Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
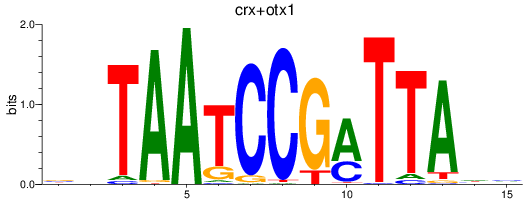
Results for crx+otx1
Z-value: 1.17
Transcription factors associated with crx+otx1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
crx
|
ENSDARG00000011989 | cone-rod homeobox |
|
otx1
|
ENSDARG00000094992 | orthodenticle homeobox 1 |
|
crx
|
ENSDARG00000113850 | cone-rod homeobox |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| otx1 | dr11_v1_chr17_+_24320861_24320861 | -0.71 | 6.0e-04 | Click! |
| crx | dr11_v1_chr5_+_36932718_36932718 | 0.07 | 7.7e-01 | Click! |
Activity profile of crx+otx1 motif
Sorted Z-values of crx+otx1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr21_+_13366353 | 4.21 |
ENSDART00000151630
|
si:ch73-62l21.1
|
si:ch73-62l21.1 |
| chr4_+_68562464 | 3.41 |
ENSDART00000192954
|
BX548011.4
|
|
| chr1_+_5576151 | 3.17 |
ENSDART00000109756
|
cpo
|
carboxypeptidase O |
| chr4_-_16853464 | 3.12 |
ENSDART00000125743
ENSDART00000164570 |
slc25a3a
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 3a |
| chr6_+_54538948 | 3.10 |
ENSDART00000149270
|
tulp1b
|
tubby like protein 1b |
| chr1_-_58766424 | 2.56 |
ENSDART00000191010
|
CABZ01096573.1
|
|
| chr17_+_30448452 | 2.54 |
ENSDART00000153939
|
lpin1
|
lipin 1 |
| chr19_-_5207361 | 2.53 |
ENSDART00000174611
|
stx1b
|
syntaxin 1B |
| chr10_+_17026870 | 2.51 |
ENSDART00000184529
ENSDART00000157480 |
CR855996.2
|
|
| chr6_-_16406210 | 2.46 |
ENSDART00000012023
|
faimb
|
Fas apoptotic inhibitory molecule b |
| chr10_-_35108683 | 2.42 |
ENSDART00000049633
|
zgc:110006
|
zgc:110006 |
| chr25_-_19090479 | 2.27 |
ENSDART00000027465
ENSDART00000177670 |
cacna2d4b
|
calcium channel, voltage-dependent, alpha 2/delta subunit 4b |
| chr7_+_19903924 | 2.26 |
ENSDART00000159112
|
si:ch211-285j22.3
|
si:ch211-285j22.3 |
| chr8_-_46572298 | 2.20 |
ENSDART00000030470
|
sult1st3
|
sulfotransferase family 1, cytosolic sulfotransferase 3 |
| chr11_-_37693019 | 2.20 |
ENSDART00000102898
|
zgc:158258
|
zgc:158258 |
| chr3_+_29980603 | 2.08 |
ENSDART00000151509
|
kcna7
|
potassium voltage-gated channel, shaker-related subfamily, member 7 |
| chr17_-_51202339 | 2.01 |
ENSDART00000167117
|
si:ch1073-469d17.2
|
si:ch1073-469d17.2 |
| chr7_-_39378903 | 2.00 |
ENSDART00000173659
|
slc8b1
|
solute carrier family 8 (sodium/lithium/calcium exchanger), member B1 |
| chr3_+_62000822 | 1.95 |
ENSDART00000106680
|
rai1
|
retinoic acid induced 1 |
| chr24_+_35975398 | 1.92 |
ENSDART00000173058
|
obscnb
|
obscurin, cytoskeletal calmodulin and titin-interacting RhoGEF b |
| chr18_+_10784730 | 1.90 |
ENSDART00000028938
|
mical3a
|
microtubule associated monooxygenase, calponin and LIM domain containing 3a |
| chr24_-_38079261 | 1.88 |
ENSDART00000105662
|
crp1
|
C-reactive protein 1 |
| chr4_+_72797711 | 1.87 |
ENSDART00000190934
ENSDART00000163236 |
MYRFL
|
myelin regulatory factor-like |
| chr17_+_50701748 | 1.87 |
ENSDART00000191938
ENSDART00000183220 ENSDART00000049464 |
fermt2
|
fermitin family member 2 |
| chr18_+_50089000 | 1.86 |
ENSDART00000058936
|
scamp5b
|
secretory carrier membrane protein 5b |
| chr4_+_7841627 | 1.85 |
ENSDART00000037997
|
ucmaa
|
upper zone of growth plate and cartilage matrix associated a |
| chr22_+_17203752 | 1.85 |
ENSDART00000143376
|
rab3b
|
RAB3B, member RAS oncogene family |
| chr5_-_69180587 | 1.83 |
