000138148 |
itgb5
|
integrin, beta 5 |
| chr5_-_37116265 | 2.86 |
ENSDART00000057613
|
il13ra2
|
interleukin 13 receptor, alpha 2 |
| chr21_+_6291027 | 2.83 |
ENSDART00000180467
ENSDART00000184952 ENSDART00000184006 |
fnbp1b
|
formin binding protein 1b |
| chr4_-_38507880 | 2.82 |
ENSDART00000183225
|
si:ch211-209n20.3
|
si:ch211-209n20.3 |
| chr4_+_25651720 | 2.79 |
ENSDART00000100693
ENSDART00000100717 |
acot16
|
acyl-CoA thioesterase 16 |
| chr13_+_912123 | 2.77 |
ENSDART00000169931
|
pcsk2
|
proprotein convertase subtilisin/kexin type 2 |
| chr18_+_17428258 | 2.73 |
ENSDART00000010452
|
zgc:91860
|
zgc:91860 |
| chr20_+_54336137 | 2.72 |
ENSDART00000113792
|
cipcb
|
CLOCK-interacting pacemaker b |
| chr15_+_37559570 | 2.72 |
ENSDART00000085522
|
hspb6
|
heat shock protein, alpha-crystallin-related, b6 |
| chr21_-_30648106 | 2.67 |
ENSDART00000160800
ENSDART00000177022 |
phka1b
|
phosphorylase kinase, alpha 1b (muscle) |
| chr18_+_50278858 | 2.66 |
ENSDART00000014582
|
si:dkey-105e17.1
|
si:dkey-105e17.1 |
| chr6_-_27123327 | 2.65 |
ENSDART00000073881
|
agxta
|
alanine-glyoxylate aminotransferase a |
| chr1_+_51615672 | 2.64 |
ENSDART00000165117
|
zgc:165656
|
zgc:165656 |
| chr5_+_51594209 | 2.61 |
ENSDART00000164668
ENSDART00000058403 ENSDART00000055857 |
ckmt2b
|
creatine kinase, mitochondrial 2b (sarcomeric) |
| chr10_-_24689725 | 2.61 |
ENSDART00000079566
|
si:ch211-287a12.9
|
si:ch211-287a12.9 |
| chr21_+_1378250 | 2.60 |
ENSDART00000186912
|
TCF4
|
transcription factor 4 |
| chr20_-_9273669 | 2.56 |
ENSDART00000175069
|
syt14b
|
synaptotagmin XIVb |
| chr16_-_22781446 | 2.54 |
ENSDART00000144107
|
lenep
|
lens epithelial protein |
| chr8_+_7359294 | 2.51 |
ENSDART00000121708
|
pcsk1nl
|
proprotein convertase subtilisin/kexin type 1 inhibitor, like |
| chr10_+_375042 | 2.49 |
ENSDART00000171854
|
si:ch1073-303d10.1
|
si:ch1073-303d10.1 |
| chr18_+_8346920 | 2.49 |
ENSDART00000083421
|
cpt1b
|
carnitine palmitoyltransferase 1B (muscle) |
| chr1_-_59176949 | 2.46 |
ENSDART00000128742
|
CABZ01118678.1
|
|
| chr22_+_37874691 | 2.43 |
ENSDART00000028565
|
ahsg1
|
alpha-2-HS-glycoprotein 1 |
| chr12_+_16440708 | 2.42 |
ENSDART00000113810
|
ankrd1b
|
ankyrin repeat domain 1b (cardiac muscle) |
| chr20_+_28266892 | 2.39 |
ENSDART00000103330
|
chac1
|
ChaC, cation transport regulator homolog 1 (E. coli) |
| chr10_-_17988779 | 2.39 |
ENSDART00000132206
ENSDART00000144841 |
si:dkey-242g16.2
|
si:dkey-242g16.2 |
| chr20_-_34801181 | 2.33 |
ENSDART00000048375
ENSDART00000132426 |
stmn4
|
stathmin-like 4 |
| chr19_-_205104 | 2.31 |
ENSDART00000011890
|
zbtb22a
|
zinc finger and BTB domain containing 22a |
| chr8_-_13315567 | 2.29 |
ENSDART00000132685
ENSDART00000168635 |
kcnn1b
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 1b |
| chr5_-_23362602 | 2.29 |
ENSDART00000137120
|
gria3a
|
glutamate receptor, ionotropic, AMPA 3a |
| chr22_-_16400484 | 2.29 |
ENSDART00000135987
|
lama3
|
laminin, alpha 3 |
| chr9_+_23009608 | 2.29 |
ENSDART00000079879
|
si:dkey-91i10.3
|
si:dkey-91i10.3 |
| chr7_-_29340427 | 2.27 |
ENSDART00000052584
|
trpm1a
|
transient receptor potential cation channel, subfamily M, member 1a |
| chr20_+_39283849 | 2.27 |
ENSDART00000002481
ENSDART00000146683 |
scara3
|
scavenger receptor class A, member 3 |
| chr4_-_11163112 | 2.25 |
ENSDART00000188854
|
prmt8b
|
protein arginine methyltransferase 8b |
| chr4_-_73709502 | 2.25 |
ENSDART00000170308
|
BX855614.3
|
|
| chr19_-_32804535 | 2.23 |
ENSDART00000175613
ENSDART00000052098 |
nt5c1aa
|
5'-nucleotidase, cytosolic IAa |
| chr16_-_22225295 | 2.21 |
ENSDART00000163519
|
LO017682.1
|
|
| chr24_-_24271629 | 2.21 |
