Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
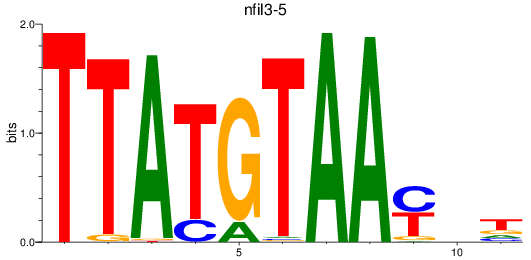
Results for nfil3-5
Z-value: 2.16
Transcription factors associated with nfil3-5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nfil3-5
|
ENSDARG00000094965 | nuclear factor, interleukin 3 regulated, member 5 |
|
nfil3-5
|
ENSDARG00000113345 | nuclear factor, interleukin 3 regulated, member 5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nfil3-5 | dr11_v1_chr6_+_8172227_8172227 | 0.36 | 1.3e-01 | Click! |
Activity profile of nfil3-5 motif
Sorted Z-values of nfil3-5 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_-_44953349 | 9.29 |
ENSDART00000135156
|
vil1
|
villin 1 |
| chr20_-_1151265 | 9.11 |
ENSDART00000012376
|
gabrr1
|
gamma-aminobutyric acid (GABA) A receptor, rho 1 |
| chr17_+_27456804 | 9.06 |
ENSDART00000017756
ENSDART00000181461 ENSDART00000180178 |
ctsl.1
|
cathepsin L.1 |
| chr25_+_34984333 | 8.34 |
ENSDART00000154760
|
ccdc136b
|
coiled-coil domain containing 136b |
| chr3_+_16612574 | 7.45 |
ENSDART00000104481
|
slc17a7a
|
solute carrier family 17 (vesicular glutamate transporter), member 7a |
| chr9_-_48736388 | 7.19 |
ENSDART00000022074
|
dhrs9
|
dehydrogenase/reductase (SDR family) member 9 |
| chr6_-_345503 | 7.13 |
ENSDART00000168901
|
pde6ha
|
phosphodiesterase 6H, cGMP-specific, cone, gamma, paralog a |
| chr5_+_37837245 | 7.11 |
ENSDART00000171617
|
epd
|
ependymin |
| chr25_+_15354095 | 6.70 |
ENSDART00000090397
|
kiaa1549la
|
KIAA1549-like a |
| chr2_+_24177190 | 6.45 |
ENSDART00000099546
|
map4l
|
microtubule associated protein 4 like |
| chr21_-_22115136 | 6.36 |
ENSDART00000134715
ENSDART00000089246 ENSDART00000139789 |
elmod1
|
ELMO/CED-12 domain containing 1 |
| chr20_-_25533739 | 6.23 |
ENSDART00000063064
|
cyp2ad6
|
cytochrome P450, family 2, subfamily AD, polypeptide 6 |
| chr22_+_20720808 | 6.21 |
ENSDART00000171321
|
si:dkey-211f22.5
|
si:dkey-211f22.5 |
| chr5_+_51594209 | 6.20 |
ENSDART00000164668
ENSDART00000058403 ENSDART00000055857 |
ckmt2b
|
creatine kinase, mitochondrial 2b (sarcomeric) |
| chr5_-_41494831 | 6.12 |
ENSDART00000051081
|
eef2l2
|
eukaryotic translation elongation factor 2, like 2 |
| chr21_-_43550120 | 5.96 |
ENSDART00000151627
|
si:ch73-362m14.2
|
si:ch73-362m14.2 |
| chr3_+_15296824 | 5.76 |
ENSDART00000043801
|
cabp5b
|
calcium binding protein 5b |
| chr5_+_51597677 | 5.70 |
ENSDART00000048210
ENSDART00000184797 |
ckmt2b
|
creatine kinase, mitochondrial 2b (sarcomeric) |
| chr20_+_1121458 | 5.62 |
ENSDART00000064472
|
pnrc1
|
proline-rich nuclear receptor coactivator 1 |
| chr2_+_10134345 | 5.60 |
ENSDART00000100725
|
ahsg2
|
alpha-2-HS-glycoprotein 2 |
| chr21_-_22114625 | 5.57 |
ENSDART00000177426
ENSDART00000135410 |
elmod1
|
ELMO/CED-12 domain containing 1 |
| chr20_-_44576949 | 5.53 |
ENSDART00000148639
|
ubxn2a
|
UBX domain protein 2A |
| chr7_-_26457208 | 5.31 |
ENSDART00000173519
|
zgc:172079
|
zgc:172079 |
| chr12_+_15002757 | 5.21 |
ENSDART00000135036
|
mylpfb
|
myosin light chain, phosphorylatable, fast skeletal muscle b |
| chr9_-_44953664 | 5.05 |
ENSDART00000188558
ENSDART00000185210 |
vil1
|
villin 1 |
| chr10_-_24371312 | 4.96 |
ENSDART00000149362
|
pitpnab
|
phosphatidylinositol transfer protein, alpha b |
| chr5_+_4332220 | 4.90 |
ENSDART00000051699
|
sat1a.1
|
spermidine/spermine N1-acetyltransferase 1a, duplicate 1 |
| chr19_-_20093341 | 4.78 |
ENSDART00000129917
|
mpp6b
|
membrane protein, palmitoylated 6b (MAGUK p55 subfamily member 6) |
| chr16_-_45069882 | 4.70 |
ENSDART00000058384
|
gapdhs
|
glyceraldehyde-3-phosphate dehydrogenase, spermatogenic |
| chr20_-_52939501 | 4.56 |
ENSDART00000166508
|
fdft1
|
farnesyl-diphosphate farnesyltransferase 1 |
| chr11_-_10770053 | 4.56 |
ENSDART00000179213
|
slc4a10a
|
solute carrier family 4, sodium bicarbonate transporter, member 10a |
| chr12_-_15205087 | 4.53 |
ENSDART00000010068
|
sult1st6
|
sulfotransferase family 1, cytosolic sulfotransferase 6 |
| chr8_-_39903803 | 4.51 |
ENSDART00000012391
|
cabp1a
|
calcium binding protein 1a |
| chr15_+_37559570 | 4.48 |
ENSDART00000085522
|
hspb6
|
heat shock protein, alpha-crystallin-related, b6 |
| chr5_+_1278092 | 4.48 |
ENSDART00000147972
ENSDART00000159783 |
dnm1a
|
dynamin 1a |
| chr17_-_20979077 | 4.45 |
ENSDART00000006676
|
phyhipla
|
phytanoyl-CoA 2-hydroxylase interacting protein-like a |
| chr8_-_32497815 | 4.33 |
ENSDART00000122359
|
si:dkey-164f24.2
|
si:dkey-164f24.2 |
| chr18_-_898870 | 4.32 |
ENSDART00000151777
ENSDART00000062654 |
parp6a
|
poly (ADP-ribose) polymerase family, member 6a |
| chr1_-_8101495 | 4.28 |
ENSDART00000161938
|
si:dkeyp-9d4.3
|
si:dkeyp-9d4.3 |
| chr14_+_4151379 | 4.22 |
ENSDART00000160431
|
dhrs13l1
|
dehydrogenase/reductase (SDR family) member 13 like 1 |
| chr3_+_23092762 | 4.16 |
ENSDART00000142884
ENSDART00000024136 |
gngt2a
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 2a |
| chr1_-_14233815 | 4.14 |
ENSDART00000044896
|
camk2d2
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II delta 2 |
| chr21_-_42007213 | 4.14 |
ENSDART00000188804
ENSDART00000092821 ENSDART00000165743 |
gabrg2
|
gamma-aminobutyric acid (GABA) A receptor, gamma 2 |
| chr7_+_38717624 | 4.12 |
ENSDART00000132522
|
syt13
|
synaptotagmin XIII |
| chr7_-_6604623 | 4.11 |
ENSDART00000172874
|
kcnj10a
|
potassium inwardly-rectifying channel, subfamily J, member 10a |
| chr8_-_37263524 | 4.04 |
ENSDART00000061327
|
rh50
|
Rh50-like protein |
| chr2_+_38039857 | 4.04 |
ENSDART00000159951
|
casq1a
|
calsequestrin 1a |
| chr17_+_15845765 | 4.03 |
ENSDART00000130881
ENSDART00000074936 |
gabrr2a
|
gamma-aminobutyric acid (GABA) A receptor, rho 2a |
| chr19_-_657439 | 3.98 |
ENSDART00000167100
|
slc6a18
|
solute carrier family 6 (neutral amino acid transporter), member 18 |
| chr22_-_354592 | 3.96 |
ENSDART00000155769
|
tmem240b
|
transmembrane protein 240b |
| chr16_+_46111849 | 3.92 |
ENSDART00000172232
|
sv2a
|
synaptic vesicle glycoprotein 2A |
| chr9_-_18742704 | 3.91 |
ENSDART00000145401
|
tsc22d1
|
TSC22 domain family, member 1 |
| chr6_-_37403444 | 3.87 |
ENSDART00000136314
|
cthl
|
cystathionase (cystathionine gamma-lyase), like |
| chr23_+_19590006 | 3.81 |
ENSDART00000021231
|
slmapb
|
sarcolemma associated protein b |
| chr23_+_37323962 | 3.78 |
ENSDART00000102881
|
fam43b
|
family with sequence similarity 43, member B |
| chr19_+_4990320 | 3.77 |
ENSDART00000147056
|
zgc:91968
|
zgc:91968 |
| chr17_-_23709347 | 3.74 |
ENSDART00000124661
|
papss2a
|