ENSDART00000156681
ENSDART00000160753 |
zgc:171967
|
zgc:171967 |
| chr16_+_4055331 | 1.83 |
ENSDART00000128978
|
CR391998.1
|
|
| chr18_-_8579907 | 1.79 |
ENSDART00000147284
|
FRMD4A
|
si:ch211-220f12.1 |
| chr21_-_28340977 | 1.79 |
ENSDART00000141629
|
nrxn2a
|
neurexin 2a |
| chr1_+_23162124 | 1.78 |
ENSDART00000188428
|
si:dkey-92j12.5
|
si:dkey-92j12.5 |
| chr7_+_19904136 | 1.77 |
ENSDART00000173452
|
si:ch211-285j22.3
|
si:ch211-285j22.3 |
| chr20_+_46560258 | 1.71 |
ENSDART00000122115
|
si:ch211-153j24.3
|
si:ch211-153j24.3 |
| chr12_+_2660733 | 1.68 |
ENSDART00000152235
|
irbpl
|
interphotoreceptor retinoid-binding protein like |
| chr23_+_7710721 | 1.68 |
ENSDART00000186852
ENSDART00000161193 |
kif3b
|
kinesin family member 3B |
| chr3_+_15296824 | 1.65 |
ENSDART00000043801
|
cabp5b
|
calcium binding protein 5b |
| chr23_+_5465806 | 1.61 |
ENSDART00000149434
ENSDART00000148506 |
tulp1a
|
tubby like protein 1a |
| chr24_-_31140356 | 1.61 |
ENSDART00000167837
|
tmem56a
|
transmembrane protein 56a |
| chr1_-_58009216 | 1.60 |
ENSDART00000143829
|
nxnl1
|
nucleoredoxin like 1 |
| chr9_-_55744713 | 1.58 |
ENSDART00000157570
|
asmt
|
acetylserotonin O-methyltransferase |
| chr11_+_6295370 | 1.55 |
ENSDART00000139882
|
ranbp3a
|
RAN binding protein 3a |
| chr19_+_14921000 | 1.47 |
ENSDART00000144052
|
oprd1a
|
opioid receptor, delta 1a |
| chr23_+_39963599 | 1.45 |
ENSDART00000166539
|
fyco1a
|
FYVE and coiled-coil domain containing 1a |
| chr18_+_3634652 | 1.44 |
ENSDART00000159913
|
lrch3
|
leucine-rich repeats and calponin homology (CH) domain containing 3 |
| chr6_-_28980756 | 1.44 |
ENSDART00000014661
|
glmnb
|
glomulin, FKBP associated protein b |
| chr1_-_24349759 | 1.42 |
ENSDART00000142740
ENSDART00000177989 |
lrba
|
LPS-responsive vesicle trafficking, beach and anchor containing |
| chr12_-_35883814 | 1.40 |
ENSDART00000177986
ENSDART00000129888 |
cep131
|
centrosomal protein 131 |
| chr19_+_7152966 | 1.40 |
ENSDART00000080348
|
brd2a
|
bromodomain containing 2a |
| chr2_-_51757328 | 1.40 |
ENSDART00000189286
|
si:ch211-9d9.1
|
si:ch211-9d9.1 |
| chr7_+_67486807 | 1.37 |
ENSDART00000159989
|
cpne7
|
copine VII |
| chr18_-_8030073 | 1.35 |
ENSDART00000151490
|
shank3a
|
SH3 and multiple ankyrin repeat domains 3a |
| chr3_+_22327738 | 1.35 |
ENSDART00000055675
|
gh1
|
growth hormone 1 |
| chr8_+_2656231 | 1.34 |
ENSDART00000160833
|
fam102aa
|
family with sequence similarity 102, member Aa |
| chr25_-_204019 | 1.30 |
ENSDART00000188440
ENSDART00000191735 |
FP236318.2
|
|
| chr11_-_3366782 | 1.29 |
ENSDART00000127982
|
suox
|
sulfite oxidase |
| chr20_+_2134816 | 1.28 |
ENSDART00000039249
|
l3mbtl3
|
l(3)mbt-like 3 (Drosophila) |
| chr16_+_46497149 | 1.27 |
ENSDART00000135151
ENSDART00000058324 |
rpz4
|
rapunzel 4 |
| chr17_-_29194219 | 1.27 |
ENSDART00000157340
|
sptbn5
|
spectrin, beta, non-erythrocytic 5 |
| chr16_+_43219363 | 1.25 |
ENSDART00000183610
|
adam22
|
ADAM metallopeptidase domain 22 |
| chr5_-_69180227 | 1.24 |
ENSDART00000154816
|
zgc:171967
|
zgc:171967 |
| chr13_-_40499296 | 1.24 |
ENSDART00000158338
|
CNNM1
|
Danio rerio cyclin and CBS domain divalent metal cation transport mediator 1 (cnnm1), mRNA. |
| chr1_-_43712120 | 1.22 |
ENSDART00000074600
|
SLC9B2
|
si:dkey-162b23.4 |
| chr2_+_20430366 | 1.22 |
ENSDART00000155108
|
si:ch211-153l6.6
|
si:ch211-153l6.6 |