ENSDART00000135060
|
rps6ka3b
|
ribosomal protein S6 kinase, polypeptide 3b |
| chr1_+_19433004 | 2.16 |
ENSDART00000133959
|
clockb
|
clock circadian regulator b |
| chr2_+_13056802 | 2.14 |
ENSDART00000142649
|
prkag2b
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit b |
| chr9_-_22076368 | 2.13 |
ENSDART00000128486
|
crygm2a
|
crystallin, gamma M2a |
| chr25_+_10416583 | 2.10 |
ENSDART00000073907
|
ehf
|
ets homologous factor |
| chr4_-_20043484 | 2.09 |
ENSDART00000167780
|
zgc:193726
|
zgc:193726 |
| chr16_+_19677484 | 2.08 |
ENSDART00000150588
|
tmem196b
|
transmembrane protein 196b |
| chr20_-_3166168 | 2.07 |
ENSDART00000134137
|
si:ch73-212j7.3
|
si:ch73-212j7.3 |
| chr8_-_37263524 | 2.06 |
ENSDART00000061327
|
rh50
|
Rh50-like protein |
| chr5_-_41494831 | 2.04 |
ENSDART00000051081
|
eef2l2
|
eukaryotic translation elongation factor 2, like 2 |
| chr12_+_19401854 | 2.04 |
ENSDART00000153415
|
si:dkey-16i5.8
|
si:dkey-16i5.8 |
| chr6_+_40354424 | 2.03 |
ENSDART00000047416
|
slc4a8
|
solute carrier family 4, sodium bicarbonate cotransporter, member 8 |
| chr5_+_28830643 | 2.03 |
ENSDART00000051448
|
serpina7
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 7 |
| chr8_-_13315304 | 2.02 |
ENSDART00000142596
|
kcnn1b
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 1b |
| chr24_-_38110779 | 2.01 |
ENSDART00000147783
|
crp
|
c-reactive protein, pentraxin-related |
| chr2_+_25929619 | 2.01 |
ENSDART00000137746
|
slc7a14a
|
solute carrier family 7, member 14a |
| chr3_-_60571218 | 1.99 |
ENSDART00000178981
|
si:ch73-366l1.5
|
si:ch73-366l1.5 |
| chr4_-_19921410 | 1.96 |
ENSDART00000184359
ENSDART00000179965 |
BX323820.1
|
|
| chr23_-_1557195 | 1.95 |
ENSDART00000136436
|
epm2a
|
epilepsy, progressive myoclonus type 2A, Lafora disease (laforin) |
| chr10_+_41765944 | 1.92 |
ENSDART00000171484
|
rnf34b
|
ring finger protein 34b |
| chr5_-_15524782 | 1.91 |
ENSDART00000189781
|
CU694999.1
|
|
| chr8_-_50525360 | 1.90 |
ENSDART00000175648
|
CABZ01060030.1
|
|
| chr4_-_789645 | 1.90 |
ENSDART00000164441
|
mapre3b
|
microtubule-associated protein, RP/EB family, member 3b |
| chr8_+_8459192 | 1.89 |
ENSDART00000140942
ENSDART00000014939 |
comta
|
catechol-O-methyltransferase a |
| chr7_-_30087048 | 1.88 |
ENSDART00000112743
|
nmbb
|
neuromedin Bb |
| chr6_+_4255319 | 1.85 |
ENSDART00000170351
|
nbeal1
|
neurobeachin-like 1 |
| chr20_-_6812688 | 1.84 |
ENSDART00000170934
|
igfbp1a
|
insulin-like growth factor binding protein 1a |
| chr5_+_9417409 | 1.84 |
ENSDART00000125421
ENSDART00000130265 |
ugt2b5
|
UDP glucuronosyltransferase 2 family, polypeptide B5 |
| chr17_-_277046 | 1.82 |
ENSDART00000182587
|
LO018437.1
|
|
| chr7_+_18364176 | 1.80 |
ENSDART00000171606
ENSDART00000186368 |
cd248a
|
CD248 molecule, endosialin a |
| chr5_+_32206378 | 1.79 |
ENSDART00000126873
ENSDART00000051361 |
myhz2
|
myosin, heavy polypeptide 2, fast muscle specific |
| chr11_+_34523132 | 1.77 |
ENSDART00000192257
|
zmat3
|
zinc finger, matrin-type 3 |
| chr23_+_9088191 | 1.76 |
ENSDART00000030811
|
cables2b
|
Cdk5 and Abl enzyme substrate 2b |
| chr3_+_55288200 | 1.73 |
ENSDART00000157002
ENSDART00000155600 |
si:dkey-114l24.2
|
si:dkey-114l24.2 |
| chr23_+_37444933 | 1.72 |
ENSDART00000074385
|
AL954146.1
|
|
| chr17_-_45247151 | 1.70 |
ENSDART00000186230
|
ttbk2a
|
tau tubulin kinase 2a |
| chr10_-_16868211 | 1.70 |
ENSDART00000171755
|
stoml2
|
stomatin (EPB72)-like 2 |
| chr3_-_48716422 | 1.69 |
ENSDART00000164979
|
si:ch211-114m9.1
|
si:ch211-114m9.1 |
| chr7_-_18168493 | 1.68 |
ENSDART00000127428
|
peli3
|
pellino E3 ubiquitin protein ligase family member 3 |
| chr20_-_29475172 | 1.67 |
ENSDART00000183164
|
scg5
|
secretogranin V |
| chr8_-_46700278 | 1.66 |
ENSDART00000143780
|
gpr153
|
G protein-coupled receptor 153 |
| chr15_-_12011202 | 1.64 |
ENSDART00000160427
ENSDART00000168715 |
si:dkey-202l22.6
|
si:dkey-202l22.6 |
| chr21_-_131236 | 1.62 |
ENSDART00000160005
|
si:ch1073-398f15.1
|
si:ch1073-398f15.1 |
| chr10_+_34315719 | 1.60 |
ENSDART00000135303
|
stard13a
|
StAR-related lipid transfer (START) domain containing 13a |
| chr21_+_30563115 | 1.59 |
ENSDART00000028566
|
si:ch211-200p22.4
|
si:ch211-200p22.4 |
| chr3_-_35800221 | 1.58 |
ENSDART00000031390
|
caskin1
|
CASK interacting protein 1 |
| chr3_-_50124413 | 1.58 |
ENSDART00000189920
|
cldnk
|
claudin k |
| chr11_+_11175814 | 1.56 |
ENSDART00000183864
|
CR847973.1
|
|
| chr7_+_18017756 | 1.55 |
ENSDART00000173717
|
ehbp1l1a
|
EH domain binding protein 1-like 1a |
| chr11_-_19097746 | 1.55 |
ENSDART00000103973
|
rhol
|
rhodopsin, like |
| chr10_+_33754967 | 1.54 |
ENSDART00000153442
|
rxfp2a
|
relaxin/insulin-like family peptide receptor 2a |
| chr7_+_6969909 | 1.54 |
ENSDART00000189886
|
actn3b
|
actinin alpha 3b |
| chr22_+_26813917 | 1.52 |
ENSDART00000147719
|
slx4
|
SLX4 structure-specific endonuclease subunit homolog (S. cerevisiae) |
| chr17_-_32698503 | 1.47 |
ENSDART00000157175
ENSDART00000077439 |
kcnk2a
|
potassium channel, subfamily K, member 2a |
| chr6_-_8580857 | 1.47 |
ENSDART00000138858
ENSDART00000041142 |
myh11a
|
myosin, heavy chain 11a, smooth muscle |
| chr11_+_37144328 | 1.47 |
ENSDART00000162830
|
wnk2
|
WNK lysine deficient protein kinase 2 |
| chr12_-_44151296 | 1.47 |
ENSDART00000168734
|
si:ch73-329n5.3
|
si:ch73-329n5.3 |
| chr24_+_35564668 | 1.46 |
ENSDART00000122734
|
cebpd
|
CCAAT/enhancer binding protein (C/EBP), delta |
| chr4_+_19534833 | 1.46 |
ENSDART00000140028
|
lrrc4.1
|
leucine rich repeat containing 4.1 |
| chr9_+_45428041 | 1.46 |
ENSDART00000193087
|
adarb1b
|
adenosine deaminase, RNA-specific, B1b |
| chr4_-_2162688 | 1.41 |
ENSDART00000148900
|
kcnc2
|
potassium voltage-gated channel, Shaw-related subfamily, member 2 |
| chr13_+_13821054 | 1.41 |
ENSDART00000127417
|
slc4a11
|
solute carrier family 4, sodium borate transporter, member 11 |
| chr24_+_13316737 | 1.40 |
ENSDART00000191658
|
SBSPON
|
somatomedin B and thrombospondin type 1 domain containing |
| chr18_-_35736591 | 1.38 |
ENSDART00000036015
|
ryr1b
|
ryanodine receptor 1b (skeletal) |
| chr16_-_50175069 | 1.38 |
ENSDART00000192979
|
lim2.5
|
lens intrinsic membrane protein 2.5 |
| chr23_+_44732863 | 1.36 |
ENSDART00000160044
ENSDART00000172268 |
atp1b2a
|
ATPase Na+/K+ transporting subunit beta 2a |
| chr1_-_55008882 | 1.36 |
ENSDART00000083572
|
zgc:136864
|
zgc:136864 |
| chr11_-_34147205 | 1.33 |
ENSDART00000173216
|
atp13a3
|
ATPase 13A3 |
| chr18_+_17428506 | 1.32 |
ENSDART00000100223
|
zgc:91860
|
zgc:91860 |
| chr15_-_12011390 | 1.32 |
ENSDART00000187403
|
si:dkey-202l22.6
|
si:dkey-202l22.6 |
| chr15_-_23908605 | 1.30 |
ENSDART00000185578
|
usp32
|
ubiquitin specific peptidase 32 |
| chr1_-_1631399 | 1.29 |
ENSDART00000176787
|
clic6
|
chloride intracellular channel 6 |
| chr4_+_76735113 | 1.29 |
ENSDART00000075602
|
ms4a17a.6
|
membrane-spanning 4-domains, subfamily A, member 17A.6 |
| chr2_-_48396061 | 1.28 |
ENSDART00000161657
|
si:ch211-195j11.30
|
si:ch211-195j11.30 |
| chr25_-_29363934 | 1.28 |
ENSDART00000166889
|
nptna
|
neuroplastin a |
| chr23_+_4583747 | 1.28 |
ENSDART00000160350
|
LO017700.1
|
|
| chr2_+_47708853 | 1.27 |
ENSDART00000124307
|
mbnl1
|
muscleblind-like splicing regulator 1 |
| chr11_-_7050825 | 1.26 |
ENSDART00000179749
|
si:ch211-253b8.5
|
si:ch211-253b8.5 |
| chr13_-_37619159 | 1.26 |
ENSDART00000186348
|
zgc:152791
|
zgc:152791 |
| chr17_+_45737992 | 1.25 |
ENSDART00000135073
ENSDART00000143525 ENSDART00000158165 ENSDART00000184167 ENSDART00000109525 |
aspg
|
asparaginase homolog (S. cerevisiae) |
| chr10_+_37500234 | 1.24 |
ENSDART00000132096
ENSDART00000099473 |
msi2a
|
musashi RNA-binding protein 2a |
| chr12_+_32073660 | 1.22 |
ENSDART00000153245
ENSDART00000153268 |
stxbp4
|
syntaxin binding protein 4 |
| chr24_-_9960290 | 1.21 |
ENSDART00000143390
ENSDART00000092975 ENSDART00000184953 |
vps41
|
vacuolar protein sorting 41 homolog (S. cerevisiae) |
| chr9_+_42157578 | 1.18 |
ENSDART00000142888
|
lrrc3
|
leucine rich repeat containing 3 |
| chr5_+_32162684 | 1.17 |
ENSDART00000134472
|
taok3b
|
TAO kinase 3b |
| chr17_-_45552602 | 1.16 |
ENSDART00000154844
ENSDART00000034432 |
susd4
|
sushi domain containing 4 |
| chr2_+_30144979 | 1.16 |
ENSDART00000129365
ENSDART00000130344 |
kcnb2
|
potassium voltage-gated channel, Shab-related subfamily, member 2 |
| chr5_+_70345518 | 1.15 |
ENSDART00000167029
|
pappaa
|
pregnancy-associated plasma protein A, pappalysin 1a |
| chr3_+_20012891 | 1.15 |
ENSDART00000078974
|
tmub2
|
transmembrane and ubiquitin-like domain containing 2 |
| chr3_-_36440705 | 1.14 |
ENSDART00000162875
|
rogdi
|
rogdi homolog (Drosophila) |
| chr5_+_28830388 | 1.14 |
ENSDART00000149150
|
serpina7
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 7 |
| chr7_+_53152108 | 1.14 |
ENSDART00000171350
|
cdh29
|
cadherin 29 |
| chr20_+_34915945 | 1.14 |
ENSDART00000153064
|
snap25a
|
synaptosomal-associated protein, 25a |
| chr17_+_29345606 | 1.14 |
ENSDART00000086164
|
kctd3
|
potassium channel tetramerization domain containing 3 |
| chr13_+_4505232 | 1.13 |
ENSDART00000007500
ENSDART00000161684 |
pde10a
|
phosphodiesterase 10A |
| chr3_+_53228684 | 1.13 |
ENSDART00000156490
|
pet100
|
PET100 homolog |
| chr23_+_23183449 | 1.13 |
ENSDART00000132296
|
klhl17
|
kelch-like family member 17 |
| chr17_+_23578316 | 1.13 |
ENSDART00000149281
|
slc16a12a
|
solute carrier family 16, member 12a |
| chr5_+_65536095 | 1.12 |
ENSDART00000189898
|
si:dkey-21e5.1
|
si:dkey-21e5.1 |
| chr8_-_45835056 | 1.12 |
ENSDART00000022242
|
ogdha
|
oxoglutarate (alpha-ketoglutarate) dehydrogenase a (lipoamide) |
| chr25_+_19149241 | 1.12 |
ENSDART00000184982
ENSDART00000067324 |
mfge8b
|
milk fat globule-EGF factor 8 protein b |
| chr10_+_26667475 | 1.12 |
ENSDART00000133281
ENSDART00000147013 |
si:ch73-52f15.5
|
si:ch73-52f15.5 |
| chr1_+_32521469 | 1.11 |
ENSDART00000113818
ENSDART00000152580 |
nlgn4a
|
neuroligin 4a |
| chr17_+_6276559 | 1.11 |
ENSDART00000131075
|
dusp23b
|
dual specificity phosphatase 23b |
| chr19_-_41213718 | 1.09 |
ENSDART00000077121
|
pdk4
|
pyruvate dehydrogenase kinase, isozyme 4 |
| chr8_+_31777633 | 1.08 |
ENSDART00000142519
|
oxct1a
|
3-oxoacid CoA transferase 1a |
| chr1_-_48922273 | 1.08 |
ENSDART00000150254
|
si:ch211-112b1.2
|
si:ch211-112b1.2 |
| chr3_-_35602233 | 1.07 |
ENSDART00000055269
|
gng13b
|
guanine nucleotide binding protein (G protein), gamma 13b |
| chr23_+_6752828 | 1.06 |
ENSDART00000105179
|
zgc:158254
|
zgc:158254 |
| chr7_-_19168375 | 1.06 |
ENSDART00000112447
|
il13ra1
|
interleukin 13 receptor, alpha 1 |
| chr13_-_11378355 | 1.06 |
ENSDART00000164566
|
akt3a
|
v-akt murine thymoma viral oncogene homolog 3a |
| chr9_-_21918963 | 1.05 |
ENSDART00000090782
|
lmo7a
|
LIM domain 7a |
| chr5_-_3627110 | 1.05 |
ENSDART00000156071
|
si:zfos-375h5.1
|
si:zfos-375h5.1 |
| chr3_-_58543658 | 1.03 |
ENSDART00000042386
|
unm_sa1261
|
un-named sa1261 |
Network of associatons between targets according to the STRING database.
First level regulatory network of nr3c1_ar
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.4 | 10.2 | GO:0071236 | cellular response to antibiotic(GO:0071236) cellular response to toxic substance(GO:0097237) |
| 1.3 | 8.0 | GO:0006545 | glycine biosynthetic process(GO:0006545) |
| 1.0 | 30.8 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.8 | 5.3 | GO:2001270 | negative regulation of execution phase of apoptosis(GO:1900118) regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001270) negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.7 | 2.7 | GO:0042754 | negative regulation of circadian rhythm(GO:0042754) |
| 0.6 | 12.3 | GO:0030007 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.5 | 2.3 | GO:0060401 | cytosolic calcium ion transport(GO:0060401) calcium ion transport into cytosol(GO:0060402) |
| 0.4 | 1.7 | GO:0099563 | modification of synaptic structure(GO:0099563) |
| 0.4 | 8.5 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.4 | 2.0 | GO:0010754 | negative regulation of cGMP-mediated signaling(GO:0010754) |
| 0.4 | 3.1 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.4 | 1.2 | GO:0006957 | complement activation, alternative pathway(GO:0006957) regulation of protein activation cascade(GO:2000257) |
| 0.4 | 1.5 | GO:0061010 | gall bladder development(GO:0061010) |
| 0.3 | 1.7 | GO:0008592 | regulation of Toll signaling pathway(GO:0008592) |
| 0.3 | 4.4 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.3 | 1.4 | GO:0031443 | fast-twitch skeletal muscle fiber contraction(GO:0031443) |
| 0.3 | 1.9 | GO:0019614 | catechol-containing compound catabolic process(GO:0019614) catecholamine catabolic process(GO:0042424) |
| 0.3 | 2.4 | GO:0010955 | negative regulation of protein processing(GO:0010955) negative regulation of protein maturation(GO:1903318) |
| 0.2 | 5.4 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.2 | 11.0 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.2 | 3.1 | GO:0006603 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.2 | 0.9 | GO:2000171 | negative regulation of dendrite development(GO:2000171) |
| 0.2 | 1.1 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.2 | 2.6 | GO:0002031 | G-protein coupled receptor internalization(GO:0002031) |
| 0.2 | 1.9 | GO:0035372 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.2 | 3.0 | GO:0048009 | insulin-like growth factor receptor signaling pathway(GO:0048009) |
| 0.2 | 10.3 | GO:0006368 | transcription elongation from RNA polymerase II promoter(GO:0006368) |
| 0.2 | 0.8 | GO:0050999 | regulation of nitric-oxide synthase activity(GO:0050999) positive regulation of nitric-oxide synthase activity(GO:0051000) beta-amyloid clearance(GO:0097242) regulation of beta-amyloid clearance(GO:1900221) |
| 0.2 | 0.6 | GO:0001562 | response to protozoan(GO:0001562) defense response to protozoan(GO:0042832) |
| 0.2 | 2.8 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.2 | 5.9 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.2 | 4.2 | GO:0006595 | polyamine metabolic process(GO:0006595) |
| 0.2 | 2.2 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.2 | 1.5 | GO:0030859 | polarized epithelial cell differentiation(GO:0030859) |
| 0.2 | 0.5 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.2 | 0.8 | GO:0014005 | microglia development(GO:0014005) |
| 0.2 | 1.2 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.1 | 3.2 | GO:0005979 | regulation of glycogen biosynthetic process(GO:0005979) regulation of glucan biosynthetic process(GO:0010962) |
| 0.1 | 2.5 | GO:0009437 | carnitine metabolic process(GO:0009437) |
| 0.1 | 2.2 | GO:0018216 | peptidyl-arginine methylation(GO:0018216) |
| 0.1 | 0.4 | GO:0006097 | glyoxylate cycle(GO:0006097) |
| 0.1 | 1.5 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.1 | 1.5 | GO:0007190 | activation of adenylate cyclase activity(GO:0007190) positive regulation of cAMP metabolic process(GO:0030816) positive regulation of cAMP biosynthetic process(GO:0030819) positive regulation of adenylate cyclase activity(GO:0045762) |
| 0.1 | 0.5 | GO:0097037 | heme export(GO:0097037) |
| 0.1 | 1.9 | GO:0046887 | positive regulation of hormone secretion(GO:0046887) |
| 0.1 | 0.2 | GO:2000108 | positive regulation of leukocyte apoptotic process(GO:2000108) |
| 0.1 | 3.6 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.1 | 6.9 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.1 | 0.7 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.1 | 0.8 | GO:0030431 | sleep(GO:0030431) |
| 0.1 | 0.5 | GO:0016081 | synaptic vesicle docking(GO:0016081) presynaptic dense core granule exocytosis(GO:0099525) |
| 0.1 | 1.1 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.1 | 22.3 | GO:0070588 | calcium ion transmembrane transport(GO:0070588) |
| 0.1 | 2.0 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.1 | 0.5 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.1 | 2.6 | GO:0046883 | regulation of hormone secretion(GO:0046883) |
| 0.1 | 0.3 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.1 | 1.1 | GO:0097034 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.1 | 1.9 | GO:0016082 | synaptic vesicle priming(GO:0016082) |
| 0.1 | 0.3 | GO:0003228 | atrial cardiac muscle tissue development(GO:0003228) |
| 0.1 | 4.8 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.1 | 1.5 | GO:0014823 | response to activity(GO:0014823) |
| 0.1 | 0.6 | GO:0031063 | regulation of histone deacetylation(GO:0031063) |
| 0.1 | 0.5 | GO:0045905 | positive regulation of translational termination(GO:0045905) |
| 0.1 | 2.4 | GO:0033627 | cell adhesion mediated by integrin(GO:0033627) |
| 0.1 | 1.5 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.1 | 1.6 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.1 | 0.3 | GO:1903405 | nuclear body organization(GO:0030575) Cajal body organization(GO:0030576) protein localization to nuclear body(GO:1903405) protein localization to Cajal body(GO:1904867) protein localization to nucleoplasm(GO:1990173) |
| 0.1 | 1.5 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.1 | 2.1 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.1 | 4.5 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.1 | 1.1 | GO:0019471 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) |
| 0.1 | 1.6 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 0.1 | 0.4 | GO:0050957 | neuromuscular process controlling balance(GO:0050885) equilibrioception(GO:0050957) |
| 0.0 | 0.7 | GO:0099645 | protein localization to postsynaptic specialization membrane(GO:0099633) neurotransmitter receptor localization to postsynaptic specialization membrane(GO:0099645) |
| 0.0 | 0.8 | GO:0010765 | positive regulation of sodium ion transport(GO:0010765) positive regulation of sodium ion transmembrane transport(GO:1902307) regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.0 | 0.3 | GO:0010165 | response to X-ray(GO:0010165) |
| 0.0 | 0.2 | GO:0048210 | Golgi vesicle fusion to target membrane(GO:0048210) |
| 0.0 | 0.7 | GO:0048016 | calcineurin-NFAT signaling cascade(GO:0033173) inositol phosphate-mediated signaling(GO:0048016) |
| 0.0 | 1.1 | GO:0010906 | regulation of glucose metabolic process(GO:0010906) |
| 0.0 | 2.1 | GO:0044042 | glycogen metabolic process(GO:0005977) cellular glucan metabolic process(GO:0006073) glucan metabolic process(GO:0044042) |
| 0.0 | 0.3 | GO:0014829 | vascular smooth muscle contraction(GO:0014829) |
| 0.0 | 9.6 | GO:0007601 | visual perception(GO:0007601) |
| 0.0 | 0.5 | GO:0097369 | sodium ion import(GO:0097369) inorganic cation import into cell(GO:0098659) sodium ion import across plasma membrane(GO:0098719) inorganic ion import into cell(GO:0099587) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.3 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.0 | 0.6 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.0 | 2.0 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.0 | 0.5 | GO:0035094 | response to nicotine(GO:0035094) |
| 0.0 | 2.0 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 1.1 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.0 | 8.1 | GO:0009617 | response to bacterium(GO:0009617) |
| 0.0 | 0.5 | GO:1901222 | regulation of NIK/NF-kappaB signaling(GO:1901222) |
| 0.0 | 1.6 | GO:0043297 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.0 | 5.7 | GO:0071805 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.0 | 0.3 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.0 | 0.1 | GO:0007414 | axonal defasciculation(GO:0007414) |
| 0.0 | 0.9 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
| 0.0 | 0.3 | GO:0071850 | mitotic cell cycle arrest(GO:0071850) |
| 0.0 | 0.2 | GO:0044209 | AMP salvage(GO:0044209) |
| 0.0 | 2.3 | GO:0016358 | dendrite development(GO:0016358) |
| 0.0 | 1.0 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.0 | 0.4 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.5 | GO:0006144 | purine nucleobase metabolic process(GO:0006144) |
| 0.0 | 1.7 | GO:0050673 | epithelial cell proliferation(GO:0050673) |
| 0.0 | 0.1 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 0.0 | 0.3 | GO:0099560 | synaptic membrane adhesion(GO:0099560) |
| 0.0 | 0.8 | GO:0046330 | positive regulation of JNK cascade(GO:0046330) |
| 0.0 | 0.4 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.0 | 6.5 | GO:0043062 | extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |
| 0.0 | 0.4 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.0 | 0.4 | GO:0071482 | cellular response to light stimulus(GO:0071482) |
| 0.0 | 0.4 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 1.1 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.0 | 0.3 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.1 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.0 | 0.4 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.0 | 0.9 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 1.3 | GO:0034765 | regulation of ion transmembrane transport(GO:0034765) |
| 0.0 | 3.9 | GO:0006470 | protein dephosphorylation(GO:0006470) |
| 0.0 | 1.1 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.0 | 1.6 | GO:0001503 | ossification(GO:0001503) |
| 0.0 | 1.3 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 0.6 | GO:0006469 | negative regulation of protein kinase activity(GO:0006469) |
| 0.0 | 0.3 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 | 0.6 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.0 | 0.1 | GO:0001993 | regulation of systemic arterial blood pressure by norepinephrine-epinephrine(GO:0001993) |
| 0.0 | 0.9 | GO:0006694 | steroid biosynthetic process(GO:0006694) |
| 0.0 | 0.9 | GO:0031018 | endocrine pancreas development(GO:0031018) |
| 0.0 | 2.1 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.2 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.1 | GO:0046633 | negative regulation of interferon-gamma production(GO:0032689) CD4-positive, alpha-beta T cell proliferation(GO:0035739) alpha-beta T cell proliferation(GO:0046633) regulation of alpha-beta T cell activation(GO:0046634) negative regulation of alpha-beta T cell activation(GO:0046636) regulation of alpha-beta T cell proliferation(GO:0046640) negative regulation of alpha-beta T cell proliferation(GO:0046642) regulation of CD4-positive, alpha-beta T cell activation(GO:2000514) negative regulation of CD4-positive, alpha-beta T cell activation(GO:2000515) regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000561) negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 10.8 | GO:0030891 | VCB complex(GO:0030891) |
| 0.7 | 2.2 | GO:0098753 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) anchored component of the cytoplasmic side of the plasma membrane(GO:0098753) |
| 0.6 | 4.8 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.6 | 13.8 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.4 | 2.2 | GO:1990513 | CLOCK-BMAL transcription complex(GO:1990513) |
| 0.3 | 1.1 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.3 | 1.5 | GO:0033557 | Slx1-Slx4 complex(GO:0033557) |
| 0.2 | 6.7 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.2 | 8.9 | GO:0031970 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.2 | 3.1 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.2 | 0.8 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.2 | 1.1 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.2 | 2.1 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.2 | 2.0 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.2 | 0.5 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.1 | 1.6 | GO:0098888 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) extrinsic component of presynaptic membrane(GO:0098888) extrinsic component of presynaptic endocytic zone(GO:0098894) |
| 0.1 | 2.4 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 1.4 | GO:0090533 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) |
| 0.1 | 0.5 | GO:0005915 | cell-cell adherens junction(GO:0005913) zonula adherens(GO:0005915) |
| 0.1 | 8.4 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.1 | 1.1 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.1 | 10.6 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.1 | 2.6 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.1 | 4.7 | GO:0016342 | catenin complex(GO:0016342) |
| 0.1 | 1.4 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.1 | 3.4 | GO:0008305 | integrin complex(GO:0008305) |
| 0.1 | 1.9 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 0.5 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.1 | 1.6 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.1 | 0.8 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.1 | 2.3 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 0.8 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.9 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 1.7 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.0 | 0.3 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.0 | 4.7 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.0 | 1.2 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 0.4 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.0 | 1.8 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 1.5 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.6 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 1.5 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.6 | GO:0016282 | eukaryotic 43S preinitiation complex(GO:0016282) |
| 0.0 | 1.9 | GO:0030141 | secretory granule(GO:0030141) |
| 0.0 | 2.6 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 2.1 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 7.3 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 0.4 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.4 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 4.7 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 1.5 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.3 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.1 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.0 | 1.1 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.1 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.0 | 0.1 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.4 | GO:0098978 | glutamatergic synapse(GO:0098978) |
| 0.0 | 0.1 | GO:0031526 | brush border membrane(GO:0031526) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.7 | 30.8 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 1.7 | 8.5 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 1.2 | 5.0 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 1.1 | 10.9 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.8 | 3.1 | GO:0101006 | inorganic diphosphatase activity(GO:0004427) protein histidine phosphatase activity(GO:0101006) |
| 0.7 | 12.4 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.7 | 5.1 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.6 | 2.4 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.6 | 1.8 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.6 | 2.4 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.5 | 5.9 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.5 | 4.3 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.5 | 1.9 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.4 | 3.0 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.4 | 2.0 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.4 | 2.0 | GO:2001070 | starch binding(GO:2001070) |
| 0.4 | 2.5 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.3 | 2.2 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 0.3 | 2.2 | GO:0070888 | E-box binding(GO:0070888) |
| 0.3 | 16.7 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.3 | 8.2 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.3 | 8.4 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.3 | 1.4 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) calcium-induced calcium release activity(GO:0048763) |
| 0.3 | 1.9 | GO:0031705 | bombesin receptor binding(GO:0031705) |
| 0.2 | 3.1 | GO:0005326 | neurotransmitter transporter activity(GO:0005326) |
| 0.2 | 3.1 | GO:0016775 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.2 | 1.1 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 0.2 | 10.0 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.2 | 2.2 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.2 | 2.6 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.2 | 4.5 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.2 | 2.4 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.2 | 1.8 | GO:0031995 | insulin-like growth factor I binding(GO:0031994) insulin-like growth factor II binding(GO:0031995) |
| 0.2 | 0.5 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.2 | 4.4 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.2 | 0.5 | GO:0016657 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.2 | 3.4 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.2 | 1.3 | GO:0004067 | asparaginase activity(GO:0004067) |
| 0.2 | 1.5 | GO:0016918 | retinal binding(GO:0016918) |
| 0.2 | 2.3 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.1 | 1.1 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.1 | 0.6 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.1 | 0.8 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.1 | 0.8 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.1 | 1.1 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.1 | 1.5 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.1 | 0.4 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) |
| 0.1 | 1.6 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 2.2 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.1 | 0.9 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.1 | 1.5 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.1 | 2.3 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.1 | 0.5 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.1 | 1.1 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.1 | 0.5 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.1 | 9.4 | GO:0008201 | heparin binding(GO:0008201) |
| 0.1 | 0.4 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.1 | 0.7 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.1 | 1.1 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.1 | 0.5 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.1 | 0.5 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.1 | 2.1 | GO:0016208 | AMP binding(GO:0016208) |
| 0.1 | 0.5 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.1 | 0.4 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.1 | 0.6 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 1.4 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.1 | 1.5 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.1 | 0.9 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.1 | 2.4 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.1 | 7.3 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) |
| 0.1 | 2.8 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.1 | 0.3 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.1 | 0.5 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.1 | 3.0 | GO:0005178 | integrin binding(GO:0005178) |
| 0.1 | 0.8 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.1 | 1.9 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 1.1 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.1 | 0.6 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.8 | GO:0019870 | chloride channel inhibitor activity(GO:0019869) potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 1.6 | GO:0098632 | protein binding involved in cell adhesion(GO:0098631) protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 2.2 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.6 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 4.7 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 8.2 | GO:0042277 | peptide binding(GO:0042277) |
| 0.0 | 2.0 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.5 | GO:0015385 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.3 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.0 | 0.4 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 3.2 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.4 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.0 | 3.9 | GO:0019955 | cytokine binding(GO:0019955) |
| 0.0 | 0.4 | GO:0008320 | protein transmembrane transporter activity(GO:0008320) |
| 0.0 | 0.6 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.2 | GO:0004001 | adenosine kinase activity(GO:0004001) |
| 0.0 | 1.1 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.3 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.1 | GO:0051380 | norepinephrine binding(GO:0051380) |
| 0.0 | 0.3 | GO:0031013 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.0 | 0.2 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.2 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 3.5 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 3.1 | GO:0015293 | symporter activity(GO:0015293) |
| 0.0 | 0.7 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 2.3 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.5 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 1.1 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 2.3 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.0 | 0.8 | GO:0008373 | sialyltransferase activity(GO:0008373) |
| 0.0 | 0.7 | GO:0019902 | phosphatase binding(GO:0019902) |
| 0.0 | 2.4 | GO:0004497 | monooxygenase activity(GO:0004497) |
| 0.0 | 0.3 | GO:0042805 | actinin binding(GO:0042805) |
| 0.0 | 0.8 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.2 | GO:0072542 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.0 | 2.6 | GO:0001664 | G-protein coupled receptor binding(GO:0001664) |
| 0.0 | 0.8 | GO:0016627 | oxidoreductase activity, acting on the CH-CH group of donors(GO:0016627) |
| 0.0 | 0.3 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 3.9 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.4 | 31.6 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.3 | 3.0 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.2 | 2.3 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 3.3 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.1 | 2.5 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 1.2 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.1 | 0.4 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 2.9 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.7 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 0.6 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.5 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.2 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.3 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 1.5 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 3.5 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.2 | 2.0 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.2 | 0.9 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.2 | 2.5 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.1 | 2.8 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.1 | 3.0 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 2.0 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.1 | 1.1 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 2.6 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 0.1 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.1 | 1.6 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 0.6 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 2.7 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.0 | 0.5 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 1.3 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.8 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.4 | REACTOME ACTIVATED POINT MUTANTS OF FGFR2 | Genes involved in Activated point mutants of FGFR2 |
| 0.0 | 0.6 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.0 | 0.2 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 1.6 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.4 | REACTOME PYRUVATE METABOLISM AND CITRIC ACID TCA CYCLE | Genes involved in Pyruvate metabolism and Citric Acid (TCA) cycle |