3'-phosphoadenosine 5'-phosphosulfate synthase 2a |
| chr19_+_5674907 | 3.73 |
ENSDART00000042189
|
pdk2b
|
pyruvate dehydrogenase kinase, isozyme 2b |
| chr14_-_21219659 | 3.72 |
ENSDART00000089867
|
ppp2r2cb
|
protein phosphatase 2, regulatory subunit B, gamma b |
| chr19_+_4990496 | 3.71 |
ENSDART00000151050
ENSDART00000017535 |
zgc:91968
|
zgc:91968 |
| chr21_+_43178831 | 3.65 |
ENSDART00000151512
|
aff4
|
AF4/FMR2 family, member 4 |
| chr23_+_45579497 | 3.64 |
ENSDART00000110381
|
egr4
|
early growth response 4 |
| chr5_+_62340799 | 3.62 |
ENSDART00000074117
|
aspa
|
aspartoacylase |
| chr8_+_8459192 | 3.60 |
ENSDART00000140942
ENSDART00000014939 |
comta
|
catechol-O-methyltransferase a |
| chr11_+_714386 | 3.57 |
ENSDART00000167824
ENSDART00000187110 |
timp4.3
|
TIMP metallopeptidase inhibitor 4, tandem duplicate 3 |
| chr21_-_42202792 | 3.57 |
ENSDART00000124708
|
gabra6b
|
gamma-aminobutyric acid (GABA) A receptor, alpha 6b |
| chr1_+_11881559 | 3.54 |
ENSDART00000166981
|
snx8b
|
sorting nexin 8b |
| chr18_-_7539166 | 3.54 |
ENSDART00000133541
|
si:dkey-30c15.2
|
si:dkey-30c15.2 |
| chr13_+_27314795 | 3.53 |
ENSDART00000128726
|
eef1a1a
|
eukaryotic translation elongation factor 1 alpha 1a |
| chr17_-_14613711 | 3.53 |
ENSDART00000157345
|
sdsl
|
serine dehydratase-like |
| chr11_+_3585934 | 3.52 |
ENSDART00000055694
|
cdab
|
cytidine deaminase b |
| chr3_+_7763114 | 3.50 |
ENSDART00000057434
|
hook2
|
hook microtubule-tethering protein 2 |
| chr23_+_33963619 | 3.48 |
ENSDART00000140666
ENSDART00000084792 |
plpbp
|
pyridoxal phosphate binding protein |
| chr24_-_17023392 | 3.47 |
ENSDART00000106058
|
ptgdsb.2
|
prostaglandin D2 synthase b, tandem duplicate 2 |
| chr6_-_13187168 | 3.46 |
ENSDART00000193286
ENSDART00000188350 ENSDART00000150036 ENSDART00000149940 |
adam23a
|
ADAM metallopeptidase domain 23a |
| chr1_-_14234076 | 3.41 |
ENSDART00000040049
|
camk2d2
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II delta 2 |
| chr16_+_39146696 | 3.40 |
ENSDART00000121756
ENSDART00000084381 |
sybu
|
syntabulin (syntaxin-interacting) |
| chr6_-_43449013 | 3.36 |
ENSDART00000122423
|
eevs
|
2-epi-5-epi-valiolone synthase |
| chr24_-_38079261 | 3.30 |
ENSDART00000105662
|
crp1
|
C-reactive protein 1 |
| chr10_-_17284055 | 3.30 |
ENSDART00000167464
|
GNAZ
|
G protein subunit alpha z |
| chr2_+_7192966 | 3.27 |
ENSDART00000142735
|
si:ch211-13f8.1
|
si:ch211-13f8.1 |
| chr3_+_12732382 | 3.25 |
ENSDART00000158403
|
cyp2k19
|
cytochrome P450, family 2, subfamily k, polypeptide 19 |
| chr11_+_25619892 | 3.25 |
ENSDART00000146647
ENSDART00000033702 |
comtb
|
catechol-O-methyltransferase b |
| chr11_-_33960318 | 3.24 |
ENSDART00000087597
|
col6a2
|
collagen, type VI, alpha 2 |
| chr23_+_19590598 | 3.24 |
ENSDART00000170149
|
slmapb
|
sarcolemma associated protein b |
| chr23_-_30960506 | 3.22 |
ENSDART00000142661
|
osbpl2a
|
oxysterol binding protein-like 2a |
| chr12_+_46960579 | 3.17 |
ENSDART00000149032
|
oat
|
ornithine aminotransferase |
| chr18_+_8346920 | 3.15 |
ENSDART00000083421
|
cpt1b
|
carnitine palmitoyltransferase 1B (muscle) |
| chr9_-_16109001 | 3.12 |
ENSDART00000053473
|
upp2
|
uridine phosphorylase 2 |
| chr3_-_61162750 | 3.10 |
ENSDART00000055064
|
pvalb8
|
parvalbumin 8 |
| chr23_+_4022620 | 3.08 |
ENSDART00000055099
|
b4galt5
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 5 |
| chr4_-_14915268 | 3.08 |
ENSDART00000067040
|
si:dkey-180p18.9
|
si:dkey-180p18.9 |
| chr25_-_9805269 | 3.05 |
ENSDART00000192048
|
lrrc4c
|
leucine rich repeat containing 4C |
| chr17_-_43466317 | 3.04 |
ENSDART00000155313
|
hspa4l
|
heat shock protein 4 like |
| chr4_+_3389866 | 3.04 |
ENSDART00000153834
|
sypl1
|
synaptophysin-like 1 |
| chr11_+_30161168 | 3.03 |
ENSDART00000157385
|
cdkl5
|
cyclin-dependent kinase-like 5 |
| chr1_+_54673846 | 3.02 |
ENSDART00000145018
|
gprc5bb
|
G protein-coupled receptor, class C, group 5, member Bb |
| chr21_-_43949208 | 3.02 |
ENSDART00000150983
|
camk2a
|
calcium/calmodulin-dependent protein kinase II alpha |
| chr3_-_28665291 | 3.00 |
ENSDART00000151670
|
fbxl16
|
F-box and leucine-rich repeat protein 16 |
| chr3_-_35602233 | 2.99 |
ENSDART00000055269
|
gng13b
|
guanine nucleotide binding protein (G protein), gamma 13b |
| chr6_+_28051978 | 2.95 |
ENSDART00000143218
|
si:ch73-194h10.2
|
si:ch73-194h10.2 |
| chr4_-_22671469 | 2.94 |
ENSDART00000050753
|
cd36
|
CD36 molecule (thrombospondin receptor) |
| chr11_+_30161699 | 2.93 |
ENSDART00000190504
|
cdkl5
|
cyclin-dependent kinase-like 5 |
| chr9_+_46644633 | 2.91 |
ENSDART00000160285
|
slc4a3
|
solute carrier family 4 (anion exchanger), member 3 |
| chr13_-_27675212 | 2.89 |
ENSDART00000141035
|
rims1a
|
regulating synaptic membrane exocytosis 1a |
| chr11_+_6819050 | 2.88 |
ENSDART00000104289
|
rab3ab
|
RAB3A, member RAS oncogene family, b |
| chr7_-_32833153 | 2.86 |
ENSDART00000099871
ENSDART00000099872 |
slc17a6b
|
solute carrier family 17 (vesicular glutamate transporter), member 6b |
| chr19_-_28367413 | 2.84 |
ENSDART00000079092
|
si:dkey-261i16.5
|
si:dkey-261i16.5 |
| chr5_-_22001003 | 2.80 |
ENSDART00000134393
ENSDART00000143878 |
arhgef9a
|
Cdc42 guanine nucleotide exchange factor (GEF) 9a |
| chr13_+_25396896 | 2.79 |
ENSDART00000041257
|
gsto2
|
glutathione S-transferase omega 2 |
| chr5_-_21970881 | 2.77 |
ENSDART00000182907
|
arhgef9a
|
Cdc42 guanine nucleotide exchange factor (GEF) 9a |
| chr12_-_7806007 | 2.74 |
ENSDART00000190359
|
ank3b
|
ankyrin 3b |
| chr1_+_44127292 | 2.72 |
ENSDART00000160542
|
cabp2a
|
calcium binding protein 2a |
| chr23_+_18944388 | 2.71 |
ENSDART00000104487
|
cox4i2
|
cytochrome c oxidase subunit IV isoform 2 |
| chr13_-_37254777 | 2.65 |
ENSDART00000139734
|
syne2b
|
spectrin repeat containing, nuclear envelope 2b |
| chr24_-_39772045 | 2.65 |
ENSDART00000087441
|
GFOD1
|
si:ch211-276f18.2 |
| chr3_-_33880951 | 2.64 |
ENSDART00000013228
|
cacna1aa
|
calcium channel, voltage-dependent, P/Q type, alpha 1A subunit, a |
| chr10_+_24627683 | 2.64 |
ENSDART00000112652
|
slc46a3
|
solute carrier family 46, member 3 |
| chr4_-_77432218 | 2.60 |
ENSDART00000158683
|
slco1d1
|
solute carrier organic anion transporter family, member 1D1 |
| chr24_-_10897511 | 2.57 |
ENSDART00000145593
ENSDART00000102484 ENSDART00000066784 |
fam49bb
|
family with sequence similarity 49, member Bb |
| chr19_-_5254699 | 2.54 |
ENSDART00000081951
|
stx1b
|
syntaxin 1B |
| chr3_+_37827373 | 2.52 |
ENSDART00000039517
|
asic2
|
acid-sensing (proton-gated) ion channel 2 |