| chr3_-_3366590 | 1.22 |
ENSDART00000109428
ENSDART00000175329 |
si:dkey-46g23.1
|
si:dkey-46g23.1 |
| chr2_-_26499842 | 1.21 |
ENSDART00000186929
|
BX569796.1
|
|
| chr10_+_4875262 | 1.21 |
ENSDART00000165942
|
palm2
|
paralemmin 2 |
| chr11_+_29770966 | 1.19 |
ENSDART00000088624
ENSDART00000124471 |
rpgrb
|
retinitis pigmentosa GTPase regulator b |
| chr14_+_11909966 | 1.19 |
ENSDART00000171829
|
frmpd3
|
FERM and PDZ domain containing 3 |
| chr21_-_39058490 | 1.15 |
ENSDART00000114885
|
aldh3a2b
|
aldehyde dehydrogenase 3 family, member A2b |
| chr17_-_7371564 | 1.15 |
ENSDART00000060336
|
rab32b
|
RAB32b, member RAS oncogene family |
| chr2_+_49417900 | 1.13 |
ENSDART00000122742
ENSDART00000160783 |
rorcb
|
RAR-related orphan receptor C b |
| chr20_+_13969414 | 1.07 |
ENSDART00000049864
|
rd3
|
retinal degeneration 3 |
| chr21_+_31838386 | 1.06 |
ENSDART00000135591
|
si:ch211-12m10.1
|
si:ch211-12m10.1 |
| chr9_-_1459985 | 1.05 |
ENSDART00000124777
|
osbpl6
|
oxysterol binding protein-like 6 |
| chr10_+_40235959 | 1.05 |
ENSDART00000145862
|
gramd1ba
|
GRAM domain containing 1Ba |
| chr9_+_10014817 | 1.04 |
ENSDART00000132065
|
nxph2a
|
neurexophilin 2a |
| chr23_+_1181248 | 1.02 |
ENSDART00000170942
|
utrn
|
utrophin |
| chr24_+_9475809 | 1.02 |
ENSDART00000132688
|
si:ch211-285f17.1
|
si:ch211-285f17.1 |
| chr13_-_36050303 | 1.02 |
ENSDART00000134955
ENSDART00000139087 |
lgmn
|
legumain |
| chr12_+_23850661 | 1.01 |
ENSDART00000152921
|
svila
|
supervillin a |
| chr22_+_5176693 | 1.00 |
ENSDART00000160927
|
cers1
|
ceramide synthase 1 |
| chr22_+_5176255 | 0.98 |
ENSDART00000092647
|
cers1
|
ceramide synthase 1 |
| chr11_-_15090564 | 0.95 |
ENSDART00000162079
|
slc1a8a
|
solute carrier family 1 (glutamate transporter), member 8a |
| chr19_+_48176745 | 0.95 |
ENSDART00000164963
|
prdm1b
|
PR domain containing 1b, with ZNF domain |
| chr17_-_31058900 | 0.95 |
ENSDART00000134998
ENSDART00000104307 ENSDART00000172721 |
eml1
|
echinoderm microtubule associated protein like 1 |
| chr8_-_19649617 | 0.90 |
ENSDART00000189033
|
fam78bb
|
family with sequence similarity 78, member B b |
| chr9_+_10014514 | 0.90 |
ENSDART00000185590
|
nxph2a
|
neurexophilin 2a |
| chr3_-_13068189 | 0.88 |
ENSDART00000167180
|
prkar1b
|
protein kinase, cAMP-dependent, regulatory, type I, beta |
| chr8_-_51806584 | 0.85 |
ENSDART00000146976
|
si:ch73-313l22.2
|
si:ch73-313l22.2 |
| chr9_+_17982737 | 0.82 |
ENSDART00000192569
|
akap11
|
A kinase (PRKA) anchor protein 11 |
| chr21_-_40562705 | 0.80 |
ENSDART00000158289
ENSDART00000171997 |
taok1b
|
TAO kinase 1b |
| chr14_-_6402769 | 0.79 |
ENSDART00000121552
|
slc44a1b
|
solute carrier family 44 (choline transporter), member 1b |
| chr5_+_9434288 | 0.78 |
ENSDART00000162089
|
ugt2a7
|
UDP glucuronosyltransferase 2 family, polypeptide A7 |
| chr13_+_733027 | 0.78 |
ENSDART00000149547
|
nrxn1b
|
neurexin 1b |
| chr8_+_28724692 | 0.78 |
ENSDART00000140115
|
ccser1
|
coiled-coil serine-rich protein 1 |
| chr3_-_30218504 | 0.77 |
ENSDART00000103487
|
zgc:195001
|
zgc:195001 |
| chr23_+_45512825 | 0.77 |
ENSDART00000064846
|
prelid1b
|
PRELI domain containing 1b |
| chr13_+_2442841 | 0.75 |
ENSDART00000114456
ENSDART00000137124 ENSDART00000193737 ENSDART00000189722 ENSDART00000187485 |
arfgef3
|
ARFGEF family member 3 |
| chr14_-_31694274 | 0.75 |
ENSDART00000173353
|
map7d3
|
MAP7 domain containing 3 |
| chr2_+_30721070 | 0.74 |
ENSDART00000099052
|
si:dkey-94e7.2
|
si:dkey-94e7.2 |
| chr14_+_2487672 | 0.72 |
ENSDART00000170629
ENSDART00000123063 |
fgf18a
|
fibroblast growth factor 18a |
| chr7_-_38340674 | 0.72 |
ENSDART00000075782
|
slc7a10a
|
solute carrier family 7 (neutral amino acid transporter light chain, asc system), member 10a |
| chr10_-_42898220 | 0.72 |
ENSDART00000099270
|
CU326366.2
|
|
| chr15_+_46605482 | 0.71 |
ENSDART00000184449
|
CABZ01044048.1
|
|
| chr22_+_30047245 | 0.71 |
ENSDART00000142857
ENSDART00000141247 ENSDART00000140015 ENSDART00000040538 |
add3a
|
adducin 3 (gamma) a |
| chr21_-_2209012 | 0.69 |
ENSDART00000158345
|
zgc:162971
|
zgc:162971 |
| chr4_+_33537372 | 0.69 |
ENSDART00000181450
|
si:dkey-84h14.2
|
si:dkey-84h14.2 |
| chr9_-_5318873 | 0.69 |
ENSDART00000129308
|
ACVR1C
|
activin A receptor type 1C |
| chr9_-_46276626 | 0.69 |
ENSDART00000165238
|
hdac4
|
histone deacetylase 4 |
| chr12_+_49100365 | 0.68 |
ENSDART00000171905
|
CABZ01075125.1
|
|
| chr25_-_10629940 | 0.67 |
ENSDART00000154483
ENSDART00000155231 |
ppp6r3
|
protein phosphatase 6, regulatory subunit 3 |
| chr6_-_10168822 | 0.66 |
ENSDART00000151016
|
b3galt1a
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 1a |
| chr9_-_135774 | 0.66 |
ENSDART00000160435
|
FQ377903.1
|
|
| chr4_+_12111154 | 0.66 |
ENSDART00000036779
|
tmem178b
|
transmembrane protein 178B |
| chr4_-_908821 | 0.66 |
ENSDART00000168266
|
mrap2b
|
melanocortin 2 receptor accessory protein 2b |
| chr25_-_4733008 | 0.65 |
ENSDART00000191884
|
drd4a
|
dopamine receptor D4a |
| chr21_-_20765338 | 0.64 |
ENSDART00000135940
|
ghrb
|
growth hormone receptor b |
| chr9_-_2945008 | 0.63 |
ENSDART00000183452
|
zak
|
sterile alpha motif and leucine zipper containing kinase AZK |
| chr1_+_55264712 | 0.62 |
ENSDART00000193162
|
BX284640.1
|
|
| chr18_+_19006063 | 0.60 |
ENSDART00000135729
|
slc24a1
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 1 |
| chr9_+_1313418 | 0.59 |
ENSDART00000191361
|
casp8l2
|
caspase 8, apoptosis-related cysteine peptidase, like 2 |
| chr19_+_34169055 | 0.58 |
ENSDART00000135592
ENSDART00000186043 |
poc1bl
|
POC1 centriolar protein homolog B (Chlamydomonas), like |
| chr25_+_35891342 | 0.58 |
ENSDART00000147093
|
lsm14aa
|
LSM14A mRNA processing body assembly factor a |
| chr5_-_40910749 | 0.58 |
ENSDART00000083467
ENSDART00000133183 |
parp8
|
poly (ADP-ribose) polymerase family, member 8 |
| chr2_-_37537887 | 0.57 |
ENSDART00000143496
ENSDART00000025841 |
arhgef18a
|
rho/rac guanine nucleotide exchange factor (GEF) 18a |
| chr13_+_12528043 | 0.56 |
ENSDART00000057761
|
rrh
|
retinal pigment epithelium-derived rhodopsin homolog |
| chr23_-_42810664 | 0.56 |
ENSDART00000102328
|
pfkfb2a
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2a |
| chr19_-_35340027 | 0.56 |
ENSDART00000146394
|
macf1a
|
microtubule-actin crosslinking factor 1a |
| chr15_-_26931541 | 0.55 |
ENSDART00000027563
|
ccdc9
|
coiled-coil domain containing 9 |
| chr2_-_32551178 | 0.55 |
ENSDART00000145603
|
smarcd3a
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3a |
| chr10_+_39476600 | 0.54 |
ENSDART00000135756
|
kirrel3a
|
kirre like nephrin family adhesion molecule 3a |
| chr13_+_1015749 | 0.53 |
ENSDART00000190982
|
prokr1b
|
prokineticin receptor 1b |
| chr13_-_8815530 | 0.53 |
ENSDART00000122498
|
si:ch73-105b23.6
|
si:ch73-105b23.6 |
| chr4_+_28442223 | 0.53 |
ENSDART00000150879
|
znf1067
|
zinc finger protein 1067 |
| chr17_+_10593398 | 0.52 |
ENSDART00000168897
ENSDART00000193989 ENSDART00000191664 ENSDART00000167188 |
mapkbp1
|
mitogen-activated protein kinase binding protein 1 |
| chr21_+_45496442 | 0.51 |
ENSDART00000164880
|
si:dkey-223p19.1
|
si:dkey-223p19.1 |
| chr21_+_36581384 | 0.51 |
ENSDART00000183924
|
BX936439.1
|
|
| chr3_+_23063718 | 0.50 |
ENSDART00000140225
ENSDART00000184431 |
b4galnt2.1
|
beta-1,4-N-acetyl-galactosaminyl transferase 2, tandem duplicate 1 |
| chr12_-_33659328 | 0.49 |
ENSDART00000153457
|
tmem94
|
transmembrane protein 94 |
| chr7_+_22330466 | 0.49 |
ENSDART00000187347
ENSDART00000174483 |
fgf11a
|
fibroblast growth factor 11a |
| chr10_+_39476432 | 0.48 |
ENSDART00000190375
ENSDART00000183954 |
kirrel3a
|
kirre like nephrin family adhesion molecule 3a |
| chr10_-_40968095 | 0.47 |
ENSDART00000184104
|
npffr1l1
|
neuropeptide FF receptor 1 like 1 |
| chr18_-_6975175 | 0.47 |
ENSDART00000134194
|
si:dkey-266m15.6
|
si:dkey-266m15.6 |
| chr16_+_54592907 | 0.46 |
ENSDART00000172113
|
dennd4b
|
DENN/MADD domain containing 4B |
| chr8_+_4838114 | 0.45 |
ENSDART00000146667
|
es1
|
es1 protein |
| chr7_+_20030888 | 0.45 |
ENSDART00000192808
|
slc16a13
|
solute carrier family 16, member 13 (monocarboxylic acid transporter 13) |
| chr20_-_30920356 | 0.44 |
ENSDART00000022951
|
kif25
|
kinesin family member 25 |
| chr23_+_16935494 | 0.43 |
ENSDART00000143120
|
si:dkey-147f3.4
|
si:dkey-147f3.4 |
| chr24_-_21689146 | 0.43 |
ENSDART00000105917
|
urad
|
ureidoimidazoline (2-oxo-4-hydroxy-4-carboxy-5-) decarboxylase |
| chr14_+_52413846 | 0.43 |
ENSDART00000160952
|
noa1
|
nitric oxide associated 1 |
| chr7_+_39011852 | 0.41 |
ENSDART00000093009
|
dgkza
|
diacylglycerol kinase, zeta a |
| chr10_-_322769 | 0.40 |
ENSDART00000165244
|
akt2l
|
v-akt murine thymoma viral oncogene homolog 2, like |
| chr4_-_49133107 | 0.39 |
ENSDART00000150806
|
znf1146
|
zinc finger protein 1146 |
| chr20_+_53577502 | 0.39 |
ENSDART00000126983
|
myh6
|
myosin, heavy chain 6, cardiac muscle, alpha |
| chr25_+_35913614 | 0.39 |
ENSDART00000022437
|
gpia
|
glucose-6-phosphate isomerase a |
| chr18_+_27439680 | 0.38 |
ENSDART00000014726
|
tp53i11b
|
tumor protein p53 inducible protein 11b |
| chr11_+_19080400 | 0.38 |
ENSDART00000044423
|
magi1b
|
membrane associated guanylate kinase, WW and PDZ domain containing 1b |
| chr5_-_23715027 | 0.37 |
ENSDART00000139020
|
gbgt1l1
|
globoside alpha-1,3-N-acetylgalactosaminyltransferase 1, like 1 |
| chr9_-_394088 | 0.37 |
ENSDART00000169014
|
si:dkey-11f4.7
|
si:dkey-11f4.7 |
| chr9_+_7030016 | 0.36 |
ENSDART00000148047
ENSDART00000148181 |
inpp4aa
|
inositol polyphosphate-4-phosphatase type I Aa |
| chr15_-_34468599 | 0.35 |
ENSDART00000192984
|
meox2a
|
mesenchyme homeobox 2a |
| chr11_-_15090118 | 0.35 |
ENSDART00000171118
|
slc1a8a
|
solute carrier family 1 (glutamate transporter), member 8a |
| chr20_-_35012093 | 0.35 |
ENSDART00000062761
|
cnstb
|
consortin, connexin sorting protein b |
| chr14_+_30340251 | 0.34 |
ENSDART00000148448
|
mtus1a
|
microtubule associated tumor suppressor 1a |
| chr15_+_25683069 | 0.33 |
ENSDART00000148190
|
hic1
|
hypermethylated in cancer 1 |
| chr23_-_437467 | 0.33 |
ENSDART00000192106
|
tspan2b
|
tetraspanin 2b |
| chr10_+_6013076 | 0.32 |
ENSDART00000167613
ENSDART00000159216 |
hmgcs1
|
3-hydroxy-3-methylglutaryl-CoA synthase 1 (soluble) |
| chr9_+_1365747 | 0.31 |
ENSDART00000140917
ENSDART00000036605 |
prkra
|
protein kinase, interferon-inducible double stranded RNA dependent activator |
| chr6_-_39218609 | 0.28 |
ENSDART00000133305
|
os9
|
osteosarcoma amplified 9, endoplasmic reticulum lectin |
| chr23_-_35064785 | 0.28 |
ENSDART00000172240
|
BX294434.1
|
|
| chr20_+_54666222 | 0.27 |
ENSDART00000166592
|
CABZ01087948.1
|
|
| chr15_+_887032 | 0.27 |
ENSDART00000156395
|
si:dkey-7i4.9
|
si:dkey-7i4.9 |
| chr21_-_38853737 | 0.25 |
ENSDART00000184100
|
tlr22
|
toll-like receptor 22 |
| chr4_+_50401133 | 0.25 |
ENSDART00000150644
|
si:dkey-156k2.7
|
si:dkey-156k2.7 |
| chr5_-_27994679 | 0.24 |
ENSDART00000132740
|
ppp3cca
|
protein phosphatase 3, catalytic subunit, gamma isozyme, a |
| chr2_+_30721466 | 0.24 |
ENSDART00000128982
|
si:dkey-94e7.2
|
si:dkey-94e7.2 |
| chr19_+_43884120 | 0.23 |
ENSDART00000139684
ENSDART00000142312 |
lypla2
|
lysophospholipase II |
| chr15_+_20355465 | 0.23 |
ENSDART00000143092
ENSDART00000062666 |
il15l
|
interleukin 15, like |
| chr4_-_73561848 | 0.22 |
ENSDART00000174210
|
CU570689.2
|
|
| chr1_+_52633367 | 0.21 |
ENSDART00000134658
|
slc44a1a
|
solute carrier family 44 (choline transporter), member 1a |
| chr4_+_22480169 | 0.20 |
ENSDART00000146272
ENSDART00000066904 |
ndufb2
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 2 |
| chr7_+_39011355 | 0.20 |
ENSDART00000173855
|
dgkza
|
diacylglycerol kinase, zeta a |
| chr24_+_10039165 | 0.20 |
ENSDART00000144186
|
pou6f2
|
POU class 6 homeobox 2 |
| chr7_+_13702248 | 0.19 |
ENSDART00000172811
|
pdcd7
|
programmed cell death 7 |
| chr7_-_54320088 | 0.18 |
ENSDART00000172396
|
fadd
|
Fas (tnfrsf6)-associated via death domain |
| chr25_+_16731802 | 0.18 |
ENSDART00000188715
|
BX004976.1
|
|
| chr3_-_20058417 | 0.17 |
ENSDART00000165695
|
ubtf
|
upstream binding transcription factor, RNA polymerase I |
| chr20_-_2134620 | 0.17 |
ENSDART00000064375
|
tmem244
|
transmembrane protein 244 |
| chr23_-_45501177 | 0.15 |
ENSDART00000150103
|
col24a1
|
collagen type XXIV alpha 1 |
| chr4_+_11053301 | 0.15 |
ENSDART00000140362
|
ccdc59
|
coiled-coil domain containing 59 |
| chr13_-_29406534 | 0.14 |
ENSDART00000100877
|
zgc:153142
|
zgc:153142 |
| chr14_-_36412473 | 0.12 |
ENSDART00000128244
ENSDART00000138376 |
asb5a
|
ankyrin repeat and SOCS box containing 5a |
| chr13_+_30228077 | 0.12 |
ENSDART00000100813
ENSDART00000147729 ENSDART00000133404 |
rps24
|
ribosomal protein S24 |
| chr17_+_14886828 | 0.11 |
ENSDART00000010507
ENSDART00000131052 |
ptger2a
|
prostaglandin E receptor 2a (subtype EP2) |
| chr16_+_53519048 | 0.10 |
ENSDART00000124691
|
smpd5
|
sphingomyelin phosphodiesterase 5 |
| chr10_+_8690936 | 0.09 |
ENSDART00000144218
|
si:dkey-27b3.4
|
si:dkey-27b3.4 |
| chr18_-_24989580 | 0.07 |
ENSDART00000163449
|
chd2
|
chromodomain helicase DNA binding protein 2 |
| chr9_-_3519253 | 0.06 |
ENSDART00000169586
|
dcaf17
|
ddb1 and cul4 associated factor 17 |
| chr2_+_24080694 | 0.05 |
ENSDART00000024058
|
kcnh2a
|
potassium voltage-gated channel, subfamily H (eag-related), member 2a |
Network of associatons between targets according to the STRING database.
First level regulatory network of crx+otx1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.6 | GO:0030186 | melatonin metabolic process(GO:0030186) |
| 0.5 | 2.5 | GO:0098967 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.4 | 1.5 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.3 | 1.4 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.3 | 1.3 | GO:0050996 | positive regulation of lipid catabolic process(GO:0050996) |
| 0.3 | 1.0 | GO:0045668 | negative regulation of osteoblast differentiation(GO:0045668) |
| 0.2 | 0.7 | GO:0042941 | D-amino acid transport(GO:0042940) D-alanine transport(GO:0042941) D-serine transport(GO:0042942) |
| 0.2 | 3.8 | GO:0060999 | positive regulation of dendritic spine development(GO:0060999) |
| 0.2 | 2.5 | GO:1902042 | regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902041) negative regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902042) |
| 0.2 | 1.7 | GO:0043584 | nose development(GO:0043584) |
| 0.2 | 0.6 | GO:0060292 | long term synaptic depression(GO:0060292) |
| 0.2 | 1.4 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.2 | 0.7 | GO:0090299 | regulation of neural crest formation(GO:0090299) |
| 0.2 | 1.9 | GO:0045667 | regulation of osteoblast differentiation(GO:0045667) |
| 0.1 | 0.4 | GO:0046415 | urate catabolic process(GO:0019628) urate metabolic process(GO:0046415) |
| 0.1 | 2.3 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.1 | 1.6 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.1 | 3.1 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.1 | 0.5 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.1 | 2.2 | GO:0006584 | catecholamine metabolic process(GO:0006584) catechol-containing compound metabolic process(GO:0009712) |
| 0.1 | 1.2 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.1 | 0.3 | GO:0042998 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.1 | 0.3 | GO:0010142 | farnesyl diphosphate biosynthetic process, mevalonate pathway(GO:0010142) isoprenoid biosynthetic process via mevalonate(GO:1902767) |
| 0.1 | 4.7 | GO:0061512 | protein localization to cilium(GO:0061512) |
| 0.1 | 0.6 | GO:0060832 | oocyte animal/vegetal axis specification(GO:0060832) |
| 0.1 | 0.9 | GO:0036368 | cone photoresponse recovery(GO:0036368) |
| 0.1 | 1.2 | GO:0040033 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.1 | 2.5 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.1 | 0.6 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.1 | 0.9 | GO:0006336 | DNA replication-independent nucleosome assembly(GO:0006336) |
| 0.1 | 1.4 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.1 | 0.4 | GO:0055014 | atrial cardiac muscle cell development(GO:0055014) |
| 0.1 | 0.5 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.1 | 0.5 | GO:0006337 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.1 | 0.7 | GO:0097009 | energy homeostasis(GO:0097009) |
| 0.1 | 4.9 | GO:0030042 | actin filament depolymerization(GO:0030042) |
| 0.1 | 1.1 | GO:0032438 | melanosome organization(GO:0032438) |
| 0.1 | 1.9 | GO:0055008 | cardiac muscle tissue morphogenesis(GO:0055008) |
| 0.1 | 0.3 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.1 | 0.9 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.1 | 0.6 | GO:0071378 | growth hormone receptor signaling pathway(GO:0060396) response to growth hormone(GO:0060416) cellular response to growth hormone stimulus(GO:0071378) |
| 0.1 | 0.5 | GO:0030431 | sleep(GO:0030431) |
| 0.1 | 0.7 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.0 | 1.6 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
| 0.0 | 0.7 | GO:0030309 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.0 | 0.2 | GO:0036462 | TRAIL-activated apoptotic signaling pathway(GO:0036462) |
| 0.0 | 0.3 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) |
| 0.0 | 1.9 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.0 | 0.4 | GO:0071260 | cellular response to mechanical stimulus(GO:0071260) |
| 0.0 | 1.7 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.3 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.0 | 0.6 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.5 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.8 | GO:0097194 | execution phase of apoptosis(GO:0097194) |
| 0.0 | 2.1 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.0 | 0.5 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.6 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.1 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) collagen-activated signaling pathway(GO:0038065) |
| 0.0 | 1.4 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.1 | GO:0034695 | response to prostaglandin E(GO:0034695) cellular response to prostaglandin E stimulus(GO:0071380) |
| 0.0 | 1.2 | GO:0006081 | cellular aldehyde metabolic process(GO:0006081) |
| 0.0 | 0.2 | GO:0042531 | positive regulation of tyrosine phosphorylation of STAT protein(GO:0042531) |
| 0.0 | 1.8 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.4 | GO:0051156 | glucose 6-phosphate metabolic process(GO:0051156) |
| 0.0 | 0.4 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 0.2 | GO:0098734 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.0 | 0.7 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 2.1 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.0 | 0.6 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.0 | 0.6 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.8 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 0.0 | GO:0070257 | regulation of mucus secretion(GO:0070255) positive regulation of mucus secretion(GO:0070257) |
| 0.0 | 1.5 | GO:0030239 | myofibril assembly(GO:0030239) |
| 0.0 | 0.8 | GO:0007416 | synapse assembly(GO:0007416) |
| 0.0 | 0.2 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.9 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.2 | 0.6 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 0.1 | 1.4 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.1 | 3.2 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 2.5 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.1 | 0.9 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 1.9 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 1.9 | GO:0031430 | M band(GO:0031430) |
| 0.1 | 1.1 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.1 | 0.6 | GO:0032019 | mitochondrial cloud(GO:0032019) |
| 0.0 | 0.1 | GO:0005592 | collagen type XI trimer(GO:0005592) |
| 0.0 | 2.2 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 2.1 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.6 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.5 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 2.3 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 1.1 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 2.1 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.0 | 2.1 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 1.4 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.9 | GO:0031970 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.0 | 1.7 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 2.1 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.0 | 0.2 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.8 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:0038046 | enkephalin receptor activity(GO:0038046) |
| 0.5 | 1.4 | GO:0055105 | ubiquitin-protein transferase inhibitor activity(GO:0055105) |
| 0.4 | 1.3 | GO:0005131 | growth hormone receptor binding(GO:0005131) growth hormone activity(GO:0070186) |
| 0.2 | 2.2 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.2 | 3.1 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.2 | 1.4 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.2 | 0.5 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.2 | 0.7 | GO:0031782 | type 3 melanocortin receptor binding(GO:0031781) type 4 melanocortin receptor binding(GO:0031782) |
| 0.2 | 0.6 | GO:0004903 | growth hormone receptor activity(GO:0004903) |
| 0.2 | 2.0 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.1 | 2.0 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.1 | 0.9 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.1 | 3.2 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 1.2 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.1 | 0.8 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.1 | 0.3 | GO:0004421 | hydroxymethylglutaryl-CoA synthase activity(GO:0004421) |
| 0.1 | 1.3 | GO:0016670 | oxidoreductase activity, acting on a sulfur group of donors, oxygen as acceptor(GO:0016670) |
| 0.1 | 0.7 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.1 | 0.3 | GO:0071253 | connexin binding(GO:0071253) |
| 0.1 | 2.5 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.1 | 1.7 | GO:0019840 | retinoid binding(GO:0005501) isoprenoid binding(GO:0019840) |
| 0.1 | 0.6 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.1 | 0.4 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.0 | 0.7 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.7 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.0 | 0.7 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.6 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 1.1 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 2.5 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 1.9 | GO:0071949 | FAD binding(GO:0071949) |
| 0.0 | 1.9 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.4 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 2.3 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.6 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.8 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 0.6 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.0 | 1.0 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.9 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 2.1 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.5 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.7 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.0 | 0.9 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.5 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.0 | 2.7 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.0 | 5.5 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 2.1 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.0 | 0.2 | GO:0002020 | protease binding(GO:0002020) |
| 0.0 | 0.8 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.6 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.4 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.2 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.6 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.1 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.7 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.9 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.6 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 0.3 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.5 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.5 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 2.5 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.1 | 1.9 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 1.3 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.1 | 0.6 | REACTOME OPSINS | Genes involved in Opsins |
| 0.1 | 1.0 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.1 | 1.7 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 1.3 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 0.7 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 2.1 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 0.8 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.1 | 1.3 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.2 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.7 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.4 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.2 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.3 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |