Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
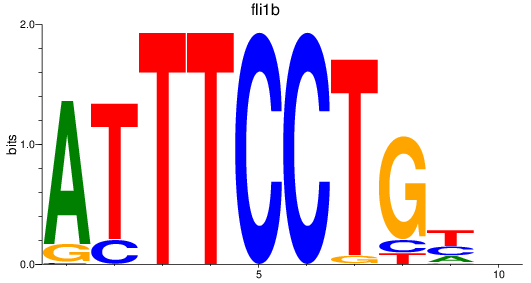
Results for fli1b
Z-value: 1.28
Transcription factors associated with fli1b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
fli1b
|
ENSDARG00000040080 | Fli-1 proto-oncogene, ETS transcription factor b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| fli1b | dr11_v1_chr16_+_42018367_42018367 | 0.65 | 2.7e-03 | Click! |
Activity profile of fli1b motif
Sorted Z-values of fli1b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_8129536 | 3.73 |
ENSDART00000158112
ENSDART00000011183 |
prkacba
|
protein kinase, cAMP-dependent, catalytic, beta a |
| chr15_-_4528326 | 2.91 |
ENSDART00000158122
ENSDART00000155619 ENSDART00000128602 |
tfdp2
|
transcription factor Dp-2 |
| chr5_-_54712159 | 2.89 |
ENSDART00000149207
|
ccnb1
|
cyclin B1 |
| chr13_+_33651416 | 2.79 |
ENSDART00000180221
|
BX005372.1
|
|
| chr10_+_22775253 | 2.56 |
ENSDART00000190141
|
tmem88a
|
transmembrane protein 88 a |
| chr19_-_2231146 | 2.53 |
ENSDART00000181909
ENSDART00000043595 |
twist1a
|
twist family bHLH transcription factor 1a |
| chr5_+_46424437 | 2.38 |
ENSDART00000186511
|
vcana
|
versican a |
| chr18_+_40381102 | 2.31 |
ENSDART00000136588
|
sema6dl
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D, like |
| chr20_-_36800002 | 2.24 |
ENSDART00000015190
|
ptrhd1
|
peptidyl-tRNA hydrolase domain containing 1 |
| chr12_+_28955766 | 2.12 |
ENSDART00000123417
ENSDART00000139347 |
znf668
|
zinc finger protein 668 |
| chr1_-_13968153 | 2.11 |
ENSDART00000103383
|
elf2b
|
E74-like factor 2b (ets domain transcription factor) |
| chr22_+_10163901 | 2.10 |
ENSDART00000190468
|
rpp14
|
ribonuclease P/MRP 14 subunit |
| chr7_+_22981909 | 2.07 |
ENSDART00000122449
|
ccnb3
|
cyclin B3 |
| chr16_-_42013858 | 2.04 |
ENSDART00000045403
|
etv2
|
ets variant 2 |
| chr14_-_41478265 | 2.03 |
ENSDART00000149886
ENSDART00000016002 |
tspan7
|
tetraspanin 7 |
| chr16_+_40043673 | 1.94 |
ENSDART00000102552
ENSDART00000125484 |
trmt11
|
tRNA methyltransferase 11 homolog (S. cerevisiae) |
| chr12_-_48312647 | 1.93 |
ENSDART00000114415
|
ascc1
|
activating signal cointegrator 1 complex subunit 1 |
| chr7_+_41340520 | 1.91 |
ENSDART00000005127
|
garem
|
GRB2 associated, regulator of MAPK1 |
| chr20_+_35282682 | 1.90 |
ENSDART00000187199
|
fam49a
|
family with sequence similarity 49, member A |
| chr12_-_35830625 | 1.88 |
ENSDART00000180028
|
CU459056.1
|
|
| chr16_-_34195002 | 1.85 |
ENSDART00000054026
|
rcc1
|
regulator of chromosome condensation 1 |
| chr17_+_10242166 | 1.81 |
ENSDART00000170420
|
clec14a
|
C-type lectin domain containing 14A |
| chr24_-_20192996 | 1.79 |
ENSDART00000011143
|
myd88
|
myeloid differentiation primary response 88 |
| chr8_-_25814263 | 1.75 |
ENSDART00000143397
|
taf10
|
TAF10 RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr6_+_21202639 | 1.74 |
ENSDART00000083126
|
cidec
|
cell death-inducing DFFA-like effector c |
| chr7_+_20017211 | 1.74 |
ENSDART00000100808
|
bcl6b
|
B-cell CLL/lymphoma 6, member B |
| chr2_-_15040345 | 1.72 |
ENSDART00000109657
|
si:dkey-10f21.4
|
si:dkey-10f21.4 |
| chr8_-_31107537 | 1.71 |
ENSDART00000098925
|
vgll4l
|
vestigial like 4 like |
| chr13_-_33700461 | 1.71 |
ENSDART00000160520
|
mad2l1bp
|
MAD2L1 binding protein |
| chr23_-_28025943 | 1.71 |
ENSDART00000181146
|
sp5l
|
Sp5 transcription factor-like |
| chr9_-_24218059 | 1.70 |
ENSDART00000007914
|
nabp1a
|
nucleic acid binding protein 1a |
| chr22_-_16291041 | 1.66 |
ENSDART00000021666
|
rtca
|
RNA 3'-terminal phosphate cyclase |
| chr14_-_26482096 | 1.65 |
ENSDART00000187280
|
smad5
|
SMAD family member 5 |
| chr22_-_23748284 | 1.61 |
ENSDART00000162005
|
cfhl2
|
complement factor H like 2 |
| chr23_-_21535040 | 1.61 |
ENSDART00000010647
|
rcc2
|
regulator of chromosome condensation 2 |
| chr7_+_41314862 | 1.60 |
ENSDART00000185198
|
zgc:165532
|
zgc:165532 |
| chr16_+_42018041 | 1.60 |
ENSDART00000134010
ENSDART00000102789 |
fli1b
|
Fli-1 proto-oncogene, ETS transcription factor b |
| chr18_+_38288877 | 1.59 |
ENSDART00000134247
|
lmo2
|
LIM domain only 2 (rhombotin-like 1) |
| chr6_-_33924883 | 1.59 |
ENSDART00000132762
ENSDART00000148142 ENSDART00000142213 |
akr1a1b
|
aldo-keto reductase family 1, member A1b (aldehyde reductase) |
| chr1_-_45213565 | 1.59 |
ENSDART00000145757
|
ddx39aa
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 39Aa |
| chr19_-_33274978 | 1.59 |
ENSDART00000020301
ENSDART00000114714 |
fam92a1
|
family with sequence similarity 92, member A1 |
| chr12_+_27462225 | 1.58 |
ENSDART00000105661
|
meox1
|
mesenchyme homeobox 1 |
| chr7_+_16509201 | 1.57 |
ENSDART00000173777
|
zdhhc13
|
zinc finger, DHHC-type containing 13 |
| chr5_-_69716501 | 1.56 |
ENSDART00000158956
|
mob1a
|
MOB kinase activator 1A |
| chr4_+_13909398 | 1.55 |
ENSDART00000187959
ENSDART00000184926 |
pphln1
|
periphilin 1 |
| chr7_-_38689562 | 1.55 |
ENSDART00000167209
|
aplnr2
|
apelin receptor 2 |
| chr9_-_24218367 | 1.55 |
ENSDART00000135356
|
nabp1a
|
nucleic acid binding protein 1a |
| chr7_-_66133786 | 1.54 |
ENSDART00000154961
|
btbd10b
|
BTB (POZ) domain containing 10b |
| chr18_+_17021391 | 1.53 |
ENSDART00000100160
|
chtf8
|
CTF8, chromosome transmission fidelity factor 8 homolog (S. cerevisiae) |
| chr8_-_7093507 | 1.52 |
ENSDART00000045669
|
si:dkey-222n6.2
|
si:dkey-222n6.2 |
| chr8_+_18010978 | 1.50 |
ENSDART00000039887
ENSDART00000144532 |
ssbp3b
|
single stranded DNA binding protein 3b |
| chr3_+_19665319 | 1.50 |
ENSDART00000007857
ENSDART00000193509 |
mettl2a
|
methyltransferase like 2A |
| chr9_+_2041535 | 1.50 |
ENSDART00000093187
|
lnpa
|
limb and neural patterns a |
| chr11_-_36350876 | 1.50 |
ENSDART00000146495
ENSDART00000020655 |
psma5
|
proteasome subunit alpha 5 |
| chr8_+_26373149 | 1.50 |
ENSDART00000132787
|
hyal3
|
hyaluronoglucosaminidase 3 |
| chr7_-_69025306 | 1.49 |
ENSDART00000180796
|
CABZ01057488.2
|
|
| chr19_-_22843480 | 1.47 |
ENSDART00000052503
|
nudcd1
|
NudC domain containing 1 |
| chr1_+_43686251 | 1.44 |
ENSDART00000074604
ENSDART00000137791 |
cisd2
|
CDGSH iron sulfur domain 2 |
| chr21_+_5080789 | 1.44 |
ENSDART00000024199
|
atp5fa1
|
ATP synthase F1 subunit alpha |
| chr9_-_31524907 | 1.44 |
ENSDART00000142904
ENSDART00000127214 ENSDART00000133427 ENSDART00000146268 ENSDART00000182541 ENSDART00000184736 |
tmtc4
|
transmembrane and tetratricopeptide repeat containing 4 |
| chr25_+_18556588 | 1.42 |
ENSDART00000073726
|
cav2
|
caveolin 2 |
| chr1_+_49686408 | 1.42 |
ENSDART00000140824
|
si:ch211-149l1.2
|
si:ch211-149l1.2 |
| chr1_+_22851261 | 1.42 |
ENSDART00000193925
|
gtf2e2
|
general transcription factor IIE, polypeptide 2, beta |
| chr20_+_35255403 | 1.41 |
ENSDART00000002773
ENSDART00000137915 |
fam49a
|
family with sequence similarity 49, member A |
| chr12_+_27232173 | 1.41 |
ENSDART00000193714
|
tmem106a
|
transmembrane protein 106A |
| chr6_-_40851499 | 1.39 |
ENSDART00000182591
|
rft1
|
RFT1 homolog |
| chr19_+_1873059 | 1.39 |
ENSDART00000145246
|
snrpd1
|
small nuclear ribonucleoprotein D1 polypeptide |
| chr15_-_18176694 | 1.38 |
ENSDART00000189840
|
tmprss5
|
transmembrane protease, serine 5 |
| chr7_+_41812636 | 1.38 |
ENSDART00000174333
|
orc6
|
origin recognition complex, subunit 6 |
| chr23_+_44374041 | 1.38 |
ENSDART00000136056
|
ephb4b
|
eph receptor B4b |
| chr6_-_33925381 | 1.37 |
ENSDART00000137268
ENSDART00000145019 ENSDART00000141822 |
akr1a1b
|
aldo-keto reductase family 1, member A1b (aldehyde reductase) |
| chr8_+_48943009 | 1.37 |
ENSDART00000180763
|
rer1
|
retention in endoplasmic reticulum sorting receptor 1 |
| chr19_+_10661520 | 1.36 |
ENSDART00000091813
ENSDART00000165653 |
ago3b
|
argonaute RISC catalytic component 3b |
| chr20_-_2619316 | 1.35 |
ENSDART00000185777
|
bub1
|
BUB1 mitotic checkpoint serine/threonine kinase |
| chr5_-_39171302 | 1.35 |
ENSDART00000020808
|
paqr3a
|
progestin and adipoQ receptor family member IIIa |
| chr7_+_22981441 | 1.34 |
ENSDART00000182887
|
ccnb3
|
cyclin B3 |
| chr7_-_17816175 | 1.34 |
ENSDART00000091272
ENSDART00000173757 |
ecsit
|
ECSIT signalling integrator |
| chr7_+_19374683 | 1.34 |
ENSDART00000162700
|
snrpf
|
small nuclear ribonucleoprotein polypeptide F |
| chr13_+_39532050 | 1.34 |
ENSDART00000019379
|
marveld1
|
MARVEL domain containing 1 |
| chr1_-_52498146 | 1.34 |
ENSDART00000122217
|
acy3.2
|
aspartoacylase (aminocyclase) 3, tandem duplicate 2 |
| chr13_+_5978809 | 1.33 |
ENSDART00000102563
ENSDART00000121598 |
phf10
|
PHD finger protein 10 |
| chr8_-_52715911 | 1.32 |
ENSDART00000168241
|
tubb2b
|
tubulin, beta 2b |
| chr2_-_22363460 | 1.32 |
ENSDART00000158486
|
selenof
|
selenoprotein F |
| chr8_+_18010568 | 1.32 |
ENSDART00000121984
|
ssbp3b
|
single stranded DNA binding protein 3b |
| chr6_-_10728057 | 1.32 |
ENSDART00000002247
|
sp3b
|
Sp3b transcription factor |
| chr4_-_13973213 | 1.31 |
ENSDART00000067177
ENSDART00000144127 |
prickle1b
|
prickle homolog 1b |
| chr2_-_25140022 | 1.31 |
ENSDART00000134543
|
nceh1a
|
neutral cholesterol ester hydrolase 1a |
| chr11_+_30647545 | 1.30 |
ENSDART00000114792
|
gb:eh507706
|
expressed sequence EH507706 |
| chr5_+_69716458 | 1.30 |
ENSDART00000159594
|
mthfd2
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 2, methenyltetrahydrofolate cyclohydrolase |
| chr10_-_40826657 | 1.29 |
ENSDART00000076304
|
pcna
|
proliferating cell nuclear antigen |
| chr22_+_5135884 | 1.28 |
ENSDART00000141276
|
mydgf
|
myeloid-derived growth factor |
| chr5_+_15495351 | 1.28 |
ENSDART00000111646
ENSDART00000114446 |
suds3
|
SDS3 homolog, SIN3A corepressor complex component |
| chr16_+_11818126 | 1.28 |
ENSDART00000145727
|
cxcr3.2
|
chemokine (C-X-C motif) receptor 3, tandem duplicate 2 |
| chr1_-_22851481 | 1.26 |
ENSDART00000054386
|
qdprb1
|
quinoid dihydropteridine reductase b1 |
| chr14_-_3381303 | 1.26 |
ENSDART00000171601
|
im:7150988
|
im:7150988 |
| chr21_-_4793686 | 1.25 |
ENSDART00000158232
|
notch1a
|
notch 1a |
| chr10_+_8875195 | 1.25 |
ENSDART00000141045
|
itga2.3
|
integrin, alpha 2 (CD49B, alpha 2 subunit of VLA-2 receptor), tandem duplicate 3 |
| chr23_+_30730121 | 1.25 |
ENSDART00000134141
|
asxl1
|
additional sex combs like transcriptional regulator 1 |
| chr3_+_17939828 | 1.23 |
ENSDART00000185047
|
cnp
|
2',3'-cyclic nucleotide 3' phosphodiesterase |
| chr1_-_44484 | 1.22 |
ENSDART00000171547
ENSDART00000164075 ENSDART00000168091 |
tmem39a
|
transmembrane protein 39A |
| chr7_+_56453646 | 1.22 |
ENSDART00000112483
|
slc22a31
|
solute carrier family 22, member 31 |
| chr13_-_35908275 | 1.21 |
ENSDART00000013961
|
mycla
|
MYCL proto-oncogene, bHLH transcription factor a |
| chr2_+_38271392 | 1.21 |
ENSDART00000042100
|
homeza
|
homeobox and leucine zipper encoding a |
| chr5_-_7513082 | 1.20 |
ENSDART00000158913
|
bmpr1ba
|
bone morphogenetic protein receptor, type IBa |
| chr6_+_40775800 | 1.20 |
ENSDART00000085090
|
si:ch211-157b11.8
|
si:ch211-157b11.8 |
| chr9_+_54179306 | 1.20 |
ENSDART00000189829
|
tmsb4x
|
thymosin, beta 4 x |
| chr10_+_9553935 | 1.20 |
ENSDART00000028855
|
si:ch211-243g18.2
|
si:ch211-243g18.2 |
| chr16_+_35401543 | 1.20 |
ENSDART00000171608
|
rab42b
|
RAB42, member RAS oncogene family |
| chr25_+_5015019 | 1.19 |
ENSDART00000127600
|
hdac10
|
histone deacetylase 10 |
| chr10_-_11012000 | 1.19 |
ENSDART00000132995
|
ak3
|
adenylate kinase 3 |
| chr12_-_49168398 | 1.19 |
ENSDART00000186608
|
FO704624.1
|
|
| chr19_-_18152407 | 1.19 |
ENSDART00000193264
ENSDART00000016135 |
nfe2l3
|
nuclear factor, erythroid 2-like 3 |
| chr13_+_30903816 | 1.18 |
ENSDART00000191727
|
ercc6
|
excision repair cross-complementation group 6 |
| chr13_-_35459928 | 1.18 |
ENSDART00000144109
|
slx4ip
|
SLX4 interacting protein |
| chr5_+_37087583 | 1.18 |
ENSDART00000049900
|
tagln2
|
transgelin 2 |
| chr13_+_28495419 | 1.18 |
ENSDART00000025583
|
fgf8a
|
fibroblast growth factor 8a |
| chr2_+_48288461 | 1.18 |
ENSDART00000141495
|
hes6
|
hes family bHLH transcription factor 6 |
| chr1_+_59146298 | 1.18 |
ENSDART00000191885
ENSDART00000152747 |
gpr108
|
G protein-coupled receptor 108 |
| chr20_+_25340814 | 1.17 |
ENSDART00000063028
|
ctgfa
|
connective tissue growth factor a |
| chr13_+_32144370 | 1.17 |
ENSDART00000020270
|
osr1
|
odd-skipped related transciption factor 1 |
| chr6_+_3717613 | 1.17 |
ENSDART00000184330
|
ssb
|
Sjogren syndrome antigen B (autoantigen La) |
| chr21_+_29077509 | 1.17 |
ENSDART00000128561
|
ecscr
|
endothelial cell surface expressed chemotaxis and apoptosis regulator |
| chr21_+_8239544 | 1.17 |
ENSDART00000122773
|
nr6a1b
|
nuclear receptor subfamily 6, group A, member 1b |
| chr14_+_41345175 | 1.16 |
ENSDART00000086104
|
nox1
|
NADPH oxidase 1 |
| chr2_+_3823813 | 1.16 |
ENSDART00000103596
ENSDART00000161880 ENSDART00000185408 |
npc1
|
Niemann-Pick disease, type C1 |
| chr15_+_30310843 | 1.16 |
ENSDART00000112784
|
lyrm9
|
LYR motif containing 9 |
| chr21_+_19834072 | 1.15 |
ENSDART00000147555
|
ccdc80l2
|
coiled-coil domain containing 80 like 2 |
| chr19_+_1872794 | 1.15 |
ENSDART00000013217
|
snrpd1
|
small nuclear ribonucleoprotein D1 polypeptide |
| chr8_-_17997845 | 1.15 |
ENSDART00000121660
|
acot11b
|
acyl-CoA thioesterase 11b |
| chr19_-_34742440 | 1.14 |
ENSDART00000122625
ENSDART00000175621 |
elp2
|
elongator acetyltransferase complex subunit 2 |
| chr16_+_38201840 | 1.14 |
ENSDART00000044971
|
myo1eb
|
myosin IE, b |
| chr5_+_30741730 | 1.14 |
ENSDART00000098246
ENSDART00000186992 ENSDART00000182533 |
ftr83
|
finTRIM family, member 83 |
| chr18_+_41527877 | 1.14 |
ENSDART00000146972
|
selenot1b
|
selenoprotein T, 1b |
| chr12_+_27536270 | 1.14 |
ENSDART00000133719
|
etv4
|
ets variant 4 |
| chr12_+_27537357 | 1.14 |
ENSDART00000136212
|
etv4
|
ets variant 4 |
| chr19_+_47476908 | 1.13 |
ENSDART00000157886
|
zgc:114119
|
zgc:114119 |
| chr16_-_31435020 | 1.13 |
ENSDART00000138508
|
zgc:194210
|
zgc:194210 |
| chr25_-_25575717 | 1.13 |
ENSDART00000067138
|
hic1l
|
hypermethylated in cancer 1 like |
| chr24_-_2423791 | 1.13 |
ENSDART00000190402
|
rreb1a
|
ras responsive element binding protein 1a |
| chr7_+_41812817 | 1.12 |
ENSDART00000174165
|
orc6
|
origin recognition complex, subunit 6 |
| chr18_+_7543347 | 1.11 |
ENSDART00000103467
|
arf5
|
ADP-ribosylation factor 5 |
| chr14_-_38828057 | 1.10 |
ENSDART00000186088
|
spdl1
|
spindle apparatus coiled-coil protein 1 |
| chr14_-_11456724 | 1.10 |
ENSDART00000110424
|
si:ch211-153b23.4
|
si:ch211-153b23.4 |
| chr23_-_45318760 | 1.10 |
ENSDART00000166883
|
ccdc171
|
coiled-coil domain containing 171 |
| chr6_+_40554551 | 1.10 |
ENSDART00000017859
ENSDART00000155928 |
ddi2
|
DNA-damage inducible protein 2 |
| chr21_+_11923701 | 1.09 |
ENSDART00000109292
|
ubap2a
|
ubiquitin associated protein 2a |
| chr3_-_23512285 | 1.09 |
ENSDART00000159151
|
BX682558.1
|
|
| chr25_-_21031007 | 1.09 |
ENSDART00000138985
|
gnaia
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide a |
| chr22_-_23668356 | 1.09 |
ENSDART00000167106
ENSDART00000159622 ENSDART00000163228 |
cfh
|
complement factor H |
| chr5_+_22133153 | 1.09 |
ENSDART00000016214
|
msna
|
moesin a |
| chr4_-_5826320 | 1.09 |
ENSDART00000165354
|
foxm1
|
forkhead box M1 |
| chr21_-_22325124 | 1.09 |
ENSDART00000142100
|
gdpd4b
|
glycerophosphodiester phosphodiesterase domain containing 4b |
| chr5_-_30704390 | 1.07 |
ENSDART00000016709
|
ift22
|
intraflagellar transport 22 homolog (Chlamydomonas) |
| chr17_-_19535328 | 1.07 |
ENSDART00000077809
|
cyp26c1
|
cytochrome P450, family 26, subfamily C, polypeptide 1 |
| chr14_-_26436951 | 1.07 |
ENSDART00000140173
|
si:dkeyp-110e4.6
|
si:dkeyp-110e4.6 |
| chr16_+_40563533 | 1.06 |
ENSDART00000190368
|
tp53inp1
|
tumor protein p53 inducible nuclear protein 1 |
| chr20_+_13883131 | 1.06 |
ENSDART00000003248
ENSDART00000152611 |
nek2
|
NIMA-related kinase 2 |
| chr15_-_23475051 | 1.06 |
ENSDART00000152460
|
nlrx1
|
NLR family member X1 |
| chr3_-_40528333 | 1.06 |
ENSDART00000193047
|
actb2
|
actin, beta 2 |
| chr21_+_30470809 | 1.05 |
ENSDART00000171648
ENSDART00000126678 ENSDART00000003335 ENSDART00000133478 ENSDART00000140811 ENSDART00000147375 |
snx12
|
sorting nexin 12 |
| chr17_+_24809221 | 1.05 |
ENSDART00000082251
ENSDART00000147871 ENSDART00000130871 |
spdya
|
speedy/RINGO cell cycle regulator family member A |
| chr17_-_10000339 | 1.05 |
ENSDART00000162893
|
snx6
|
sorting nexin 6 |
| chr5_-_32396929 | 1.04 |
ENSDART00000023977
|
fbxw2
|
F-box and WD repeat domain containing 2 |
| chr3_-_45281350 | 1.04 |
ENSDART00000020168
|
kctd5a
|
potassium channel tetramerization domain containing 5a |
| chr13_-_35765028 | 1.04 |
ENSDART00000157391
|
erlec1
|
endoplasmic reticulum lectin 1 |
| chr17_+_24064014 | 1.04 |
ENSDART00000182782
ENSDART00000139063 ENSDART00000132755 |
b3gnt2b
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 2b |
| chr7_-_26601307 | 1.04 |
ENSDART00000188934
|
plscr3b
|
phospholipid scramblase 3b |
| chr13_-_42749916 | 1.02 |
ENSDART00000140019
|
capn2a
|
calpain 2, (m/II) large subunit a |
| chr13_-_7575216 | 1.02 |
ENSDART00000159443
|
pitx3
|
paired-like homeodomain 3 |
| chr17_-_33716688 | 1.02 |
ENSDART00000043651
|
dnal1
|
dynein, axonemal, light chain 1 |
| chr19_+_42086862 | 1.02 |
ENSDART00000151605
|
nfyc
|
nuclear transcription factor Y, gamma |
| chr8_+_19489854 | 1.01 |
ENSDART00000184671
ENSDART00000011258 |
npl
|
N-acetylneuraminate pyruvate lyase (dihydrodipicolinate synthase) |
| chr7_+_17543487 | 1.01 |
ENSDART00000126652
|
CU672228.2
|
Danio rerio novel immune-type receptor 1l (nitr1l), mRNA. |
| chr3_-_36839115 | 1.01 |
ENSDART00000154553
|
ramp2
|
receptor (G protein-coupled) activity modifying protein 2 |
| chr15_+_20239141 | 1.01 |
ENSDART00000101152
ENSDART00000152473 |
spint2
|
serine peptidase inhibitor, Kunitz type, 2 |
| chr8_+_30117297 | 1.01 |
ENSDART00000077554
|
fancc
|
Fanconi anemia, complementation group C |
| chr24_-_31090948 | 1.01 |
ENSDART00000176799
|
hccsb
|
holocytochrome c synthase b |
| chr22_-_14115292 | 1.01 |
ENSDART00000105717
ENSDART00000165670 |
aox5
|
aldehyde oxidase 5 |
| chr10_-_23099809 | 1.01 |
ENSDART00000148333
ENSDART00000079703 ENSDART00000162444 |
nle1
|
notchless homolog 1 (Drosophila) |
| chr12_-_4301234 | 1.00 |
ENSDART00000152377
ENSDART00000152521 |
ca15b
|
carbonic anhydrase XVb |
| chr9_+_48819280 | 1.00 |
ENSDART00000112555
|
spc25
|
SPC25, NDC80 kinetochore complex component, homolog (S. cerevisiae) |
| chr13_-_18195942 | 1.00 |
ENSDART00000079902
|
slc25a16
|
solute carrier family 25 (mitochondrial carrier; Graves disease autoantigen), member 16 |
| chr21_-_34972872 | 1.00 |
ENSDART00000023838
|
lipia
|
lipase, member Ia |
| chr1_+_36194761 | 0.99 |
ENSDART00000053773
ENSDART00000147458 |
lsm6
|
LSM6 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr6_-_55423220 | 0.98 |
ENSDART00000158929
|
ctsa
|
cathepsin A |
| chr20_+_19066858 | 0.98 |
ENSDART00000192086
|
sox7
|
SRY (sex determining region Y)-box 7 |
| chr1_+_16127825 | 0.98 |
ENSDART00000122503
|
tusc3
|
tumor suppressor candidate 3 |
| chr1_+_22851745 | 0.98 |
ENSDART00000138235
ENSDART00000016488 |
gtf2e2
|
general transcription factor IIE, polypeptide 2, beta |
| chr9_+_56232548 | 0.97 |
ENSDART00000099276
|
cnot11
|
CCR4-NOT transcription complex, subunit 11 |
| chr10_+_5954787 | 0.97 |
ENSDART00000161887
ENSDART00000160345 ENSDART00000190046 |
map3k1
|
mitogen-activated protein kinase kinase kinase 1, E3 ubiquitin protein ligase |
| chr9_+_34952203 | 0.97 |
ENSDART00000121828
ENSDART00000142347 |
tfdp1a
|
transcription factor Dp-1, a |
| chr18_-_24988645 | 0.97 |
ENSDART00000136434
ENSDART00000085735 |
chd2
|
chromodomain helicase DNA binding protein 2 |
| chr21_+_27340682 | 0.96 |
ENSDART00000011305
|
dpp3
|
dipeptidyl-peptidase 3 |
| chr23_-_9859989 | 0.95 |
ENSDART00000005015
|
prkcbp1l
|
protein kinase C binding protein 1, like |
| chr13_+_24402406 | 0.94 |
ENSDART00000043002
|
rab1ab
|
RAB1A, member RAS oncogene family b |
| chr5_-_31875645 | 0.94 |
ENSDART00000098160
|
tmem119b
|
transmembrane protein 119b |
| chr7_-_17816921 | 0.94 |
ENSDART00000149821
|
ecsit
|
ECSIT signalling integrator |
| chr19_+_46222918 | 0.94 |
ENSDART00000158703
|
vps28
|
vacuolar protein sorting 28 (yeast) |
| chr4_-_11064073 | 0.94 |
ENSDART00000150760
|
si:dkey-21h14.8
|
si:dkey-21h14.8 |
| chr6_+_27992886 | 0.93 |
ENSDART00000160354
|
amotl2a
|
angiomotin like 2a |
Network of associatons between targets according to the STRING database.
First level regulatory network of fli1b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.6 | GO:1905208 | negative regulation of striated muscle cell differentiation(GO:0051154) negative regulation of cardiac muscle tissue development(GO:0055026) cardiac muscle cell fate commitment(GO:0060923) canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:0061317) regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901295) negative regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901296) negative regulation of cardiocyte differentiation(GO:1905208) negative regulation of cardiac muscle cell differentiation(GO:2000726) |
| 0.8 | 2.4 | GO:0003093 | renal system process involved in regulation of blood volume(GO:0001977) renal system process involved in regulation of systemic arterial blood pressure(GO:0003071) regulation of glomerular filtration(GO:0003093) |
| 0.6 | 2.4 | GO:0042117 | monocyte activation(GO:0042117) |
| 0.5 | 1.6 | GO:2000425 | regulation of apoptotic cell clearance(GO:2000425) |
| 0.5 | 1.5 | GO:1903373 | regulation of endoplasmic reticulum tubular network organization(GO:1903371) positive regulation of endoplasmic reticulum tubular network organization(GO:1903373) |
| 0.5 | 0.9 | GO:0051148 | negative regulation of muscle cell differentiation(GO:0051148) |
| 0.4 | 4.0 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.4 | 1.3 | GO:0002522 | leukocyte migration involved in immune response(GO:0002522) |
| 0.4 | 1.3 | GO:0021531 | spinal cord radial glial cell differentiation(GO:0021531) |
| 0.4 | 1.6 | GO:0034087 | establishment of mitotic sister chromatid cohesion(GO:0034087) mitotic sister chromatid cohesion, centromeric(GO:0071962) |
| 0.4 | 2.8 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.4 | 1.6 | GO:2000383 | ectodermal cell fate commitment(GO:0001712) ectodermal cell fate specification(GO:0001715) ectodermal cell differentiation(GO:0010668) regulation of ectodermal cell fate specification(GO:0042665) regulation of ectoderm development(GO:2000383) |
| 0.4 | 1.2 | GO:0048389 | intermediate mesoderm development(GO:0048389) |
| 0.4 | 1.4 | GO:0010259 | multicellular organism aging(GO:0010259) |
| 0.3 | 3.1 | GO:0055011 | atrial cardiac muscle cell differentiation(GO:0055011) |
| 0.3 | 1.0 | GO:0006043 | glucosamine metabolic process(GO:0006041) glucosamine catabolic process(GO:0006043) |
| 0.3 | 1.0 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.3 | 1.3 | GO:0006272 | leading strand elongation(GO:0006272) |
| 0.3 | 1.6 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.3 | 1.6 | GO:0097355 | protein localization to heterochromatin(GO:0097355) |
| 0.3 | 0.9 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.3 | 1.5 | GO:0031062 | positive regulation of histone methylation(GO:0031062) |
| 0.3 | 0.9 | GO:1903173 | fatty alcohol metabolic process(GO:1903173) |
| 0.3 | 1.2 | GO:0009447 | polyamine catabolic process(GO:0006598) putrescine catabolic process(GO:0009447) |
| 0.3 | 0.9 | GO:1990120 | messenger ribonucleoprotein complex assembly(GO:1990120) |
| 0.3 | 0.3 | GO:0071788 | endoplasmic reticulum tubular network maintenance(GO:0071788) |
| 0.3 | 1.7 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.3 | 0.8 | GO:1901890 | positive regulation of cell junction assembly(GO:1901890) |
| 0.3 | 1.6 | GO:0051561 | positive regulation of mitochondrial calcium ion concentration(GO:0051561) |
| 0.3 | 0.8 | GO:0033690 | positive regulation of osteoblast proliferation(GO:0033690) |
| 0.3 | 1.1 | GO:0048211 | Golgi vesicle docking(GO:0048211) |
| 0.3 | 1.8 | GO:1901673 | regulation of mitotic spindle assembly(GO:1901673) |
| 0.3 | 1.3 | GO:0048245 | eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) |
| 0.2 | 0.7 | GO:0050666 | regulation of sulfur amino acid metabolic process(GO:0031335) regulation of homocysteine metabolic process(GO:0050666) |
| 0.2 | 1.2 | GO:0030219 | megakaryocyte differentiation(GO:0030219) |
| 0.2 | 2.3 | GO:0061056 | sclerotome development(GO:0061056) |
| 0.2 | 1.6 | GO:0061298 | retina vasculature development in camera-type eye(GO:0061298) |
| 0.2 | 0.9 | GO:0042779 | tRNA 3'-trailer cleavage(GO:0042779) |
| 0.2 | 1.4 | GO:0001937 | negative regulation of endothelial cell proliferation(GO:0001937) |
| 0.2 | 1.0 | GO:0017003 | protein-heme linkage(GO:0017003) protein-tetrapyrrole linkage(GO:0017006) cytochrome c-heme linkage(GO:0018063) |
| 0.2 | 0.8 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.2 | 1.4 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.2 | 1.4 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.2 | 0.6 | GO:0021563 | glossopharyngeal nerve development(GO:0021563) |
| 0.2 | 1.5 | GO:0042554 | superoxide anion generation(GO:0042554) |
| 0.2 | 0.7 | GO:0050732 | negative regulation of peptidyl-tyrosine phosphorylation(GO:0050732) negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.2 | 1.6 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.2 | 3.9 | GO:0010737 | protein kinase A signaling(GO:0010737) |
| 0.2 | 0.9 | GO:0019388 | galactose catabolic process(GO:0019388) galactose catabolic process via UDP-galactose(GO:0033499) |
| 0.2 | 6.7 | GO:0010389 | regulation of G2/M transition of mitotic cell cycle(GO:0010389) |
| 0.2 | 1.2 | GO:0006172 | ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) |
| 0.2 | 1.2 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.2 | 1.7 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.2 | 0.8 | GO:0048320 | axial mesoderm formation(GO:0048320) |
| 0.2 | 3.3 | GO:0045580 | regulation of T cell differentiation(GO:0045580) |
| 0.2 | 1.5 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.2 | 2.4 | GO:0035108 | limb morphogenesis(GO:0035108) |
| 0.2 | 0.6 | GO:0042308 | regulation of protein import into nucleus(GO:0042306) negative regulation of protein import into nucleus(GO:0042308) negative regulation of nucleocytoplasmic transport(GO:0046823) negative regulation of protein localization to nucleus(GO:1900181) regulation of protein import(GO:1904589) negative regulation of protein import(GO:1904590) |
| 0.2 | 0.5 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.2 | 2.4 | GO:0007096 | regulation of exit from mitosis(GO:0007096) |
| 0.2 | 4.8 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.1 | 1.6 | GO:2000223 | regulation of BMP signaling pathway involved in heart jogging(GO:2000223) |
| 0.1 | 0.4 | GO:0000066 | mitochondrial ornithine transport(GO:0000066) mitochondrial L-ornithine transmembrane transport(GO:1990575) |
| 0.1 | 0.7 | GO:1901841 | regulation of high voltage-gated calcium channel activity(GO:1901841) negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.1 | 2.2 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.1 | 0.6 | GO:0016038 | absorption of visible light(GO:0016038) |
| 0.1 | 0.6 | GO:0015919 | peroxisomal membrane transport(GO:0015919) protein import into peroxisome membrane(GO:0045046) |
| 0.1 | 1.4 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.1 | 2.1 | GO:0061055 | myotome development(GO:0061055) |
| 0.1 | 0.5 | GO:1900060 | regulation of sphingolipid biosynthetic process(GO:0090153) negative regulation of sphingolipid biosynthetic process(GO:0090155) negative regulation of ceramide biosynthetic process(GO:1900060) regulation of membrane lipid metabolic process(GO:1905038) regulation of ceramide biosynthetic process(GO:2000303) |
| 0.1 | 1.4 | GO:0072091 | regulation of stem cell proliferation(GO:0072091) |
| 0.1 | 0.5 | GO:0015868 | intracellular nucleoside transport(GO:0015859) purine nucleoside transmembrane transport(GO:0015860) purine nucleotide transport(GO:0015865) ATP transport(GO:0015867) purine ribonucleotide transport(GO:0015868) adenine nucleotide transport(GO:0051503) mitochondrial ATP transmembrane transport(GO:1990544) |
| 0.1 | 0.5 | GO:0072116 | kidney rudiment formation(GO:0072003) pronephros formation(GO:0072116) |
| 0.1 | 1.4 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.1 | 0.4 | GO:0051177 | meiosis II(GO:0007135) meiotic sister chromatid segregation(GO:0045144) meiotic sister chromatid cohesion(GO:0051177) |
| 0.1 | 0.9 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.1 | 1.2 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.1 | 0.5 | GO:0046324 | regulation of glucose import(GO:0046324) |
| 0.1 | 1.3 | GO:0021535 | cell migration in hindbrain(GO:0021535) |
| 0.1 | 1.5 | GO:0043951 | regulation of cAMP-mediated signaling(GO:0043949) negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.1 | 0.9 | GO:0035909 | aorta morphogenesis(GO:0035909) dorsal aorta morphogenesis(GO:0035912) |
| 0.1 | 1.7 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.1 | 1.5 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.1 | 1.9 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.1 | 0.4 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.1 | 1.1 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.1 | 0.9 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.1 | 0.6 | GO:0097101 | blood vessel endothelial cell fate specification(GO:0097101) |
| 0.1 | 1.3 | GO:1902101 | positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of mitotic sister chromatid separation(GO:1901970) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.1 | 0.2 | GO:0003097 | renal water homeostasis(GO:0003091) renal water transport(GO:0003097) |
| 0.1 | 0.8 | GO:0021550 | medulla oblongata development(GO:0021550) dorsal motor nucleus of vagus nerve development(GO:0021744) |
| 0.1 | 0.7 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.1 | 1.1 | GO:0071156 | regulation of cell cycle arrest(GO:0071156) |
| 0.1 | 1.1 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.1 | 0.6 | GO:0070734 | histone H3-K27 methylation(GO:0070734) |
| 0.1 | 0.7 | GO:0046070 | dGTP catabolic process(GO:0006203) purine nucleoside triphosphate catabolic process(GO:0009146) purine deoxyribonucleoside triphosphate catabolic process(GO:0009217) dGTP metabolic process(GO:0046070) |
| 0.1 | 2.5 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.1 | 1.5 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.1 | 0.9 | GO:1902624 | positive regulation of neutrophil migration(GO:1902624) |
| 0.1 | 0.6 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.1 | 1.3 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.1 | 2.9 | GO:0060037 | pharyngeal system development(GO:0060037) |
| 0.1 | 1.6 | GO:0021984 | adenohypophysis development(GO:0021984) |
| 0.1 | 0.6 | GO:0000098 | sulfur amino acid catabolic process(GO:0000098) |
| 0.1 | 0.3 | GO:0036336 | dendritic cell chemotaxis(GO:0002407) dendritic cell migration(GO:0036336) |
| 0.1 | 0.4 | GO:2001223 | negative regulation of neuron migration(GO:2001223) |
| 0.1 | 0.5 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.1 | 0.6 | GO:0042116 | macrophage activation(GO:0042116) |
| 0.1 | 0.5 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.1 | 0.5 | GO:0071800 | podosome assembly(GO:0071800) |
| 0.1 | 0.2 | GO:0097510 | base-excision repair, AP site formation via deaminated base removal(GO:0097510) |
| 0.1 | 1.3 | GO:0001936 | regulation of endothelial cell proliferation(GO:0001936) positive regulation of endothelial cell proliferation(GO:0001938) |
| 0.1 | 0.6 | GO:0043584 | nose development(GO:0043584) |
| 0.1 | 1.0 | GO:0030311 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.1 | 0.4 | GO:0048011 | neurotrophin TRK receptor signaling pathway(GO:0048011) |
| 0.1 | 1.1 | GO:0061154 | endothelial tube morphogenesis(GO:0061154) blood vessel lumenization(GO:0072554) |
| 0.1 | 0.5 | GO:0090497 | mesenchymal cell migration(GO:0090497) |
| 0.1 | 1.2 | GO:0043535 | regulation of blood vessel endothelial cell migration(GO:0043535) |
| 0.1 | 1.0 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.1 | 0.3 | GO:0099563 | modification of synaptic structure(GO:0099563) |
| 0.1 | 1.7 | GO:0009214 | cyclic nucleotide catabolic process(GO:0009214) |
| 0.1 | 0.4 | GO:0035479 | angioblast cell migration from lateral mesoderm to midline(GO:0035479) |
| 0.1 | 1.9 | GO:0060840 | artery development(GO:0060840) |
| 0.1 | 0.2 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.1 | 1.4 | GO:0002097 | tRNA wobble base modification(GO:0002097) |
| 0.1 | 0.5 | GO:0048208 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.1 | 1.2 | GO:0060324 | face development(GO:0060324) |
| 0.1 | 1.6 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.1 | 1.4 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.1 | 1.4 | GO:0036342 | post-anal tail morphogenesis(GO:0036342) |
| 0.1 | 2.8 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.1 | 2.5 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.1 | 0.2 | GO:0002637 | regulation of immunoglobulin production(GO:0002637) positive regulation of immunoglobulin production(GO:0002639) |
| 0.1 | 0.1 | GO:0021501 | prechordal plate formation(GO:0021501) |
| 0.1 | 0.3 | GO:0036498 | IRE1-mediated unfolded protein response(GO:0036498) protein ufmylation(GO:0071569) protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.1 | 0.2 | GO:1903537 | meiotic cell cycle process involved in oocyte maturation(GO:1903537) regulation of meiotic cell cycle process involved in oocyte maturation(GO:1903538) |
| 0.1 | 0.6 | GO:0060021 | palate development(GO:0060021) |
| 0.0 | 0.3 | GO:1900028 | negative regulation of Rac protein signal transduction(GO:0035021) wound healing, spreading of epidermal cells(GO:0035313) negative regulation of ruffle assembly(GO:1900028) |
| 0.0 | 0.7 | GO:0031163 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.5 | GO:0031269 | pseudopodium organization(GO:0031268) pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.3 | GO:0097534 | lymphoid lineage cell migration(GO:0097534) lymphoid lineage cell migration into thymus(GO:0097535) |
| 0.0 | 0.7 | GO:0046051 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 0.0 | 0.4 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.0 | 1.4 | GO:0006406 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.0 | 1.1 | GO:0071910 | determination of liver left/right asymmetry(GO:0071910) |
| 0.0 | 1.5 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 1.2 | GO:0048263 | determination of dorsal identity(GO:0048263) |
| 0.0 | 0.2 | GO:2001046 | positive regulation of integrin-mediated signaling pathway(GO:2001046) |
| 0.0 | 0.8 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.0 | 0.2 | GO:1903523 | negative regulation of heart contraction(GO:0045822) negative regulation of blood circulation(GO:1903523) |
| 0.0 | 0.3 | GO:0031179 | peptide modification(GO:0031179) |
| 0.0 | 0.7 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.0 | 2.4 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 1.0 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.3 | GO:1902038 | positive regulation of hematopoietic stem cell differentiation(GO:1902038) |
| 0.0 | 0.7 | GO:0033077 | T cell differentiation in thymus(GO:0033077) thymocyte aggregation(GO:0071594) |
| 0.0 | 0.2 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 | 0.9 | GO:1902653 | cholesterol biosynthetic process(GO:0006695) secondary alcohol biosynthetic process(GO:1902653) |
| 0.0 | 0.3 | GO:0055090 | acylglycerol homeostasis(GO:0055090) triglyceride homeostasis(GO:0070328) |
| 0.0 | 0.6 | GO:0033962 | cytoplasmic mRNA processing body assembly(GO:0033962) |
| 0.0 | 0.7 | GO:0051351 | positive regulation of ligase activity(GO:0051351) |
| 0.0 | 1.0 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 1.0 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.0 | 0.3 | GO:0032099 | negative regulation of response to food(GO:0032096) negative regulation of appetite(GO:0032099) |
| 0.0 | 0.3 | GO:0010889 | regulation of sequestering of triglyceride(GO:0010889) positive regulation of sequestering of triglyceride(GO:0010890) sequestering of triglyceride(GO:0030730) |
| 0.0 | 0.8 | GO:0030518 | intracellular steroid hormone receptor signaling pathway(GO:0030518) |
| 0.0 | 1.9 | GO:1902600 | hydrogen ion transmembrane transport(GO:1902600) |
| 0.0 | 0.2 | GO:0060333 | interferon-gamma-mediated signaling pathway(GO:0060333) |
| 0.0 | 0.7 | GO:0070306 | lens fiber cell differentiation(GO:0070306) |
| 0.0 | 1.7 | GO:0006352 | DNA-templated transcription, initiation(GO:0006352) |
| 0.0 | 0.1 | GO:0000390 | spliceosomal complex disassembly(GO:0000390) |
| 0.0 | 0.1 | GO:0042560 | 10-formyltetrahydrofolate metabolic process(GO:0009256) 10-formyltetrahydrofolate catabolic process(GO:0009258) folic acid-containing compound catabolic process(GO:0009397) pteridine-containing compound catabolic process(GO:0042560) |
| 0.0 | 1.0 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.0 | 1.8 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 0.2 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.0 | 0.7 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.0 | 0.3 | GO:0031577 | mitotic spindle assembly checkpoint(GO:0007094) spindle checkpoint(GO:0031577) negative regulation of mitotic metaphase/anaphase transition(GO:0045841) spindle assembly checkpoint(GO:0071173) mitotic spindle checkpoint(GO:0071174) |
| 0.0 | 0.8 | GO:0051294 | establishment of spindle orientation(GO:0051294) |
| 0.0 | 0.8 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.0 | 0.4 | GO:0045740 | positive regulation of DNA replication(GO:0045740) |
| 0.0 | 0.7 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.0 | 2.1 | GO:0071559 | transforming growth factor beta receptor signaling pathway(GO:0007179) response to transforming growth factor beta(GO:0071559) cellular response to transforming growth factor beta stimulus(GO:0071560) |
| 0.0 | 0.2 | GO:0035476 | angioblast cell migration(GO:0035476) |
| 0.0 | 0.2 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.6 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.3 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.0 | 2.8 | GO:0035383 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.0 | 0.1 | GO:0046632 | alpha-beta T cell differentiation(GO:0046632) |
| 0.0 | 1.1 | GO:0031647 | regulation of protein stability(GO:0031647) |
| 0.0 | 1.1 | GO:0070830 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.0 | 1.3 | GO:0035825 | reciprocal DNA recombination(GO:0035825) |
| 0.0 | 0.5 | GO:0042953 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.0 | 0.4 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.6 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.0 | 0.8 | GO:0001706 | endoderm formation(GO:0001706) |
| 0.0 | 1.0 | GO:0007173 | epidermal growth factor receptor signaling pathway(GO:0007173) |
| 0.0 | 2.5 | GO:0008033 | tRNA processing(GO:0008033) |
| 0.0 | 1.1 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.5 | GO:1904894 | positive regulation of JAK-STAT cascade(GO:0046427) positive regulation of STAT cascade(GO:1904894) |
| 0.0 | 2.1 | GO:0008285 | negative regulation of cell proliferation(GO:0008285) |
| 0.0 | 0.3 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.0 | 0.5 | GO:0006879 | cellular iron ion homeostasis(GO:0006879) |
| 0.0 | 0.3 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.0 | 0.1 | GO:0042364 | water-soluble vitamin biosynthetic process(GO:0042364) |
| 0.0 | 0.9 | GO:0035082 | axoneme assembly(GO:0035082) |
| 0.0 | 0.4 | GO:0048010 | vascular endothelial growth factor receptor signaling pathway(GO:0048010) |
| 0.0 | 0.1 | GO:0061316 | canonical Wnt signaling pathway involved in heart development(GO:0061316) regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) |
| 0.0 | 0.1 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 0.0 | 0.2 | GO:0030878 | thyroid gland development(GO:0030878) |
| 0.0 | 0.2 | GO:0051121 | lipoxygenase pathway(GO:0019372) linoleic acid metabolic process(GO:0043651) hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.0 | 0.1 | GO:0019563 | glycerol catabolic process(GO:0019563) |
| 0.0 | 1.0 | GO:0031623 | receptor internalization(GO:0031623) |
| 0.0 | 0.9 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.0 | 0.3 | GO:0021988 | olfactory bulb development(GO:0021772) olfactory lobe development(GO:0021988) |
| 0.0 | 0.1 | GO:0032119 | sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 0.4 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 0.4 | GO:0031952 | regulation of protein autophosphorylation(GO:0031952) |
| 0.0 | 0.3 | GO:0016203 | muscle attachment(GO:0016203) |
| 0.0 | 0.4 | GO:0007568 | aging(GO:0007568) |
| 0.0 | 0.2 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.0 | 0.4 | GO:0048332 | mesoderm morphogenesis(GO:0048332) |
| 0.0 | 0.3 | GO:0006144 | purine nucleobase metabolic process(GO:0006144) |
| 0.0 | 0.3 | GO:0030500 | regulation of bone mineralization(GO:0030500) regulation of biomineral tissue development(GO:0070167) |
| 0.0 | 1.6 | GO:0071466 | cellular response to xenobiotic stimulus(GO:0071466) |
| 0.0 | 0.5 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.1 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 3.1 | GO:0000122 | negative regulation of transcription from RNA polymerase II promoter(GO:0000122) |
| 0.0 | 0.4 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.0 | 0.1 | GO:0006116 | NADH oxidation(GO:0006116) glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.0 | 0.1 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.0 | 0.9 | GO:0031018 | endocrine pancreas development(GO:0031018) |
| 0.0 | 0.4 | GO:0001570 | vasculogenesis(GO:0001570) |
| 0.0 | 0.3 | GO:0050727 | regulation of inflammatory response(GO:0050727) |
| 0.0 | 0.3 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 0.7 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
| 0.0 | 2.8 | GO:0032259 | methylation(GO:0032259) |
| 0.0 | 2.8 | GO:0045892 | negative regulation of transcription, DNA-templated(GO:0045892) |
| 0.0 | 0.2 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.5 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 0.2 | GO:0090481 | pyrimidine nucleotide-sugar transport(GO:0015781) pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.0 | 0.1 | GO:0046512 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.0 | 0.4 | GO:0030917 | midbrain-hindbrain boundary development(GO:0030917) |
| 0.0 | 0.7 | GO:0061640 | cytoskeleton-dependent cytokinesis(GO:0061640) |
| 0.0 | 3.2 | GO:0046777 | protein autophosphorylation(GO:0046777) |
| 0.0 | 0.2 | GO:0030851 | granulocyte differentiation(GO:0030851) |
| 0.0 | 0.3 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 2.9 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 0.5 | 3.2 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.5 | 1.5 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.5 | 1.5 | GO:0098826 | endoplasmic reticulum tubular network membrane(GO:0098826) |
| 0.5 | 3.5 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.5 | 2.4 | GO:0005673 | transcription factor TFIIE complex(GO:0005673) |
| 0.4 | 1.6 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.3 | 1.6 | GO:0031415 | NatA complex(GO:0031415) |
| 0.3 | 1.2 | GO:0035301 | Hedgehog signaling complex(GO:0035301) |
| 0.3 | 0.9 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.3 | 1.3 | GO:0097433 | dense body(GO:0097433) |
| 0.2 | 1.7 | GO:0072487 | MSL complex(GO:0072487) |
| 0.2 | 2.5 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.2 | 1.4 | GO:0045261 | proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.2 | 0.6 | GO:0097361 | CIA complex(GO:0097361) |
| 0.2 | 0.8 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.2 | 0.8 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.2 | 1.5 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.2 | 0.9 | GO:1990131 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.2 | 1.3 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 0.2 | 0.5 | GO:0042709 | succinate-CoA ligase complex(GO:0042709) |
| 0.2 | 1.9 | GO:0045095 | keratin filament(GO:0045095) |
| 0.2 | 1.4 | GO:0016442 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.2 | 1.7 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.2 | 1.0 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.2 | 1.3 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.2 | 1.6 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.2 | 0.9 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.1 | 1.5 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.1 | 0.4 | GO:1990879 | CST complex(GO:1990879) |
| 0.1 | 0.5 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.1 | 1.5 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.1 | 1.9 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 2.2 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.1 | 0.5 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.1 | 1.3 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.1 | 1.1 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.1 | 0.3 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.1 | 0.4 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.1 | 1.2 | GO:0030904 | retromer complex(GO:0030904) |
| 0.1 | 1.0 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.1 | 1.0 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.1 | 0.5 | GO:0070390 | transcription export complex 2(GO:0070390) |
| 0.1 | 4.7 | GO:0030496 | midbody(GO:0030496) |
| 0.1 | 0.9 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.1 | 0.6 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.1 | 1.2 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.1 | 1.3 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.1 | 1.6 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.1 | 1.1 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.1 | 1.1 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 1.1 | GO:0044545 | NSL complex(GO:0044545) |
| 0.1 | 1.4 | GO:0000780 | condensed nuclear chromosome kinetochore(GO:0000778) condensed nuclear chromosome, centromeric region(GO:0000780) |
| 0.1 | 1.6 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.1 | 0.7 | GO:0070449 | elongin complex(GO:0070449) |
| 0.1 | 0.6 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.1 | 1.2 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.1 | 0.7 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.1 | 0.9 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.1 | 0.2 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 0.1 | 1.0 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.1 | 0.4 | GO:0034990 | nuclear cohesin complex(GO:0000798) mitotic cohesin complex(GO:0030892) nuclear mitotic cohesin complex(GO:0034990) nuclear meiotic cohesin complex(GO:0034991) |
| 0.1 | 1.0 | GO:0033178 | proton-transporting two-sector ATPase complex, catalytic domain(GO:0033178) |
| 0.1 | 0.5 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.8 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 1.5 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 1.6 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.9 | GO:0034399 | nuclear periphery(GO:0034399) |
| 0.0 | 2.1 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 1.3 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.7 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 5.9 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 1.0 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 1.4 | GO:0005901 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.0 | 0.4 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.7 | GO:0005838 | proteasome regulatory particle(GO:0005838) |
| 0.0 | 1.1 | GO:0005940 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 0.3 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 1.2 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.6 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.6 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.3 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 0.6 | GO:0005605 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.0 | 0.5 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 1.2 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.4 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.6 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 3.2 | GO:0070160 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 1.9 | GO:0032592 | integral component of mitochondrial membrane(GO:0032592) |
| 0.0 | 1.6 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 0.5 | GO:0016282 | eukaryotic 43S preinitiation complex(GO:0016282) |
| 0.0 | 0.3 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.0 | 2.4 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.1 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 1.5 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.4 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.0 | 5.7 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.1 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.0 | 1.1 | GO:0030286 | dynein complex(GO:0030286) |
| 0.0 | 0.1 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.0 | 1.2 | GO:0031228 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 1.7 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.0 | 0.1 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.3 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.2 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 0.5 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.6 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.6 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.2 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.4 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.2 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.3 | GO:0032040 | small-subunit processome(GO:0032040) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 3.7 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.7 | 2.0 | GO:0004113 | 2',3'-cyclic-nucleotide 3'-phosphodiesterase activity(GO:0004113) |
| 0.6 | 1.8 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.6 | 3.0 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.5 | 1.9 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.4 | 1.5 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.4 | 1.4 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.3 | 1.7 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.3 | 1.0 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 0.3 | 1.0 | GO:0016623 | aldehyde oxidase activity(GO:0004031) oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.3 | 1.7 | GO:0016886 | ligase activity, forming phosphoric ester bonds(GO:0016886) |
| 0.3 | 2.2 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.3 | 1.5 | GO:0060182 | apelin receptor activity(GO:0060182) |
| 0.3 | 1.2 | GO:0047611 | acetylspermidine deacetylase activity(GO:0047611) |
| 0.3 | 2.4 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.3 | 1.1 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.3 | 3.0 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.3 | 1.3 | GO:0008430 | selenium binding(GO:0008430) |
| 0.3 | 1.3 | GO:0098973 | structural constituent of postsynaptic actin cytoskeleton(GO:0098973) |
| 0.3 | 1.3 | GO:0019863 | IgE binding(GO:0019863) immunoglobulin binding(GO:0019865) |
| 0.3 | 2.0 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.3 | 1.3 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.2 | 1.7 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.2 | 0.9 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.2 | 1.3 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.2 | 1.0 | GO:0004408 | holocytochrome-c synthase activity(GO:0004408) |
| 0.2 | 0.6 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.2 | 1.8 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.2 | 0.8 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.2 | 0.6 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.2 | 0.5 | GO:0016165 | linoleate 13S-lipoxygenase activity(GO:0016165) |
| 0.2 | 0.5 | GO:0004774 | succinate-CoA ligase activity(GO:0004774) succinate-CoA ligase (ADP-forming) activity(GO:0004775) succinate-CoA ligase (GDP-forming) activity(GO:0004776) |
| 0.2 | 0.9 | GO:1902387 | ceramide transporter activity(GO:0035620) ceramide 1-phosphate binding(GO:1902387) ceramide 1-phosphate transporter activity(GO:1902388) |
| 0.2 | 2.1 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.2 | 1.7 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.2 | 1.2 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.2 | 1.4 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.2 | 1.7 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.1 | 0.4 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.1 | 0.4 | GO:0043621 | protein self-association(GO:0043621) |
| 0.1 | 0.6 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.1 | 1.3 | GO:0004488 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) methylenetetrahydrofolate dehydrogenase (NADP+) activity(GO:0004488) |
| 0.1 | 0.9 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.1 | 0.7 | GO:0004122 | cystathionine beta-synthase activity(GO:0004122) |
| 0.1 | 1.4 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.1 | 0.9 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 0.5 | GO:0005471 | ATP:ADP antiporter activity(GO:0005471) |
| 0.1 | 0.8 | GO:1903924 | estradiol binding(GO:1903924) |
| 0.1 | 2.5 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.1 | 0.7 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.1 | 1.1 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.1 | 1.8 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.1 | 0.8 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.1 | 1.5 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.1 | 0.3 | GO:0003920 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.1 | 0.3 | GO:0015462 | protein-transmembrane transporting ATPase activity(GO:0015462) |
| 0.1 | 0.7 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.1 | 1.4 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.1 | 1.0 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.1 | 1.2 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.1 | 0.7 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.1 | 0.5 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.1 | 0.3 | GO:0052717 | tRNA-specific adenosine-34 deaminase activity(GO:0052717) |
| 0.1 | 0.3 | GO:0001635 | adrenomedullin receptor activity(GO:0001605) calcitonin gene-related peptide receptor activity(GO:0001635) |
| 0.1 | 0.8 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.1 | 1.3 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.1 | 1.2 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.1 | 1.2 | GO:0005537 | mannose binding(GO:0005537) |
| 0.1 | 0.3 | GO:0071568 | UFM1 transferase activity(GO:0071568) |
| 0.1 | 0.9 | GO:0017091 | AU-rich element binding(GO:0017091) mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.1 | 1.0 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.1 | 0.8 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.1 | 1.6 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.1 | 0.2 | GO:0004394 | heparan sulfate 2-O-sulfotransferase activity(GO:0004394) |
| 0.1 | 1.0 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.1 | 0.7 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.1 | 0.4 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.1 | 0.3 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.1 | 1.8 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.1 | 0.8 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.1 | 6.4 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.1 | 0.7 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.1 | 1.6 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.1 | 4.1 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.1 | 1.4 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.1 | 0.9 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.1 | 1.1 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.1 | 0.5 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 0.2 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.1 | 0.2 | GO:0052855 | ATP-dependent NAD(P)H-hydrate dehydratase activity(GO:0047453) ADP-dependent NAD(P)H-hydrate dehydratase activity(GO:0052855) |
| 0.1 | 0.8 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 1.0 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.1 | 1.0 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.1 | 0.9 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.1 | 0.2 | GO:0000996 | core DNA-dependent RNA polymerase binding promoter specificity activity(GO:0000996) mitochondrial RNA polymerase binding promoter specificity activity(GO:0034246) |
| 0.1 | 0.3 | GO:0042974 | retinoic acid receptor binding(GO:0042974) |
| 0.1 | 1.0 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.1 | 0.7 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.1 | 1.3 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.4 | GO:0070137 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.0 | 0.2 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.0 | 1.4 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 1.6 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.0 | 0.6 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.0 | 1.3 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.7 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.6 | GO:0055102 | phospholipase inhibitor activity(GO:0004859) lipase inhibitor activity(GO:0055102) |
| 0.0 | 1.7 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 2.8 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 4.0 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.3 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.7 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.5 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.4 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.0 | 0.3 | GO:0000048 | peptidyltransferase activity(GO:0000048) glutathione hydrolase activity(GO:0036374) |
| 0.0 | 6.3 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.7 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 1.0 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.2 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.0 | 0.7 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.1 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.0 | 0.4 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.5 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 1.0 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.2 | GO:0046592 | polyamine oxidase activity(GO:0046592) |
| 0.0 | 0.5 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.0 | 0.6 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.2 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.2 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.8 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.0 | 0.1 | GO:0016155 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) |
| 0.0 | 0.6 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 0.5 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.0 | 1.3 | GO:0051536 | iron-sulfur cluster binding(GO:0051536) metal cluster binding(GO:0051540) |
| 0.0 | 0.7 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 1.1 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 0.2 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.0 | 0.5 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) kinase inhibitor activity(GO:0019210) |
| 0.0 | 0.3 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.6 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.7 | GO:0008378 | galactosyltransferase activity(GO:0008378) |
| 0.0 | 0.5 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.0 | 0.5 | GO:0033549 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) MAP kinase phosphatase activity(GO:0033549) |
| 0.0 | 0.5 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.0 | 0.8 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.2 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.4 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 1.1 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) |
| 0.0 | 0.1 | GO:0043185 | vascular endothelial growth factor receptor 3 binding(GO:0043185) |
| 0.0 | 0.1 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.0 | 0.9 | GO:0008094 | DNA-dependent ATPase activity(GO:0008094) |
| 0.0 | 2.9 | GO:0004386 | helicase activity(GO:0004386) |
| 0.0 | 0.3 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.3 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.5 | GO:0004697 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.0 | 5.6 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.0 | 0.2 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.1 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.1 | GO:0051920 | peroxiredoxin activity(GO:0051920) |
| 0.0 | 0.2 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.0 | 0.4 | GO:0031543 | peptidyl-proline dioxygenase activity(GO:0031543) |
| 0.0 | 0.2 | GO:0043028 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) cysteine-type endopeptidase regulator activity involved in apoptotic process(GO:0043028) |
| 0.0 | 0.5 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.3 | GO:0004467 | long-chain fatty acid-CoA ligase activity(GO:0004467) |
| 0.0 | 0.8 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.1 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.0 | 2.3 | GO:0060090 | binding, bridging(GO:0060090) |
| 0.0 | 0.2 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.5 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.2 | GO:0004844 | uracil DNA N-glycosylase activity(GO:0004844) deaminated base DNA N-glycosylase activity(GO:0097506) |
| 0.0 | 0.1 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.0 | 0.6 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.6 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 24.3 | GO:0000981 | RNA polymerase II transcription factor activity, sequence-specific DNA binding(GO:0000981) |
| 0.0 | 0.6 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.6 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.0 | 1.1 | GO:0042393 | histone binding(GO:0042393) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 0.4 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.2 | 2.5 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.2 | 2.8 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.1 | 1.0 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.1 | 4.2 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.1 | 1.8 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 1.7 | PID MYC PATHWAY | C-MYC pathway |
| 0.1 | 3.3 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.1 | 2.5 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.1 | 0.7 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.1 | 2.1 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.1 | 4.4 | PID E2F PATHWAY | E2F transcription factor network |
| 0.1 | 0.4 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.1 | 0.2 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.0 | 1.2 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.8 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 1.5 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 1.9 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.2 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 1.2 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.6 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 0.5 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.6 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.4 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 1.0 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.7 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.5 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.5 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 0.9 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 0.4 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 1.0 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.4 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.9 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.3 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.0 | 1.4 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.3 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.2 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.2 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.9 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.2 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 0.4 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.2 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 0.3 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.2 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.1 | PID CD40 PATHWAY | CD40/CD40L signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 5.4 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.3 | 1.1 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.2 | 0.6 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.2 | 1.8 | REACTOME P75NTR RECRUITS SIGNALLING COMPLEXES | Genes involved in p75NTR recruits signalling complexes |
| 0.1 | 1.6 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.1 | 1.5 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.1 | 1.6 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 1.9 | REACTOME RNA POL I TRANSCRIPTION INITIATION | Genes involved in RNA Polymerase I Transcription Initiation |
| 0.1 | 4.1 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.1 | 1.3 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.1 | 0.9 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 0.4 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.1 | 2.0 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.1 | 1.3 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.1 | 0.5 | REACTOME INTERACTIONS OF VPR WITH HOST CELLULAR PROTEINS | Genes involved in Interactions of Vpr with host cellular proteins |
| 0.1 | 2.8 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.1 | 0.8 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.1 | 1.6 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.1 | 0.4 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.1 | 1.7 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.1 | 1.0 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.1 | 0.4 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.1 | 0.5 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.1 | 2.0 | REACTOME APC CDC20 MEDIATED DEGRADATION OF NEK2A | Genes involved in APC-Cdc20 mediated degradation of Nek2A |
| 0.1 | 1.4 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 4.8 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.1 | 3.2 | REACTOME RIG I MDA5 MEDIATED INDUCTION OF IFN ALPHA BETA PATHWAYS | Genes involved in RIG-I/MDA5 mediated induction of IFN-alpha/beta pathways |
| 0.1 | 1.0 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.1 | 1.0 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.1 | 2.2 | REACTOME AUTODEGRADATION OF CDH1 BY CDH1 APC C | Genes involved in Autodegradation of Cdh1 by Cdh1:APC/C |
| 0.1 | 0.5 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 1.7 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.1 | 1.0 | REACTOME LATE PHASE OF HIV LIFE CYCLE | Genes involved in Late Phase of HIV Life Cycle |
| 0.0 | 0.6 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.7 | REACTOME ADP SIGNALLING THROUGH P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 0.0 | 0.4 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.0 | 0.3 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.2 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.0 | 0.7 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.7 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.5 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.7 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.6 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 1.0 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.5 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.6 | REACTOME FGFR LIGAND BINDING AND ACTIVATION | Genes involved in FGFR ligand binding and activation |
| 0.0 | 0.7 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 1.5 | REACTOME TRAF6 MEDIATED INDUCTION OF NFKB AND MAP KINASES UPON TLR7 8 OR 9 ACTIVATION | Genes involved in TRAF6 mediated induction of NFkB and MAP kinases upon TLR7/8 or 9 activation |
| 0.0 | 1.0 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 0.3 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 0.6 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 0.3 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 0.4 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.0 | 0.4 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.2 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.7 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.1 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.1 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.2 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.7 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.2 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.2 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.0 | 1.0 | REACTOME ASPARAGINE N LINKED GLYCOSYLATION | Genes involved in Asparagine N-linked glycosylation |
| 0.0 | 0.3 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.2 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 0.2 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.6 | REACTOME DNA REPAIR | Genes involved in DNA Repair |