| chr13_+_25397098 | 2.51 |
ENSDART00000132953
|
gsto2
|
glutathione S-transferase omega 2 |
| chr12_-_26383242 | 2.49 |
ENSDART00000152941
|
usp54b
|
ubiquitin specific peptidase 54b |
| chr4_+_3482312 | 2.49 |
ENSDART00000109044
|
grm8a
|
glutamate receptor, metabotropic 8a |
| chr25_+_33192796 | 2.48 |
ENSDART00000125892
ENSDART00000121680 ENSDART00000014851 |
TPM1 (1 of many)
|
zgc:171719 |
| chr9_-_43375205 | 2.47 |
ENSDART00000138436
|
znf385b
|
zinc finger protein 385B |
| chr21_-_7687544 | 2.46 |
ENSDART00000134519
|
pde8b
|
phosphodiesterase 8B |
| chr8_-_32497581 | 2.44 |
ENSDART00000176298
ENSDART00000183340 |
si:dkey-164f24.2
|
si:dkey-164f24.2 |
| chr9_+_310331 | 2.44 |
ENSDART00000172446
ENSDART00000187731 ENSDART00000193970 |
stac3
|
SH3 and cysteine rich domain 3 |
| chr14_-_18671334 | 2.41 |
ENSDART00000182381
|
slitrk4
|
SLIT and NTRK-like family, member 4 |
| chr10_+_36650222 | 2.40 |
ENSDART00000126963
|
ucp3
|
uncoupling protein 3 |
| chr18_+_924949 | 2.40 |
ENSDART00000170888
ENSDART00000193163 |
pkma
|
pyruvate kinase M1/2a |
| chr9_+_13682133 | 2.36 |
ENSDART00000175639
|
mpp4a
|
membrane protein, palmitoylated 4a (MAGUK p55 subfamily member 4) |
| chr14_+_33458294 | 2.35 |
ENSDART00000075278
|
atp1b4
|
ATPase Na+/K+ transporting subunit beta 4 |
| chr8_+_1187928 | 2.27 |
ENSDART00000127252
|
slc35d2
|
solute carrier family 35 (UDP-GlcNAc/UDP-glucose transporter), member D2 |
| chr25_-_17528994 | 2.27 |
ENSDART00000061712
|
si:dkey-44k1.5
|
si:dkey-44k1.5 |
| chr14_+_21699414 | 2.24 |
ENSDART00000169942
|
stx3a
|
syntaxin 3A |
| chr18_-_1228688 | 2.22 |
ENSDART00000064403
|
nptnb
|
neuroplastin b |
| chr9_+_45227028 | 2.21 |
ENSDART00000185579
|
adarb1b
|
adenosine deaminase, RNA-specific, B1b |
| chr21_-_21148623 | 2.21 |
ENSDART00000184364
|
ank1b
|
ankyrin 1, erythrocytic b |
| chr6_-_35472923 | 2.21 |
ENSDART00000185907
|
rgs8
|
regulator of G protein signaling 8 |
| chr8_+_23703680 | 2.20 |
ENSDART00000141099
ENSDART00000135394 |
ppardb
|
peroxisome proliferator-activated receptor delta b |
| chr23_-_21763598 | 2.19 |
ENSDART00000145408
|
vps13d
|
vacuolar protein sorting 13 homolog D (S. cerevisiae) |
| chr9_-_135774 | 2.18 |
ENSDART00000160435
|
FQ377903.1
|
|
| chr7_+_30787903 | 2.17 |
ENSDART00000174000
|
apba2b
|
amyloid beta (A4) precursor protein-binding, family A, member 2b |
| chr4_-_5302866 | 2.13 |
ENSDART00000138590
|
SNAP91 (1 of many)
|
si:ch211-214j24.9 |
| chr20_+_38201644 | 2.12 |
ENSDART00000022694
|
ehd3
|
EH-domain containing 3 |
| chr15_-_4969525 | 2.11 |
ENSDART00000157223
|
lipt2
|
lipoyl(octanoyl) transferase 2 |
| chr5_+_15350954 | 2.10 |
ENSDART00000140990
ENSDART00000137287 ENSDART00000061653 |
pebp1
|
phosphatidylethanolamine binding protein 1 |
| chr6_+_41255485 | 2.09 |
ENSDART00000042683
ENSDART00000186013 |
cadpsb
|
Ca2+-dependent activator protein for secretion b |
| chr16_-_33104944 | 2.09 |
ENSDART00000151943
|
pnrc2
|
proline-rich nuclear receptor coactivator 2 |
| chr7_+_34297271 | 2.09 |
ENSDART00000180342
|
bbox1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chr14_-_30587814 | 2.08 |
ENSDART00000144912
ENSDART00000149714 |
tmem265
|
transmembrane protein 265 |
| chr18_+_1703984 | 2.07 |
ENSDART00000114010
|
slitrk3a
|
SLIT and NTRK-like family, member 3a |
| chr16_+_34493987 | 2.04 |
ENSDART00000138374
|
si:ch211-255i3.4
|
si:ch211-255i3.4 |
| chr3_-_40945710 | 2.02 |
ENSDART00000138719
ENSDART00000102416 |
cyp3c4
|
cytochrome P450, family 3, subfamily C, polypeptide 4 |
| chr20_-_44496245 | 2.02 |
ENSDART00000012229
|
fkbp1b
|
FK506 binding protein 1b |
| chr8_+_22478090 | 2.00 |
ENSDART00000170263
|
si:ch211-261n11.7
|
si:ch211-261n11.7 |
| chr4_+_12013043 | 1.99 |
ENSDART00000130692
|
cry1aa
|
cryptochrome circadian clock 1aa |
| chr14_-_9199968 | 1.97 |
ENSDART00000146113
|
arhgef9b
|
Cdc42 guanine nucleotide exchange factor (GEF) 9b |
| chr7_+_17716601 | 1.96 |
ENSDART00000173792
ENSDART00000080825 |
rtn3
|
reticulon 3 |
| chr4_-_5291256 | 1.94 |
ENSDART00000150864
|
SNAP91 (1 of many)
|
si:ch211-214j24.9 |
| chr13_-_22766445 | 1.91 |
ENSDART00000140151
|
si:ch211-150i13.1
|
si:ch211-150i13.1 |
| chr7_+_48761875 | 1.90 |
ENSDART00000003690
|
acana
|
aggrecan a |
| chr25_-_10564721 | 1.90 |
ENSDART00000154776
|
galn
|
galanin/GMAP prepropeptide |
| chr2_+_8261109 | 1.89 |
ENSDART00000024677
|
dia1a
|
deleted in autism 1a |
| chr25_+_33192404 | 1.87 |
ENSDART00000193592
|
TPM1 (1 of many)
|
zgc:171719 |
| chr16_+_16826504 | 1.86 |
ENSDART00000108691
|
kcnj14
|
potassium inwardly-rectifying channel, subfamily J, member 14 |
| chr25_-_17552299 | 1.84 |
ENSDART00000154327
|
si:dkey-44k1.5
|
si:dkey-44k1.5 |
| chr14_+_45645024 | 1.80 |
ENSDART00000168278
|
si:ch211-276i12.4
|
si:ch211-276i12.4 |
| chr5_-_55848358 | 1.80 |
ENSDART00000130891
|
camk4
|
calcium/calmodulin-dependent protein kinase IV |
| chr6_-_1553314 | 1.80 |
ENSDART00000077209
|
tpra1
|
transmembrane protein, adipocyte asscociated 1 |
| chr11_-_7078392 | 1.79 |
ENSDART00000112156
ENSDART00000188556 |
si:ch211-253b8.5
|
si:ch211-253b8.5 |
| chr19_+_23982466 | 1.79 |
ENSDART00000080673
|
syt11a
|
synaptotagmin XIa |
| chr1_+_51721851 | 1.79 |
ENSDART00000040397
|
prdx2
|
peroxiredoxin 2 |
| chr13_+_10945337 | 1.76 |
ENSDART00000091845
|
abcg5
|
ATP-binding cassette, sub-family G (WHITE), member 5 |
| chr11_+_25278772 | 1.75 |
ENSDART00000188630
|
cyldb
|
cylindromatosis (turban tumor syndrome), b |
| chr11_+_6295370 | 1.73 |
ENSDART00000139882
|
ranbp3a
|
RAN binding protein 3a |
| chr15_+_7992906 | 1.73 |
ENSDART00000090790
|
cadm2b
|
cell adhesion molecule 2b |
| chr7_-_66877058 | 1.72 |
ENSDART00000155954
|
adma
|
adrenomedullin a |
| chr6_+_8315050 | 1.71 |
ENSDART00000189987
|
gcdha
|
glutaryl-CoA dehydrogenase a |
| chr23_-_42093537 | 1.70 |
ENSDART00000122074
|
slc1a7b
|
solute carrier family 1 (glutamate transporter), member 7b |
| chr4_+_12013642 | 1.68 |
ENSDART00000067281
|
cry1aa
|
cryptochrome circadian clock 1aa |
| chr5_-_55848511 | 1.67 |
ENSDART00000183503
|
camk4
|
calcium/calmodulin-dependent protein kinase IV |
| chr15_+_28096152 | 1.67 |
ENSDART00000100293
ENSDART00000140092 |
crybb1l3
|
crystallin, beta B1, like 3 |
| chr23_-_5032587 | 1.64 |
ENSDART00000163903
|
kcna2b
|
potassium voltage-gated channel, shaker-related subfamily, member 2b |
| chr17_-_5610514 | 1.64 |
ENSDART00000004043
|
enpp4
|
ectonucleotide pyrophosphatase/phosphodiesterase 4 |
| chr7_-_38792543 | 1.63 |
ENSDART00000157416
|
si:dkey-23n7.10
|
si:dkey-23n7.10 |
| chr7_+_18176162 | 1.63 |
ENSDART00000109171
|
rce1a
|
Ras converting CAAX endopeptidase 1a |
| chr25_+_34581494 | 1.63 |
ENSDART00000167690
|
trpm1b
|
transient receptor potential cation channel, subfamily M, member 1b |
| chr4_-_5302162 | 1.62 |
ENSDART00000177099
|
SNAP91 (1 of many)
|
si:ch211-214j24.9 |
| chr22_-_37686966 | 1.61 |
ENSDART00000192217
|
htr2b
|
5-hydroxytryptamine (serotonin) receptor 2B, G protein-coupled |
| chr13_+_1182257 | 1.60 |
ENSDART00000033528
ENSDART00000183702 ENSDART00000147959 |
tnfaip3
|
tumor necrosis factor, alpha-induced protein 3 |
| chr21_-_22827548 | 1.60 |
ENSDART00000079161
|
angptl5
|
angiopoietin-like 5 |
| chr6_-_50404732 | 1.60 |
ENSDART00000055510
|
romo1
|
reactive oxygen species modulator 1 |
| chr7_-_69429561 | 1.60 |
ENSDART00000127351
|
atxn1l
|
ataxin 1-like |
| chr22_+_5682635 | 1.59 |
ENSDART00000140680
ENSDART00000131308 |
dnase1l4.2
|
deoxyribonuclease 1 like 4, tandem duplicate 2 |
| chr5_+_69808763 | 1.59 |
ENSDART00000143482
|
fsd1l
|
fibronectin type III and SPRY domain containing 1-like |
| chr9_-_30432778 | 1.59 |
ENSDART00000138364
|
gja8a
|
gap junction protein alpha 8 paralog a |
Network of associatons between targets according to the STRING database.
First level regulatory network of nfil3-5
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.6 | 14.3 | GO:0072673 | lamellipodium morphogenesis(GO:0072673) regulation of lamellipodium morphogenesis(GO:2000392) |
| 2.3 | 9.1 | GO:0045023 | G0 to G1 transition(GO:0045023) regulation of G0 to G1 transition(GO:0070316) negative regulation of G0 to G1 transition(GO:0070317) |
| 1.3 | 4.0 | GO:0014809 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 1.3 | 10.3 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 1.2 | 3.7 | GO:0034035 | sulfate assimilation(GO:0000103) purine ribonucleoside bisphosphate metabolic process(GO:0034035) purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 1.2 | 4.9 | GO:0009447 | polyamine catabolic process(GO:0006598) putrescine catabolic process(GO:0009447) |
| 1.2 | 7.1 | GO:0045745 | positive regulation of G-protein coupled receptor protein signaling pathway(GO:0045745) |
| 1.2 | 3.5 | GO:0006216 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 1.0 | 6.9 | GO:0019614 | catechol-containing compound catabolic process(GO:0019614) catecholamine catabolic process(GO:0042424) |
| 1.0 | 2.9 | GO:0031630 | regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031630) |
| 0.9 | 11.9 | GO:0006603 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.9 | 4.5 | GO:0098881 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.9 | 3.5 | GO:0009097 | isoleucine metabolic process(GO:0006549) isoleucine biosynthetic process(GO:0009097) |
| 0.9 | 3.4 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.8 | 3.3 | GO:0003151 | outflow tract morphogenesis(GO:0003151) |
| 0.8 | 4.1 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.8 | 2.4 | GO:0014808 | regulation of release of sequestered calcium ion into cytosol by sarcoplasmic reticulum(GO:0010880) release of sequestered calcium ion into cytosol by sarcoplasmic reticulum(GO:0014808) sarcoplasmic reticulum calcium ion transport(GO:0070296) calcium ion transport from endoplasmic reticulum to cytosol(GO:1903514) |
| 0.7 | 2.1 | GO:2000374 | oxygen metabolic process(GO:0072592) regulation of oxygen metabolic process(GO:2000374) positive regulation of oxygen metabolic process(GO:2000376) |
| 0.7 | 13.0 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.6 | 8.3 | GO:2000054 | negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 0.6 | 3.6 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.6 | 4.1 | GO:0036268 | swimming(GO:0036268) |
| 0.6 | 4.6 | GO:0045338 | farnesyl diphosphate metabolic process(GO:0045338) |
| 0.6 | 3.9 | GO:0019346 | homoserine metabolic process(GO:0009092) cysteine biosynthetic process via cystathionine(GO:0019343) cysteine biosynthetic process(GO:0019344) transsulfuration(GO:0019346) |
| 0.6 | 2.2 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.5 | 3.8 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.5 | 1.6 | GO:0080120 | CAAX-box protein processing(GO:0071586) CAAX-box protein maturation(GO:0080120) |
| 0.5 | 4.0 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.5 | 7.5 | GO:0090382 | phagosome maturation(GO:0090382) |
| 0.4 | 4.2 | GO:0044206 | pyrimidine-containing compound salvage(GO:0008655) pyrimidine ribonucleotide salvage(GO:0010138) pyrimidine nucleotide salvage(GO:0032262) pyrimidine nucleoside salvage(GO:0043097) UMP salvage(GO:0044206) |
| 0.4 | 3.7 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.4 | 4.1 | GO:0034694 | response to prostaglandin(GO:0034694) |
| 0.4 | 3.2 | GO:0006561 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.3 | 2.0 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.3 | 1.0 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.3 | 5.5 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.3 | 7.5 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.3 | 1.6 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.3 | 1.6 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) positive regulation of reactive oxygen species metabolic process(GO:2000379) |
| 0.3 | 0.9 | GO:0036076 | ligamentous ossification(GO:0036076) |
| 0.3 | 2.2 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.3 | 2.4 | GO:1990845 | adaptive thermogenesis(GO:1990845) |
| 0.3 | 1.5 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.3 | 3.2 | GO:0009437 | carnitine metabolic process(GO:0009437) |
| 0.3 | 6.0 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.3 | 1.0 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.3 | 3.4 | GO:0045762 | activation of adenylate cyclase activity(GO:0007190) positive regulation of cAMP metabolic process(GO:0030816) positive regulation of cAMP biosynthetic process(GO:0030819) positive regulation of adenylate cyclase activity(GO:0045762) |
| 0.2 | 3.0 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.2 | 1.7 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.2 | 3.1 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.2 | 1.6 | GO:0050820 | positive regulation of blood coagulation(GO:0030194) positive regulation of coagulation(GO:0050820) positive regulation of hemostasis(GO:1900048) |
| 0.2 | 0.6 | GO:0046689 | response to mercury ion(GO:0046689) detoxification of mercury ion(GO:0050787) |
| 0.2 | 2.7 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.2 | 3.9 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.2 | 2.9 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.2 | 2.0 | GO:0016560 | protein import into peroxisome matrix, docking(GO:0016560) protein to membrane docking(GO:0022615) |
| 0.2 | 3.6 | GO:0051932 | synaptic transmission, GABAergic(GO:0051932) |
| 0.2 | 1.3 | GO:0003321 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) positive regulation of blood pressure by epinephrine-norepinephrine(GO:0003321) positive regulation of heart rate(GO:0010460) |
| 0.2 | 9.5 | GO:0042738 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.2 | 0.9 | GO:0060055 | larval development(GO:0002164) larval heart development(GO:0007508) angiogenesis involved in wound healing(GO:0060055) |
| 0.2 | 1.8 | GO:0000303 | response to superoxide(GO:0000303) response to oxygen radical(GO:0000305) removal of superoxide radicals(GO:0019430) cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.2 | 7.9 | GO:0014059 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.2 | 1.8 | GO:2001238 | positive regulation of extrinsic apoptotic signaling pathway(GO:2001238) |
| 0.2 | 1.5 | GO:0006805 | xenobiotic metabolic process(GO:0006805) |
| 0.2 | 1.0 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.2 | 1.5 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 0.2 | 4.5 | GO:0051923 | sulfation(GO:0051923) |
| 0.2 | 2.1 | GO:1990504 | dense core granule exocytosis(GO:1990504) |
| 0.2 | 1.4 | GO:0043435 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.2 | 1.4 | GO:0010887 | negative regulation of cholesterol storage(GO:0010887) |
| 0.1 | 9.7 | GO:0006414 | translational elongation(GO:0006414) |
| 0.1 | 1.5 | GO:0015746 | tricarboxylic acid transport(GO:0006842) citrate transport(GO:0015746) |
| 0.1 | 1.2 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.1 | 1.3 | GO:0015810 | acidic amino acid transport(GO:0015800) aspartate transport(GO:0015810) L-glutamate transport(GO:0015813) malate-aspartate shuttle(GO:0043490) |
| 0.1 | 0.8 | GO:0042362 | fat-soluble vitamin biosynthetic process(GO:0042362) |
| 0.1 | 1.4 | GO:1904103 | regulation of convergent extension involved in gastrulation(GO:1904103) |
| 0.1 | 2.5 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.1 | 3.9 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.1 | 4.3 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.1 | 0.6 | GO:0019370 | leukotriene metabolic process(GO:0006691) leukotriene biosynthetic process(GO:0019370) |
| 0.1 | 0.5 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.1 | 1.8 | GO:0098962 | regulation of postsynaptic neurotransmitter receptor activity(GO:0098962) |
| 0.1 | 0.7 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.1 | 2.6 | GO:0034112 | positive regulation of homotypic cell-cell adhesion(GO:0034112) positive regulation of T cell activation(GO:0050870) positive regulation of leukocyte cell-cell adhesion(GO:1903039) |
| 0.1 | 0.3 | GO:0090141 | positive regulation of mitochondrial fission(GO:0090141) |
| 0.1 | 1.0 | GO:1901678 | heme transport(GO:0015886) iron coordination entity transport(GO:1901678) |
| 0.1 | 1.6 | GO:0042493 | response to drug(GO:0042493) |
| 0.1 | 3.0 | GO:0055092 | cholesterol homeostasis(GO:0042632) sterol homeostasis(GO:0055092) |
| 0.1 | 5.7 | GO:0006096 | glycolytic process(GO:0006096) |
| 0.1 | 0.7 | GO:1904086 | regulation of epiboly involved in gastrulation with mouth forming second(GO:1904086) |
| 0.1 | 3.7 | GO:0010675 | regulation of cellular carbohydrate metabolic process(GO:0010675) regulation of glucose metabolic process(GO:0010906) |
| 0.1 | 1.2 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.1 | 2.2 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.1 | 1.3 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.1 | 0.8 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.1 | 7.7 | GO:0006836 | neurotransmitter transport(GO:0006836) |
| 0.1 | 2.1 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.1 | 0.5 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.1 | 0.3 | GO:0010586 | miRNA metabolic process(GO:0010586) |
| 0.1 | 3.8 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.1 | 2.5 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.1 | 3.1 | GO:0030050 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.1 | 3.5 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.1 | 3.8 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.1 | 2.0 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.1 | 1.6 | GO:0060402 | cytosolic calcium ion transport(GO:0060401) calcium ion transport into cytosol(GO:0060402) |
| 0.1 | 13.4 | GO:0043547 | positive regulation of GTPase activity(GO:0043547) |
| 0.1 | 1.2 | GO:0033138 | positive regulation of peptidyl-serine phosphorylation(GO:0033138) |
| 0.1 | 1.9 | GO:1990573 | potassium ion import(GO:0010107) potassium ion import across plasma membrane(GO:1990573) |
| 0.1 | 6.5 | GO:0035725 | sodium ion transmembrane transport(GO:0035725) |
| 0.0 | 0.6 | GO:0008625 | extrinsic apoptotic signaling pathway via death domain receptors(GO:0008625) |
| 0.0 | 1.0 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 2.3 | GO:0006814 | sodium ion transport(GO:0006814) |
| 0.0 | 1.1 | GO:0007631 | feeding behavior(GO:0007631) |
| 0.0 | 3.0 | GO:0003146 | heart jogging(GO:0003146) |
| 0.0 | 1.0 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 3.5 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.0 | 0.4 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 2.6 | GO:0070509 | calcium ion import(GO:0070509) |
| 0.0 | 0.6 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.0 | 0.3 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
| 0.0 | 7.0 | GO:0006936 | muscle contraction(GO:0006936) |
| 0.0 | 1.0 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 1.4 | GO:0060319 | primitive erythrocyte differentiation(GO:0060319) |
| 0.0 | 1.3 | GO:0031398 | positive regulation of protein ubiquitination(GO:0031398) |
| 0.0 | 5.0 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
| 0.0 | 0.7 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 5.3 | GO:0021782 | glial cell development(GO:0021782) |
| 0.0 | 0.5 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 0.6 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.6 | GO:0051897 | positive regulation of protein kinase B signaling(GO:0051897) |
| 0.0 | 0.8 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.0 | 0.9 | GO:0006879 | cellular iron ion homeostasis(GO:0006879) |
| 0.0 | 2.8 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 0.5 | GO:0099560 | synaptic membrane adhesion(GO:0099560) |
| 0.0 | 0.2 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.0 | 1.2 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.0 | 0.1 | GO:0055071 | manganese ion homeostasis(GO:0055071) |
| 0.0 | 1.3 | GO:0032924 | activin receptor signaling pathway(GO:0032924) |
| 0.0 | 1.9 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.0 | 0.3 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 1.1 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.9 | GO:0043652 | engulfment of apoptotic cell(GO:0043652) |
| 0.0 | 3.0 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.0 | 3.0 | GO:1990778 | protein localization to plasma membrane(GO:0072659) protein localization to cell periphery(GO:1990778) |
| 0.0 | 2.7 | GO:0007163 | establishment or maintenance of cell polarity(GO:0007163) |
| 0.0 | 0.3 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.2 | GO:0048662 | regulation of smooth muscle cell proliferation(GO:0048660) negative regulation of smooth muscle cell proliferation(GO:0048662) |
| 0.0 | 0.1 | GO:1902369 | mitochondrion morphogenesis(GO:0070584) negative regulation of RNA catabolic process(GO:1902369) |
| 0.0 | 0.2 | GO:0019883 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous antigen(GO:0019883) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.0 | 0.9 | GO:0048821 | erythrocyte development(GO:0048821) |
| 0.0 | 0.4 | GO:0015671 | gas transport(GO:0015669) oxygen transport(GO:0015671) |
| 0.0 | 1.0 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 0.1 | GO:0032119 | sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 0.4 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 | 0.8 | GO:0007062 | sister chromatid cohesion(GO:0007062) |
| 0.0 | 0.2 | GO:0051966 | regulation of synaptic transmission, glutamatergic(GO:0051966) |
| 0.0 | 0.4 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.0 | 0.8 | GO:0090263 | positive regulation of canonical Wnt signaling pathway(GO:0090263) |
| 0.0 | 0.1 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.9 | 14.3 | GO:0032433 | filopodium tip(GO:0032433) |
| 2.0 | 6.0 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 1.0 | 7.1 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.7 | 10.3 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.6 | 7.5 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.6 | 20.9 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.6 | 4.0 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.5 | 3.0 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.5 | 3.6 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.2 | 4.2 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.2 | 5.7 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.2 | 2.3 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) |
| 0.2 | 3.9 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.2 | 2.7 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.2 | 0.7 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.2 | 1.4 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.2 | 9.4 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.2 | 2.4 | GO:0030315 | T-tubule(GO:0030315) |
| 0.1 | 1.6 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.1 | 2.1 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.1 | 1.6 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.1 | 2.2 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.1 | 1.6 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 3.4 | GO:0098978 | glutamatergic synapse(GO:0098978) |
| 0.1 | 2.9 | GO:0098831 | cytoskeleton of presynaptic active zone(GO:0048788) presynaptic active zone cytoplasmic component(GO:0098831) |
| 0.1 | 0.5 | GO:0005883 | neurofilament(GO:0005883) |
| 0.1 | 1.3 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.1 | 1.3 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.1 | 1.5 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.1 | 4.0 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 1.1 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.1 | 1.1 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.1 | 0.6 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.1 | 10.4 | GO:0070382 | exocytic vesicle(GO:0070382) |
| 0.1 | 2.9 | GO:0005901 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.1 | 0.3 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.1 | 7.8 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.1 | 3.1 | GO:0005902 | microvillus(GO:0005902) |
| 0.1 | 0.5 | GO:0002139 | stereocilia coupling link(GO:0002139) stereocilia ankle link(GO:0002141) stereocilia ankle link complex(GO:0002142) stereocilium tip(GO:0032426) |
| 0.1 | 0.5 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.1 | 16.1 | GO:0005764 | lysosome(GO:0005764) |
| 0.1 | 1.3 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 3.1 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.1 | 1.3 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.1 | 0.4 | GO:0016586 | RSC complex(GO:0016586) |
| 0.0 | 4.9 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 2.0 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 2.6 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 3.7 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 1.5 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 1.6 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 0.8 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.0 | 0.4 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 1.2 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 1.3 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 1.3 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.4 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.3 | GO:0000178 | exosome (RNase complex)(GO:0000178) |
| 0.0 | 0.2 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 12.8 | GO:0043005 | neuron projection(GO:0043005) |
| 0.0 | 22.6 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 2.7 | GO:0045202 | synapse(GO:0045202) |
| 0.0 | 5.2 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 0.3 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 1.9 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 17.1 | GO:0005576 | extracellular region(GO:0005576) |
| 0.0 | 2.2 | GO:0019898 | extrinsic component of membrane(GO:0019898) |
| 0.0 | 1.3 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.2 | GO:0031306 | intrinsic component of mitochondrial outer membrane(GO:0031306) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 5.3 | GO:0045174 | glutathione dehydrogenase (ascorbate) activity(GO:0045174) |
| 1.7 | 6.9 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 1.5 | 4.6 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 1.2 | 3.7 | GO:0004781 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 1.2 | 4.7 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 1.0 | 3.1 | GO:0008489 | UDP-galactose:glucosylceramide beta-1,4-galactosyltransferase activity(GO:0008489) |
| 0.9 | 11.9 | GO:0004111 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.9 | 7.2 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.8 | 10.3 | GO:0005326 | neurotransmitter transporter activity(GO:0005326) |
| 0.8 | 2.4 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.7 | 2.9 | GO:0030228 | lipoprotein particle receptor activity(GO:0030228) |
| 0.7 | 5.0 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.7 | 3.5 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.7 | 2.1 | GO:0033819 | lipoyl(octanoyl) transferase activity(GO:0033819) |
| 0.7 | 4.9 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.7 | 2.1 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.6 | 2.6 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.6 | 20.9 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.6 | 2.4 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.6 | 3.5 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.6 | 1.8 | GO:0043878 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (non-phosphorylating) activity(GO:0043878) |
| 0.6 | 2.3 | GO:0005463 | UDP-glucuronic acid transmembrane transporter activity(GO:0005461) UDP-N-acetylgalactosamine transmembrane transporter activity(GO:0005463) |
| 0.6 | 3.9 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 0.5 | 7.1 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.5 | 4.5 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.5 | 3.7 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.5 | 3.2 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.4 | 1.3 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.4 | 1.7 | GO:0004361 | glutaryl-CoA dehydrogenase activity(GO:0004361) |
| 0.4 | 4.2 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.4 | 2.4 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.4 | 3.4 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.4 | 1.5 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.4 | 1.5 | GO:0017153 | sodium:dicarboxylate symporter activity(GO:0017153) |
| 0.3 | 2.0 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.3 | 7.1 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.3 | 7.5 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.3 | 1.0 | GO:0005252 | open rectifier potassium channel activity(GO:0005252) |
| 0.3 | 1.0 | GO:0003913 | DNA photolyase activity(GO:0003913) |
| 0.3 | 4.1 | GO:0005221 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.3 | 0.9 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.3 | 14.7 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.3 | 1.5 | GO:0031841 | neuropeptide Y receptor binding(GO:0031841) type 2 neuropeptide Y receptor binding(GO:0031843) |
| 0.3 | 3.6 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.3 | 13.9 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.3 | 1.3 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.3 | 1.8 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.3 | 2.0 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.2 | 1.0 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) protein histidine phosphatase activity(GO:0101006) |
| 0.2 | 4.4 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.2 | 4.0 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.2 | 9.7 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.2 | 2.5 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.2 | 2.2 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.2 | 1.5 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.2 | 1.6 | GO:0010436 | beta-carotene 15,15'-monooxygenase activity(GO:0003834) carotenoid dioxygenase activity(GO:0010436) |
| 0.2 | 3.3 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.2 | 0.7 | GO:0004478 | methionine adenosyltransferase activity(GO:0004478) |
| 0.2 | 2.2 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.2 | 1.3 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.2 | 6.0 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.2 | 6.4 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.2 | 0.6 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.2 | 6.5 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.1 | 9.5 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.1 | 0.9 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.1 | 0.4 | GO:0030792 | arsenite methyltransferase activity(GO:0030791) methylarsonite methyltransferase activity(GO:0030792) |
| 0.1 | 0.6 | GO:0004968 | gonadotropin-releasing hormone receptor activity(GO:0004968) |
| 0.1 | 1.1 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.1 | 1.4 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.1 | 3.2 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.1 | 1.0 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.1 | 0.3 | GO:0004061 | arylformamidase activity(GO:0004061) |
| 0.1 | 1.0 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.1 | 1.6 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.1 | 1.3 | GO:0015172 | L-glutamate transmembrane transporter activity(GO:0005313) acidic amino acid transmembrane transporter activity(GO:0015172) |
| 0.1 | 1.8 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.1 | 1.6 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.1 | 1.5 | GO:0042910 | xenobiotic transporter activity(GO:0042910) |
| 0.1 | 2.6 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) cyclin-dependent protein kinase activity(GO:0097472) |
| 0.1 | 2.5 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.1 | 0.6 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.1 | 5.7 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.1 | 1.4 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.1 | 2.7 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 1.3 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.1 | 2.2 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.1 | 2.0 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.1 | 7.4 | GO:0016763 | transferase activity, transferring pentosyl groups(GO:0016763) |
| 0.1 | 2.1 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.1 | 0.9 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.1 | 0.8 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.1 | 1.0 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.1 | 1.0 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.1 | 2.2 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.1 | 2.9 | GO:0030170 | pyridoxal phosphate binding(GO:0030170) |
| 0.1 | 0.4 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.1 | 3.1 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.1 | 2.7 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.1 | 3.0 | GO:0030295 | kinase activator activity(GO:0019209) protein kinase activator activity(GO:0030295) |
| 0.1 | 10.0 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
| 0.1 | 1.2 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.1 | 1.4 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.1 | 0.8 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.1 | 8.0 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 1.6 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 16.5 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 3.6 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.0 | 1.7 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 3.0 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 1.4 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 1.6 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 5.5 | GO:0015293 | symporter activity(GO:0015293) |
| 0.0 | 0.4 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.0 | 0.1 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.0 | 1.7 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 2.6 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 1.3 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.0 | 0.9 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 7.8 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 24.2 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.4 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 1.2 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 1.6 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.0 | 3.5 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 1.0 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.5 | GO:0016411 | acylglycerol O-acyltransferase activity(GO:0016411) |
| 0.0 | 0.6 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 1.7 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.1 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 6.5 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 0.6 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.8 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.2 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 0.0 | 1.7 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.1 | GO:0005158 | insulin receptor binding(GO:0005158) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.3 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.2 | 3.0 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.1 | 4.4 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.1 | 3.5 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.1 | 3.2 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 1.4 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.1 | 1.6 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.1 | 2.1 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 2.2 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 0.8 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.1 | 2.4 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 1.0 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.7 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 0.7 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 1.0 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 1.0 | PID CMYB PATHWAY | C-MYB transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.0 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.5 | 13.2 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.4 | 1.3 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.4 | 3.1 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.3 | 3.2 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.3 | 1.6 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.3 | 3.2 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.3 | 4.0 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.3 | 3.7 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.3 | 6.0 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.3 | 1.0 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.2 | 3.2 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.2 | 3.5 | REACTOME ACTIVATION OF NMDA RECEPTOR UPON GLUTAMATE BINDING AND POSTSYNAPTIC EVENTS | Genes involved in Activation of NMDA receptor upon glutamate binding and postsynaptic events |
| 0.2 | 4.6 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.2 | 2.5 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.2 | 4.3 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.2 | 4.6 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.1 | 1.9 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.1 | 1.1 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.1 | 1.5 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 2.3 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.1 | 4.6 | REACTOME RESPIRATORY ELECTRON TRANSPORT ATP SYNTHESIS BY CHEMIOSMOTIC COUPLING AND HEAT PRODUCTION BY UNCOUPLING PROTEINS | Genes involved in Respiratory electron transport, ATP synthesis by chemiosmotic coupling, and heat production by uncoupling proteins. |
| 0.1 | 4.5 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 0.6 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.1 | 2.1 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.1 | 2.2 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 3.7 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.1 | 1.4 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.6 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.0 | 2.5 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 0.3 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 1.4 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 2.2 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 0.3 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 1.3 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 1.1 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |