Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
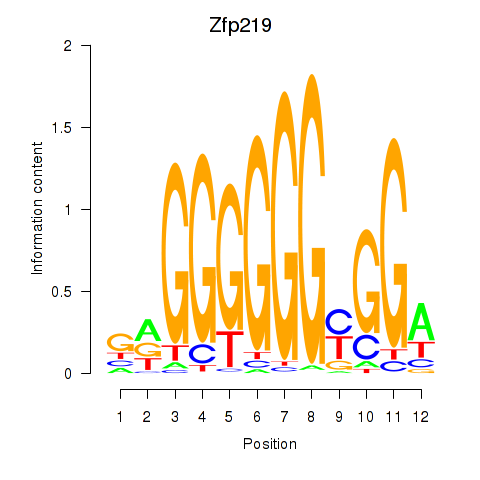
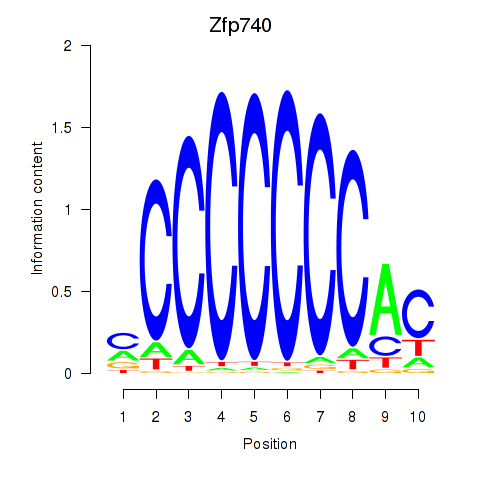
Results for Zfp219_Zfp740
Z-value: 1.34
Transcription factors associated with Zfp219_Zfp740
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zfp219
|
ENSMUSG00000049295.10 | zinc finger protein 219 |
|
Zfp740
|
ENSMUSG00000046897.10 | zinc finger protein 740 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zfp740 | mm10_v2_chr15_+_102203639_102203709 | -0.55 | 4.6e-04 | Click! |
| Zfp219 | mm10_v2_chr14_-_52020698_52020737 | -0.44 | 6.8e-03 | Click! |
Activity profile of Zfp219_Zfp740 motif
Sorted Z-values of Zfp219_Zfp740 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_+_44173271 | 6.10 |
ENSMUST00000088356.4
ENSMUST00000169582.1 |
Gm608
|
predicted gene 608 |
| chr16_+_44173239 | 5.18 |
ENSMUST00000119746.1
|
Gm608
|
predicted gene 608 |
| chr2_-_148045891 | 3.98 |
ENSMUST00000109964.1
|
Foxa2
|
forkhead box A2 |
| chr3_-_89393629 | 3.73 |
ENSMUST00000124783.1
ENSMUST00000126027.1 |
Zbtb7b
|
zinc finger and BTB domain containing 7B |
| chr4_-_114908892 | 3.22 |
ENSMUST00000068654.3
|
Foxd2
|
forkhead box D2 |
| chr6_-_72235559 | 3.18 |
ENSMUST00000042646.7
|
Atoh8
|
atonal homolog 8 (Drosophila) |
| chr11_-_69369377 | 3.10 |
ENSMUST00000092971.6
ENSMUST00000108661.1 |
Chd3
|
chromodomain helicase DNA binding protein 3 |
| chr13_-_29984219 | 3.03 |
ENSMUST00000146092.1
|
E2f3
|
E2F transcription factor 3 |
| chrX_-_38564519 | 2.98 |
ENSMUST00000016681.8
|
Cul4b
|
cullin 4B |
| chr3_-_89393294 | 2.65 |
ENSMUST00000142119.1
ENSMUST00000029677.8 ENSMUST00000148361.1 |
Zbtb7b
|
zinc finger and BTB domain containing 7B |
| chr11_-_87987528 | 2.62 |
ENSMUST00000020775.2
|
Dynll2
|
dynein light chain LC8-type 2 |
| chr11_-_102296618 | 2.28 |
ENSMUST00000107132.2
ENSMUST00000073234.2 |
Atxn7l3
|
ataxin 7-like 3 |
| chr15_+_57694651 | 1.98 |
ENSMUST00000096430.4
|
Zhx2
|
zinc fingers and homeoboxes 2 |
| chr11_-_96824008 | 1.95 |
ENSMUST00000142065.1
ENSMUST00000167110.1 ENSMUST00000169828.1 ENSMUST00000126949.1 |
Nfe2l1
|
nuclear factor, erythroid derived 2,-like 1 |
| chr11_+_97663366 | 1.86 |
ENSMUST00000044730.5
|
Mllt6
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 6 |
| chr16_+_64851991 | 1.84 |
ENSMUST00000067744.7
|
Cggbp1
|
CGG triplet repeat binding protein 1 |
| chr7_-_119895446 | 1.81 |
ENSMUST00000098080.2
|
Dcun1d3
|
DCN1, defective in cullin neddylation 1, domain containing 3 (S. cerevisiae) |
| chr7_+_24481991 | 1.81 |
ENSMUST00000068023.7
|
Cadm4
|
cell adhesion molecule 4 |
| chr3_+_129532386 | 1.78 |
ENSMUST00000071402.2
|
Elovl6
|
ELOVL family member 6, elongation of long chain fatty acids (yeast) |
| chr11_-_72266596 | 1.74 |
ENSMUST00000021161.6
ENSMUST00000140167.1 |
Slc13a5
|
solute carrier family 13 (sodium-dependent citrate transporter), member 5 |
| chr7_+_126823287 | 1.74 |
ENSMUST00000079423.5
|
Fam57b
|
family with sequence similarity 57, member B |
| chr9_+_55326913 | 1.69 |
ENSMUST00000085754.3
ENSMUST00000034862.4 |
AI118078
|
expressed sequence AI118078 |
| chr1_+_133363564 | 1.68 |
ENSMUST00000135222.2
|
Etnk2
|
ethanolamine kinase 2 |
| chr6_-_120294559 | 1.61 |
ENSMUST00000057283.7
|
B4galnt3
|
beta-1,4-N-acetyl-galactosaminyl transferase 3 |
| chr10_+_63024315 | 1.56 |
ENSMUST00000124784.1
|
Pbld2
|
phenazine biosynthesis-like protein domain containing 2 |
| chr5_-_34187670 | 1.53 |
ENSMUST00000042701.6
ENSMUST00000119171.1 |
Mxd4
|
Max dimerization protein 4 |
| chr19_+_7056731 | 1.38 |
ENSMUST00000040261.5
|
Macrod1
|
MACRO domain containing 1 |
| chr12_+_108334341 | 1.34 |
ENSMUST00000021684.4
|
Cyp46a1
|
cytochrome P450, family 46, subfamily a, polypeptide 1 |
| chr18_-_39490649 | 1.32 |
ENSMUST00000115567.1
|
Nr3c1
|
nuclear receptor subfamily 3, group C, member 1 |
| chr2_-_94264745 | 1.31 |
ENSMUST00000134563.1
|
E530001K10Rik
|
RIKEN cDNA E530001K10 gene |
| chr1_-_51915901 | 1.30 |
ENSMUST00000018561.7
ENSMUST00000114537.2 |
Myo1b
|
myosin IB |
| chr16_+_43503607 | 1.29 |
ENSMUST00000126100.1
ENSMUST00000123047.1 ENSMUST00000156981.1 |
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr4_+_97777780 | 1.28 |
ENSMUST00000107062.2
ENSMUST00000052018.5 ENSMUST00000107057.1 |
Nfia
|
nuclear factor I/A |
| chr2_+_49451486 | 1.25 |
ENSMUST00000092123.4
|
Epc2
|
enhancer of polycomb homolog 2 (Drosophila) |
| chr3_+_41564880 | 1.20 |
ENSMUST00000168086.1
|
Phf17
|
PHD finger protein 17 |
| chr19_+_42036025 | 1.19 |
ENSMUST00000026172.2
|
Ankrd2
|
ankyrin repeat domain 2 (stretch responsive muscle) |
| chr9_+_44499126 | 1.18 |
ENSMUST00000074989.5
|
Bcl9l
|
B cell CLL/lymphoma 9-like |
| chr14_+_21500879 | 1.15 |
ENSMUST00000182964.1
|
Kat6b
|
K(lysine) acetyltransferase 6B |
| chr8_+_111536492 | 1.14 |
ENSMUST00000168428.1
ENSMUST00000171182.1 |
Znrf1
|
zinc and ring finger 1 |
| chr7_-_43489967 | 1.14 |
ENSMUST00000107974.1
|
Iglon5
|
IgLON family member 5 |
| chr9_+_119402444 | 1.10 |
ENSMUST00000035093.8
ENSMUST00000165044.1 |
Acvr2b
|
activin receptor IIB |
| chr17_-_45686120 | 1.10 |
ENSMUST00000143907.1
ENSMUST00000127065.1 |
Tmem63b
|
transmembrane protein 63b |
| chr12_-_57546121 | 1.10 |
ENSMUST00000044380.6
|
Foxa1
|
forkhead box A1 |
| chrX_-_51018011 | 1.08 |
ENSMUST00000053593.7
|
Rap2c
|
RAP2C, member of RAS oncogene family |
| chr2_-_73892619 | 1.08 |
ENSMUST00000112007.1
ENSMUST00000112016.2 |
Atf2
|
activating transcription factor 2 |
| chr11_-_3774706 | 1.06 |
ENSMUST00000155197.1
|
Osbp2
|
oxysterol binding protein 2 |
| chr7_-_34654342 | 1.05 |
ENSMUST00000108069.1
|
Kctd15
|
potassium channel tetramerisation domain containing 15 |
| chr2_-_48949206 | 1.05 |
ENSMUST00000090976.3
ENSMUST00000149679.1 ENSMUST00000028098.4 |
Orc4
|
origin recognition complex, subunit 4 |
| chr2_-_73892588 | 1.04 |
ENSMUST00000154456.1
ENSMUST00000090802.4 ENSMUST00000055833.5 |
Atf2
|
activating transcription factor 2 |
| chr10_-_127620922 | 1.04 |
ENSMUST00000118455.1
|
Lrp1
|
low density lipoprotein receptor-related protein 1 |
| chr11_+_101468164 | 1.02 |
ENSMUST00000001347.6
|
Rnd2
|
Rho family GTPase 2 |
| chr2_+_48949495 | 1.01 |
ENSMUST00000112745.1
|
Mbd5
|
methyl-CpG binding domain protein 5 |
| chr10_-_24927444 | 1.00 |
ENSMUST00000020161.8
|
Arg1
|
arginase, liver |
| chr10_+_127380591 | 1.00 |
ENSMUST00000166820.1
|
R3hdm2
|
R3H domain containing 2 |
| chr17_-_27820445 | 0.99 |
ENSMUST00000114859.1
|
D17Wsu92e
|
DNA segment, Chr 17, Wayne State University 92, expressed |
| chr12_+_8771317 | 0.98 |
ENSMUST00000020911.7
|
Sdc1
|
syndecan 1 |
| chr6_+_125009665 | 0.97 |
ENSMUST00000046064.10
ENSMUST00000152752.1 ENSMUST00000088308.3 ENSMUST00000112425.1 ENSMUST00000084275.5 |
Zfp384
|
zinc finger protein 384 |
| chr11_+_76904475 | 0.97 |
ENSMUST00000142166.1
|
Tmigd1
|
transmembrane and immunoglobulin domain containing 1 |
| chr2_+_55437100 | 0.97 |
ENSMUST00000112633.2
ENSMUST00000112632.1 |
Kcnj3
|
potassium inwardly-rectifying channel, subfamily J, member 3 |
| chr10_+_80150448 | 0.97 |
ENSMUST00000153477.1
|
Midn
|
midnolin |
| chr11_+_23306884 | 0.96 |
ENSMUST00000180046.1
|
Usp34
|
ubiquitin specific peptidase 34 |
| chr6_-_116193426 | 0.96 |
ENSMUST00000088896.3
|
Tmcc1
|
transmembrane and coiled coil domains 1 |
| chr2_-_94264713 | 0.95 |
ENSMUST00000129661.1
|
E530001K10Rik
|
RIKEN cDNA E530001K10 gene |
| chrX_-_48208566 | 0.94 |
ENSMUST00000037960.4
|
Zdhhc9
|
zinc finger, DHHC domain containing 9 |
| chr11_+_77515104 | 0.93 |
ENSMUST00000094004.4
|
Abhd15
|
abhydrolase domain containing 15 |
| chr11_-_74925925 | 0.93 |
ENSMUST00000121738.1
|
Srr
|
serine racemase |
| chr18_-_56562261 | 0.92 |
ENSMUST00000066208.6
ENSMUST00000172734.1 |
Aldh7a1
|
aldehyde dehydrogenase family 7, member A1 |
| chr16_+_20673264 | 0.92 |
ENSMUST00000154950.1
ENSMUST00000115461.1 |
Eif4g1
|
eukaryotic translation initiation factor 4, gamma 1 |
| chr18_-_56562187 | 0.92 |
ENSMUST00000171844.2
|
Aldh7a1
|
aldehyde dehydrogenase family 7, member A1 |
| chr11_-_72267141 | 0.92 |
ENSMUST00000137701.1
|
Slc13a5
|
solute carrier family 13 (sodium-dependent citrate transporter), member 5 |
| chr11_-_89302545 | 0.92 |
ENSMUST00000061728.3
|
Nog
|
noggin |
| chr4_+_41135743 | 0.91 |
ENSMUST00000040008.3
|
Ube2r2
|
ubiquitin-conjugating enzyme E2R 2 |
| chr6_+_110645572 | 0.91 |
ENSMUST00000071076.6
ENSMUST00000172951.1 |
Grm7
|
glutamate receptor, metabotropic 7 |
| chr6_+_120666388 | 0.91 |
ENSMUST00000112686.1
|
Cecr2
|
cat eye syndrome chromosome region, candidate 2 |
| chr4_-_151861698 | 0.89 |
ENSMUST00000049790.7
|
Camta1
|
calmodulin binding transcription activator 1 |
| chr1_-_191397026 | 0.89 |
ENSMUST00000067976.3
|
Ppp2r5a
|
protein phosphatase 2, regulatory subunit B (B56), alpha isoform |
| chr7_+_28180226 | 0.89 |
ENSMUST00000172467.1
|
Dyrk1b
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1b |
| chr17_-_34121944 | 0.88 |
ENSMUST00000151986.1
|
Brd2
|
bromodomain containing 2 |
| chr17_-_27820534 | 0.88 |
ENSMUST00000075076.4
ENSMUST00000114863.2 |
D17Wsu92e
|
DNA segment, Chr 17, Wayne State University 92, expressed |
| chr17_-_34122311 | 0.87 |
ENSMUST00000025193.6
|
Brd2
|
bromodomain containing 2 |
| chr11_+_28853189 | 0.87 |
ENSMUST00000020759.5
|
Efemp1
|
epidermal growth factor-containing fibulin-like extracellular matrix protein 1 |
| chr10_-_7956223 | 0.87 |
ENSMUST00000146444.1
|
Tab2
|
TGF-beta activated kinase 1/MAP3K7 binding protein 2 |
| chr18_-_56562215 | 0.86 |
ENSMUST00000170309.1
|
Aldh7a1
|
aldehyde dehydrogenase family 7, member A1 |
| chr9_+_100643605 | 0.86 |
ENSMUST00000041418.6
|
Stag1
|
stromal antigen 1 |
| chr7_+_28180272 | 0.85 |
ENSMUST00000173223.1
|
Dyrk1b
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1b |
| chr14_-_51913393 | 0.85 |
ENSMUST00000004673.7
ENSMUST00000111632.3 |
Ndrg2
|
N-myc downstream regulated gene 2 |
| chr4_-_11386394 | 0.84 |
ENSMUST00000155519.1
|
Esrp1
|
epithelial splicing regulatory protein 1 |
| chr10_-_127620960 | 0.83 |
ENSMUST00000121829.1
|
Lrp1
|
low density lipoprotein receptor-related protein 1 |
| chr18_-_38211957 | 0.83 |
ENSMUST00000159405.1
ENSMUST00000160721.1 |
Pcdh1
|
protocadherin 1 |
| chr11_-_84870646 | 0.83 |
ENSMUST00000018547.2
|
Ggnbp2
|
gametogenetin binding protein 2 |
| chr15_+_102445367 | 0.83 |
ENSMUST00000023809.4
|
Amhr2
|
anti-Mullerian hormone type 2 receptor |
| chr14_-_70520254 | 0.82 |
ENSMUST00000022693.7
|
Bmp1
|
bone morphogenetic protein 1 |
| chr5_-_106696530 | 0.82 |
ENSMUST00000137285.1
ENSMUST00000124263.1 ENSMUST00000112695.1 ENSMUST00000155495.1 ENSMUST00000135108.1 |
Zfp644
|
zinc finger protein 644 |
| chr11_-_51857624 | 0.81 |
ENSMUST00000020655.7
ENSMUST00000109090.1 |
Phf15
|
PHD finger protein 15 |
| chr5_-_19226555 | 0.80 |
ENSMUST00000180594.1
|
4921504A21Rik
|
RIKEN cDNA 4921504A21 gene |
| chr9_-_98955302 | 0.80 |
ENSMUST00000181706.1
|
Foxl2os
|
forkhead box L2 opposite strand transcript |
| chr18_-_3337467 | 0.80 |
ENSMUST00000154135.1
|
Crem
|
cAMP responsive element modulator |
| chr10_+_93641041 | 0.79 |
ENSMUST00000020204.4
|
Ntn4
|
netrin 4 |
| chr11_-_97187872 | 0.79 |
ENSMUST00000001479.4
|
Kpnb1
|
karyopherin (importin) beta 1 |
| chr9_+_100643755 | 0.79 |
ENSMUST00000133388.1
|
Stag1
|
stromal antigen 1 |
| chr11_+_83473079 | 0.78 |
ENSMUST00000021018.4
|
Taf15
|
TAF15 RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr1_+_16105774 | 0.78 |
ENSMUST00000027053.7
|
Rdh10
|
retinol dehydrogenase 10 (all-trans) |
| chr18_-_3337614 | 0.77 |
ENSMUST00000150235.1
ENSMUST00000154470.1 |
Crem
|
cAMP responsive element modulator |
| chr2_-_29253001 | 0.76 |
ENSMUST00000071201.4
|
Ntng2
|
netrin G2 |
| chr4_+_152338887 | 0.76 |
ENSMUST00000005175.4
|
Chd5
|
chromodomain helicase DNA binding protein 5 |
| chr11_+_99041237 | 0.75 |
ENSMUST00000017637.6
|
Igfbp4
|
insulin-like growth factor binding protein 4 |
| chr4_+_144893077 | 0.75 |
ENSMUST00000154208.1
|
Dhrs3
|
dehydrogenase/reductase (SDR family) member 3 |
| chr12_+_8771405 | 0.75 |
ENSMUST00000171158.1
|
Sdc1
|
syndecan 1 |
| chr13_+_83721357 | 0.75 |
ENSMUST00000131907.2
|
C130071C03Rik
|
RIKEN cDNA C130071C03 gene |
| chr12_-_83597140 | 0.74 |
ENSMUST00000048319.4
|
Zfyve1
|
zinc finger, FYVE domain containing 1 |
| chr1_-_64121389 | 0.74 |
ENSMUST00000055001.3
|
Klf7
|
Kruppel-like factor 7 (ubiquitous) |
| chr7_-_127824469 | 0.73 |
ENSMUST00000106267.3
|
Stx1b
|
syntaxin 1B |
| chr10_-_86705485 | 0.73 |
ENSMUST00000020238.7
|
Hsp90b1
|
heat shock protein 90, beta (Grp94), member 1 |
| chrX_-_73716145 | 0.73 |
ENSMUST00000002091.5
|
Bcap31
|
B cell receptor associated protein 31 |
| chr11_+_51651179 | 0.72 |
ENSMUST00000170689.1
|
D930048N14Rik
|
RIKEN cDNA D930048N14 gene |
| chr1_+_131962941 | 0.72 |
ENSMUST00000177943.1
|
Slc45a3
|
solute carrier family 45, member 3 |
| chr15_+_102921103 | 0.72 |
ENSMUST00000001700.6
|
Hoxc13
|
homeobox C13 |
| chr11_+_88068242 | 0.72 |
ENSMUST00000018521.4
|
Vezf1
|
vascular endothelial zinc finger 1 |
| chr10_+_13966268 | 0.72 |
ENSMUST00000015645.4
|
Hivep2
|
human immunodeficiency virus type I enhancer binding protein 2 |
| chrX_-_36645359 | 0.72 |
ENSMUST00000051906.6
|
Akap17b
|
A kinase (PRKA) anchor protein 17B |
| chr2_-_116065798 | 0.71 |
ENSMUST00000110907.1
ENSMUST00000110908.2 |
Meis2
|
Meis homeobox 2 |
| chr1_-_168431695 | 0.71 |
ENSMUST00000176790.1
|
Pbx1
|
pre B cell leukemia homeobox 1 |
| chr17_-_70851189 | 0.71 |
ENSMUST00000059775.8
|
Tgif1
|
TGFB-induced factor homeobox 1 |
| chr18_+_64340225 | 0.71 |
ENSMUST00000175965.2
ENSMUST00000115145.3 |
Onecut2
|
one cut domain, family member 2 |
| chr15_+_102406143 | 0.70 |
ENSMUST00000170884.1
ENSMUST00000165924.1 ENSMUST00000163709.1 ENSMUST00000001326.6 |
Sp1
|
trans-acting transcription factor 1 |
| chr17_+_85620816 | 0.70 |
ENSMUST00000175898.2
|
Six3
|
sine oculis-related homeobox 3 |
| chr1_+_164151815 | 0.70 |
ENSMUST00000086040.5
|
F5
|
coagulation factor V |
| chr1_-_124045247 | 0.70 |
ENSMUST00000112603.2
|
Dpp10
|
dipeptidylpeptidase 10 |
| chr11_+_108920800 | 0.69 |
ENSMUST00000140821.1
|
Axin2
|
axin2 |
| chr19_+_32757497 | 0.69 |
ENSMUST00000013807.7
|
Pten
|
phosphatase and tensin homolog |
| chr4_-_151861762 | 0.68 |
ENSMUST00000097774.2
|
Camta1
|
calmodulin binding transcription activator 1 |
| chr9_+_68653761 | 0.68 |
ENSMUST00000034766.7
|
Rora
|
RAR-related orphan receptor alpha |
| chr2_+_18064564 | 0.67 |
ENSMUST00000114671.1
|
Mllt10
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 10 |
| chr15_-_100599983 | 0.67 |
ENSMUST00000073837.6
|
Pou6f1
|
POU domain, class 6, transcription factor 1 |
| chr12_-_56535047 | 0.66 |
ENSMUST00000178477.2
|
Nkx2-1
|
NK2 homeobox 1 |
| chr5_-_106696819 | 0.66 |
ENSMUST00000127434.1
ENSMUST00000112696.1 ENSMUST00000112698.1 |
Zfp644
|
zinc finger protein 644 |
| chr4_+_144892813 | 0.66 |
ENSMUST00000105744.1
ENSMUST00000171001.1 |
Dhrs3
|
dehydrogenase/reductase (SDR family) member 3 |
| chr4_+_133518963 | 0.66 |
ENSMUST00000149807.1
ENSMUST00000042919.9 ENSMUST00000153811.1 ENSMUST00000105901.1 ENSMUST00000121797.1 |
1810019J16Rik
|
RIKEN cDNA 1810019J16 gene |
| chr17_+_45686322 | 0.65 |
ENSMUST00000024734.7
|
Mrpl14
|
mitochondrial ribosomal protein L14 |
| chr17_-_28350747 | 0.65 |
ENSMUST00000080572.7
ENSMUST00000156862.1 |
Tead3
|
TEA domain family member 3 |
| chr2_+_164960809 | 0.64 |
ENSMUST00000124372.1
|
Slc12a5
|
solute carrier family 12, member 5 |
| chr2_+_31640037 | 0.63 |
ENSMUST00000113470.2
|
Prdm12
|
PR domain containing 12 |
| chr11_-_85139939 | 0.63 |
ENSMUST00000108075.2
|
Usp32
|
ubiquitin specific peptidase 32 |
| chr11_-_101171302 | 0.63 |
ENSMUST00000164474.1
ENSMUST00000043397.7 |
Plekhh3
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 3 |
| chr9_+_100643448 | 0.62 |
ENSMUST00000146312.1
ENSMUST00000129269.1 |
Stag1
|
stromal antigen 1 |
| chr2_+_59160884 | 0.62 |
ENSMUST00000037903.8
|
Pkp4
|
plakophilin 4 |
| chr5_-_136567242 | 0.61 |
ENSMUST00000175975.2
ENSMUST00000176216.2 ENSMUST00000176745.1 |
Cux1
|
cut-like homeobox 1 |
| chr11_+_95712673 | 0.61 |
ENSMUST00000107717.1
|
Zfp652
|
zinc finger protein 652 |
| chr12_-_31950170 | 0.61 |
ENSMUST00000176520.1
|
Hbp1
|
high mobility group box transcription factor 1 |
| chr10_+_127380799 | 0.61 |
ENSMUST00000111628.2
|
R3hdm2
|
R3H domain containing 2 |
| chr14_+_30715599 | 0.60 |
ENSMUST00000054230.4
|
Sfmbt1
|
Scm-like with four mbt domains 1 |
| chr11_-_98149551 | 0.60 |
ENSMUST00000103143.3
|
Fbxl20
|
F-box and leucine-rich repeat protein 20 |
| chr19_+_6497772 | 0.59 |
ENSMUST00000113458.1
ENSMUST00000113459.1 |
Nrxn2
|
neurexin II |
| chr2_+_29802626 | 0.59 |
ENSMUST00000080065.2
|
Slc27a4
|
solute carrier family 27 (fatty acid transporter), member 4 |
| chr1_+_74284930 | 0.59 |
ENSMUST00000113805.1
ENSMUST00000027370.6 ENSMUST00000087226.4 |
Pnkd
|
paroxysmal nonkinesiogenic dyskinesia |
| chr6_-_86526164 | 0.59 |
ENSMUST00000053015.5
|
Pcbp1
|
poly(rC) binding protein 1 |
| chr12_-_86726439 | 0.59 |
ENSMUST00000021682.8
|
Angel1
|
angel homolog 1 (Drosophila) |
| chr11_-_53470479 | 0.58 |
ENSMUST00000057722.2
|
Gm9837
|
predicted gene 9837 |
| chr11_-_116654245 | 0.58 |
ENSMUST00000021166.5
|
Cygb
|
cytoglobin |
| chr7_-_127026479 | 0.58 |
ENSMUST00000032916.4
|
Maz
|
MYC-associated zinc finger protein (purine-binding transcription factor) |
| chr17_+_85621017 | 0.57 |
ENSMUST00000162695.2
|
Six3
|
sine oculis-related homeobox 3 |
| chr11_+_88718442 | 0.57 |
ENSMUST00000138007.1
|
C030037D09Rik
|
RIKEN cDNA C030037D09 gene |
| chr2_+_18064645 | 0.57 |
ENSMUST00000114680.2
|
Mllt10
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 10 |
| chr1_+_156040884 | 0.57 |
ENSMUST00000060404.4
|
Tor1aip2
|
torsin A interacting protein 2 |
| chrX_-_160994665 | 0.57 |
ENSMUST00000087104.4
|
Cdkl5
|
cyclin-dependent kinase-like 5 |
| chr13_+_47043499 | 0.56 |
ENSMUST00000037025.8
ENSMUST00000143868.1 |
Kdm1b
|
lysine (K)-specific demethylase 1B |
| chr1_+_191906743 | 0.56 |
ENSMUST00000044954.6
|
Slc30a1
|
solute carrier family 30 (zinc transporter), member 1 |
| chr7_-_97579382 | 0.56 |
ENSMUST00000151840.1
ENSMUST00000135998.1 ENSMUST00000144858.1 ENSMUST00000146605.1 ENSMUST00000072725.5 ENSMUST00000138060.1 ENSMUST00000154853.1 ENSMUST00000136757.1 ENSMUST00000124552.1 |
Aamdc
|
adipogenesis associated Mth938 domain containing |
| chr13_-_60177357 | 0.56 |
ENSMUST00000065086.4
|
Gas1
|
growth arrest specific 1 |
| chr11_+_98795495 | 0.55 |
ENSMUST00000037915.2
|
Msl1
|
male-specific lethal 1 homolog (Drosophila) |
| chr5_+_130448801 | 0.54 |
ENSMUST00000111288.2
|
Caln1
|
calneuron 1 |
| chr5_-_24842579 | 0.54 |
ENSMUST00000030787.8
|
Rheb
|
Ras homolog enriched in brain |
| chr1_+_131970589 | 0.53 |
ENSMUST00000027695.6
|
Slc45a3
|
solute carrier family 45, member 3 |
| chr3_+_135825788 | 0.53 |
ENSMUST00000167390.1
|
Slc39a8
|
solute carrier family 39 (metal ion transporter), member 8 |
| chr15_-_100599864 | 0.53 |
ENSMUST00000177247.2
ENSMUST00000177505.2 |
Pou6f1
|
POU domain, class 6, transcription factor 1 |
| chr19_-_4175470 | 0.53 |
ENSMUST00000167055.1
|
Carns1
|
carnosine synthase 1 |
| chr12_+_76533540 | 0.52 |
ENSMUST00000075249.4
|
Plekhg3
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 3 |
| chr18_+_84088077 | 0.52 |
ENSMUST00000060223.2
|
Zadh2
|
zinc binding alcohol dehydrogenase, domain containing 2 |
| chr19_+_8839298 | 0.52 |
ENSMUST00000160556.1
|
Bscl2
|
Bernardinelli-Seip congenital lipodystrophy 2 homolog (human) |
| chr10_-_62651194 | 0.51 |
ENSMUST00000020270.4
|
Ddx50
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 50 |
| chr16_+_20733104 | 0.51 |
ENSMUST00000115423.1
ENSMUST00000007171.6 |
Chrd
|
chordin |
| chr5_+_30232581 | 0.51 |
ENSMUST00000145167.1
|
Ept1
|
ethanolaminephosphotransferase 1 (CDP-ethanolamine-specific) |
| chrX_+_77511002 | 0.50 |
ENSMUST00000088217.5
|
Tbl1x
|
transducin (beta)-like 1 X-linked |
| chr3_+_103020546 | 0.50 |
ENSMUST00000029446.8
|
Csde1
|
cold shock domain containing E1, RNA binding |
| chr16_+_20673517 | 0.50 |
ENSMUST00000115460.1
|
Eif4g1
|
eukaryotic translation initiation factor 4, gamma 1 |
| chr16_-_4213404 | 0.50 |
ENSMUST00000023165.6
|
Crebbp
|
CREB binding protein |
| chr9_+_72532609 | 0.49 |
ENSMUST00000183372.1
ENSMUST00000184015.1 |
Rfx7
|
regulatory factor X, 7 |
| chrX_+_109095359 | 0.49 |
ENSMUST00000033598.8
|
Sh3bgrl
|
SH3-binding domain glutamic acid-rich protein like |
| chr7_+_123982799 | 0.49 |
ENSMUST00000106437.1
|
Hs3st4
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 4 |
| chr7_+_105640522 | 0.49 |
ENSMUST00000106785.1
ENSMUST00000106786.1 ENSMUST00000106780.1 ENSMUST00000106784.1 |
Timm10b
|
translocase of inner mitochondrial membrane 10B |
| chr1_-_56972437 | 0.49 |
ENSMUST00000042857.7
|
Satb2
|
special AT-rich sequence binding protein 2 |
| chr3_+_66981352 | 0.49 |
ENSMUST00000162036.1
|
Rsrc1
|
arginine/serine-rich coiled-coil 1 |
| chr12_-_75177325 | 0.49 |
ENSMUST00000042299.2
|
Kcnh5
|
potassium voltage-gated channel, subfamily H (eag-related), member 5 |
| chr2_-_33371400 | 0.48 |
ENSMUST00000113164.1
ENSMUST00000091039.2 ENSMUST00000042615.6 |
Ralgps1
|
Ral GEF with PH domain and SH3 binding motif 1 |
| chr10_+_106470281 | 0.48 |
ENSMUST00000029404.9
ENSMUST00000169303.1 |
Ppfia2
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2 |
| chr11_+_35769462 | 0.48 |
ENSMUST00000018990.7
|
Pank3
|
pantothenate kinase 3 |
| chr11_+_114851142 | 0.48 |
ENSMUST00000133245.1
ENSMUST00000122967.2 |
Gprc5c
|
G protein-coupled receptor, family C, group 5, member C |
| chr2_-_28916412 | 0.48 |
ENSMUST00000050776.2
ENSMUST00000113849.1 |
Barhl1
|
BarH-like 1 (Drosophila) |
| chr7_+_105640448 | 0.48 |
ENSMUST00000058333.3
|
Timm10b
|
translocase of inner mitochondrial membrane 10B |
| chr1_+_131599239 | 0.48 |
ENSMUST00000027690.6
|
Avpr1b
|
arginine vasopressin receptor 1B |
Network of associatons between targets according to the STRING database.
First level regulatory network of Zfp219_Zfp740
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 6.4 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 1.3 | 4.0 | GO:0045014 | carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) |
| 0.7 | 2.7 | GO:0015744 | succinate transport(GO:0015744) |
| 0.6 | 1.9 | GO:2001160 | regulation of histone H3-K79 methylation(GO:2001160) |
| 0.6 | 2.3 | GO:1905167 | positive regulation of lysosomal protein catabolic process(GO:1905167) |
| 0.6 | 1.7 | GO:0048627 | myoblast development(GO:0048627) |
| 0.5 | 2.5 | GO:0032915 | positive regulation of transforming growth factor beta2 production(GO:0032915) |
| 0.5 | 1.4 | GO:0061144 | alveolar secondary septum development(GO:0061144) |
| 0.4 | 1.3 | GO:0097402 | neuroblast migration(GO:0097402) |
| 0.3 | 1.0 | GO:0060399 | positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.3 | 3.0 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.3 | 1.3 | GO:0015772 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.3 | 0.9 | GO:0060300 | regulation of cytokine activity(GO:0060300) |
| 0.3 | 0.8 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.3 | 1.8 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.3 | 1.0 | GO:0010958 | regulation of amino acid import(GO:0010958) L-arginine import(GO:0043091) arginine import(GO:0090467) |
| 0.2 | 0.7 | GO:1904154 | protein localization to endoplasmic reticulum exit site(GO:0070973) positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 0.2 | 2.6 | GO:2000576 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.2 | 3.2 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.2 | 0.7 | GO:0061181 | regulation of chondrocyte development(GO:0061181) |
| 0.2 | 0.7 | GO:0090071 | negative regulation of ribosome biogenesis(GO:0090071) |
| 0.2 | 1.4 | GO:0033132 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.2 | 0.7 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.2 | 1.3 | GO:1900170 | negative regulation of glucocorticoid mediated signaling pathway(GO:1900170) |
| 0.2 | 0.2 | GO:0070342 | brown fat cell proliferation(GO:0070342) regulation of brown fat cell proliferation(GO:0070347) |
| 0.2 | 0.8 | GO:0090361 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 0.2 | 0.6 | GO:0098974 | postsynaptic actin cytoskeleton organization(GO:0098974) |
| 0.2 | 0.2 | GO:2000137 | negative regulation of cell proliferation involved in heart morphogenesis(GO:2000137) |
| 0.2 | 0.6 | GO:1900039 | positive regulation of cellular response to hypoxia(GO:1900039) |
| 0.2 | 0.8 | GO:2001055 | positive regulation of mesenchymal cell apoptotic process(GO:2001055) |
| 0.2 | 0.8 | GO:0051305 | chromosome movement towards spindle pole(GO:0051305) |
| 0.2 | 1.0 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.2 | 0.8 | GO:0060431 | primary lung bud formation(GO:0060431) |
| 0.2 | 1.0 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.2 | 0.4 | GO:0002296 | T-helper 1 cell lineage commitment(GO:0002296) |
| 0.2 | 0.6 | GO:0071579 | regulation of zinc ion transport(GO:0071579) |
| 0.2 | 1.7 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.2 | 0.9 | GO:0070178 | D-serine metabolic process(GO:0070178) |
| 0.2 | 0.7 | GO:1903422 | negative regulation of synaptic vesicle recycling(GO:1903422) |
| 0.2 | 0.7 | GO:0003360 | brainstem development(GO:0003360) |
| 0.2 | 0.8 | GO:0033140 | negative regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033140) |
| 0.1 | 1.8 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.1 | 3.3 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.1 | 0.4 | GO:0042668 | auditory receptor cell fate determination(GO:0042668) |
| 0.1 | 0.4 | GO:0003032 | detection of oxygen(GO:0003032) cardiac jelly development(GO:1905072) |
| 0.1 | 1.0 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.1 | 0.4 | GO:0090427 | activation of meiosis(GO:0090427) |
| 0.1 | 0.1 | GO:0060741 | prostate gland stromal morphogenesis(GO:0060741) |
| 0.1 | 0.7 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.1 | 1.5 | GO:1900246 | positive regulation of RIG-I signaling pathway(GO:1900246) |
| 0.1 | 0.8 | GO:1900109 | regulation of histone H3-K9 dimethylation(GO:1900109) |
| 0.1 | 0.5 | GO:0001992 | regulation of systemic arterial blood pressure by vasopressin(GO:0001992) |
| 0.1 | 0.5 | GO:0021698 | cerebellar cortex structural organization(GO:0021698) |
| 0.1 | 0.4 | GO:0071677 | positive regulation of mononuclear cell migration(GO:0071677) |
| 0.1 | 0.5 | GO:0090126 | protein complex assembly involved in synapse maturation(GO:0090126) |
| 0.1 | 0.9 | GO:0033563 | dorsal/ventral axon guidance(GO:0033563) |
| 0.1 | 0.9 | GO:0090219 | negative regulation of lipid kinase activity(GO:0090219) |
| 0.1 | 1.1 | GO:0060836 | lymphatic endothelial cell differentiation(GO:0060836) |
| 0.1 | 0.4 | GO:0044268 | multicellular organismal protein metabolic process(GO:0044268) |
| 0.1 | 0.3 | GO:0036091 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.1 | 0.2 | GO:0050968 | detection of chemical stimulus involved in sensory perception of pain(GO:0050968) |
| 0.1 | 0.3 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.1 | 1.6 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.1 | 0.5 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.1 | 0.4 | GO:0097048 | dendritic cell apoptotic process(GO:0097048) regulation of dendritic cell apoptotic process(GO:2000668) |
| 0.1 | 1.8 | GO:0010225 | response to UV-C(GO:0010225) |
| 0.1 | 0.5 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.1 | 0.7 | GO:0070814 | hydrogen sulfide biosynthetic process(GO:0070814) |
| 0.1 | 0.6 | GO:0071630 | nucleus-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071630) |
| 0.1 | 0.3 | GO:0010845 | positive regulation of reciprocal meiotic recombination(GO:0010845) |
| 0.1 | 0.3 | GO:0045645 | regulation of eosinophil differentiation(GO:0045643) positive regulation of eosinophil differentiation(GO:0045645) |
| 0.1 | 0.6 | GO:0042760 | very long-chain fatty acid catabolic process(GO:0042760) |
| 0.1 | 1.3 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.1 | 0.3 | GO:2000724 | negative regulation of vascular associated smooth muscle cell migration(GO:1904753) regulation of cardiac vascular smooth muscle cell differentiation(GO:2000722) positive regulation of cardiac vascular smooth muscle cell differentiation(GO:2000724) |
| 0.1 | 0.3 | GO:0010615 | positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) |
| 0.1 | 0.5 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.1 | 0.4 | GO:2000973 | regulation of pro-B cell differentiation(GO:2000973) |
| 0.1 | 0.6 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.1 | 1.1 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.1 | 1.1 | GO:0043619 | regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0043619) |
| 0.1 | 0.3 | GO:0060722 | spongiotrophoblast cell proliferation(GO:0060720) cell proliferation involved in embryonic placenta development(GO:0060722) |
| 0.1 | 0.8 | GO:0035093 | spermatogenesis, exchange of chromosomal proteins(GO:0035093) histone H3-K27 trimethylation(GO:0098532) |
| 0.1 | 0.4 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.1 | 0.6 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.1 | 0.3 | GO:0097393 | post-embryonic appendage morphogenesis(GO:0035120) post-embryonic limb morphogenesis(GO:0035127) post-embryonic forelimb morphogenesis(GO:0035128) telomeric repeat-containing RNA transcription(GO:0097393) telomeric repeat-containing RNA transcription from RNA pol II promoter(GO:0097394) regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901580) negative regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901581) positive regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901582) |
| 0.1 | 0.6 | GO:0043587 | tongue morphogenesis(GO:0043587) |
| 0.1 | 0.5 | GO:0060178 | regulation of exocyst localization(GO:0060178) |
| 0.1 | 2.2 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.1 | 0.2 | GO:1902162 | regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) |
| 0.1 | 0.6 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.1 | 0.5 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.1 | 0.2 | GO:1903406 | regulation of sodium:potassium-exchanging ATPase activity(GO:1903406) |
| 0.1 | 0.4 | GO:0021853 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.1 | 0.2 | GO:1903351 | response to dopamine(GO:1903350) cellular response to dopamine(GO:1903351) |
| 0.1 | 0.4 | GO:0015886 | heme transport(GO:0015886) |
| 0.1 | 0.2 | GO:0043974 | histone H3-K27 acetylation(GO:0043974) regulation of histone H3-K27 acetylation(GO:1901674) |
| 0.1 | 0.4 | GO:0033092 | positive regulation of immature T cell proliferation in thymus(GO:0033092) |
| 0.1 | 0.7 | GO:0001842 | neural fold formation(GO:0001842) |
| 0.1 | 0.8 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.1 | 0.6 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.1 | 0.1 | GO:0007290 | spermatid nucleus elongation(GO:0007290) |
| 0.1 | 0.3 | GO:0070317 | negative regulation of G0 to G1 transition(GO:0070317) |
| 0.1 | 0.4 | GO:0060903 | positive regulation of meiosis I(GO:0060903) |
| 0.1 | 0.6 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.1 | 0.4 | GO:0051365 | leading edge cell differentiation(GO:0035026) cellular response to potassium ion starvation(GO:0051365) |
| 0.1 | 0.4 | GO:0060024 | rhythmic synaptic transmission(GO:0060024) |
| 0.1 | 0.2 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.1 | 0.2 | GO:1900045 | histone H2A K63-linked ubiquitination(GO:0070535) negative regulation of protein K63-linked ubiquitination(GO:1900045) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.1 | 0.1 | GO:0072076 | nephrogenic mesenchyme development(GO:0072076) |
| 0.1 | 1.0 | GO:0010745 | negative regulation of macrophage derived foam cell differentiation(GO:0010745) |
| 0.1 | 0.2 | GO:0014734 | skeletal muscle hypertrophy(GO:0014734) |
| 0.1 | 0.2 | GO:0034653 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) |
| 0.1 | 1.1 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.1 | 0.7 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.1 | 0.5 | GO:0007406 | negative regulation of neuroblast proliferation(GO:0007406) |
| 0.1 | 1.1 | GO:0072189 | ureter development(GO:0072189) |
| 0.1 | 0.5 | GO:0070574 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.1 | 0.5 | GO:0019896 | axonal transport of mitochondrion(GO:0019896) protein hexamerization(GO:0034214) |
| 0.1 | 0.5 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.1 | 0.3 | GO:0090472 | dibasic protein processing(GO:0090472) |
| 0.1 | 0.2 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.1 | 1.3 | GO:0098596 | vocal learning(GO:0042297) imitative learning(GO:0098596) |
| 0.1 | 0.3 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.1 | 0.4 | GO:0061087 | positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.1 | 0.6 | GO:0061727 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.1 | 0.1 | GO:0045168 | cell-cell signaling involved in cell fate commitment(GO:0045168) |
| 0.1 | 0.4 | GO:0021571 | rhombomere 5 development(GO:0021571) |
| 0.1 | 0.2 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.1 | 0.9 | GO:1903975 | regulation of glial cell migration(GO:1903975) |
| 0.1 | 0.6 | GO:0021707 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.1 | 0.7 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.1 | 1.0 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.1 | 0.4 | GO:0090234 | regulation of kinetochore assembly(GO:0090234) |
| 0.1 | 0.2 | GO:0061038 | gall bladder development(GO:0061010) uterus morphogenesis(GO:0061038) |
| 0.1 | 0.7 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.1 | 0.7 | GO:0060850 | regulation of transcription involved in cell fate commitment(GO:0060850) |
| 0.1 | 0.8 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.1 | 0.5 | GO:0071455 | response to hyperoxia(GO:0055093) cellular response to hyperoxia(GO:0071455) |
| 0.1 | 0.7 | GO:0071318 | cellular response to ATP(GO:0071318) |
| 0.1 | 0.8 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 0.1 | 0.2 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 0.1 | 0.2 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 0.1 | 0.9 | GO:0045717 | negative regulation of fatty acid biosynthetic process(GO:0045717) |
| 0.1 | 0.2 | GO:1903279 | regulation of calcium:sodium antiporter activity(GO:1903279) |
| 0.1 | 0.6 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.1 | 0.6 | GO:0042473 | outer ear morphogenesis(GO:0042473) |
| 0.1 | 0.7 | GO:0042048 | olfactory behavior(GO:0042048) |
| 0.1 | 0.7 | GO:0048935 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.1 | 0.4 | GO:1901727 | regulation of histone deacetylase activity(GO:1901725) positive regulation of histone deacetylase activity(GO:1901727) |
| 0.0 | 0.1 | GO:0051030 | snRNA transport(GO:0051030) |
| 0.0 | 0.2 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.9 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 0.2 | GO:0097494 | regulation of vesicle size(GO:0097494) |
| 0.0 | 0.2 | GO:0060598 | dichotomous subdivision of terminal units involved in mammary gland duct morphogenesis(GO:0060598) |
| 0.0 | 0.3 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 | 0.7 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.0 | 0.1 | GO:0048295 | positive regulation of isotype switching to IgE isotypes(GO:0048295) |
| 0.0 | 0.8 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 | 0.4 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.0 | 0.3 | GO:0030382 | sperm mitochondrion organization(GO:0030382) |
| 0.0 | 0.3 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 0.0 | 0.5 | GO:0090435 | protein localization to nuclear envelope(GO:0090435) |
| 0.0 | 0.3 | GO:0033184 | positive regulation of histone ubiquitination(GO:0033184) |
| 0.0 | 0.2 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.0 | 0.1 | GO:1901994 | negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 0.0 | 0.2 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.4 | GO:0032074 | negative regulation of nuclease activity(GO:0032074) |
| 0.0 | 1.2 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 1.6 | GO:0006687 | glycosphingolipid metabolic process(GO:0006687) |
| 0.0 | 0.2 | GO:0042706 | eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate commitment(GO:0046552) |
| 0.0 | 0.1 | GO:1904098 | regulation of protein O-linked glycosylation(GO:1904098) positive regulation of protein O-linked glycosylation(GO:1904100) |
| 0.0 | 0.7 | GO:0039694 | viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) |
| 0.0 | 0.2 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.0 | 0.6 | GO:0032927 | positive regulation of activin receptor signaling pathway(GO:0032927) |
| 0.0 | 0.4 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.0 | 0.1 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.0 | 0.1 | GO:0003273 | cell migration involved in endocardial cushion formation(GO:0003273) |
| 0.0 | 0.4 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.0 | 2.4 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.0 | 0.2 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.3 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 0.2 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.0 | 0.2 | GO:0003263 | cardioblast proliferation(GO:0003263) regulation of cardioblast proliferation(GO:0003264) positive regulation of cell proliferation involved in heart morphogenesis(GO:2000138) |
| 0.0 | 0.2 | GO:0032484 | Ral protein signal transduction(GO:0032484) |
| 0.0 | 0.2 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.0 | 1.9 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 0.1 | GO:0021892 | cerebral cortex GABAergic interneuron differentiation(GO:0021892) |
| 0.0 | 0.3 | GO:2000074 | regulation of type B pancreatic cell development(GO:2000074) |
| 0.0 | 0.2 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.0 | 0.1 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 0.0 | 0.1 | GO:0009816 | defense response, incompatible interaction(GO:0009814) defense response to bacterium, incompatible interaction(GO:0009816) regulation of defense response to bacterium, incompatible interaction(GO:1902477) |
| 0.0 | 1.9 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.0 | 0.3 | GO:0043691 | reverse cholesterol transport(GO:0043691) |
| 0.0 | 0.2 | GO:0003199 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) |
| 0.0 | 1.8 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.4 | GO:0002076 | osteoblast development(GO:0002076) |
| 0.0 | 0.4 | GO:0046543 | development of secondary female sexual characteristics(GO:0046543) |
| 0.0 | 0.1 | GO:0048715 | negative regulation of oligodendrocyte differentiation(GO:0048715) |
| 0.0 | 0.5 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.3 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.0 | 0.3 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.0 | 0.3 | GO:0061470 | interleukin-21 production(GO:0032625) T follicular helper cell differentiation(GO:0061470) interleukin-21 secretion(GO:0072619) |
| 0.0 | 0.3 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.8 | GO:0061036 | positive regulation of cartilage development(GO:0061036) |
| 0.0 | 1.0 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 0.3 | GO:0033601 | positive regulation of mammary gland epithelial cell proliferation(GO:0033601) |
| 0.0 | 0.8 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.3 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.0 | 0.3 | GO:0033299 | secretion of lysosomal enzymes(GO:0033299) |
| 0.0 | 0.3 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.0 | 0.1 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.0 | 0.1 | GO:0042795 | snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.0 | 0.2 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 1.2 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 0.0 | GO:0060197 | cloacal septation(GO:0060197) |
| 0.0 | 0.5 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.7 | GO:0061157 | mRNA destabilization(GO:0061157) |
| 0.0 | 0.1 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.0 | 0.2 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.5 | GO:0030325 | adrenal gland development(GO:0030325) |
| 0.0 | 0.4 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.0 | 0.2 | GO:0035280 | miRNA loading onto RISC involved in gene silencing by miRNA(GO:0035280) |
| 0.0 | 0.5 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 0.3 | GO:0090261 | positive regulation of inclusion body assembly(GO:0090261) |
| 0.0 | 0.2 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.0 | 0.1 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.0 | 0.9 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.1 | GO:1902309 | negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.0 | 0.4 | GO:0021860 | pyramidal neuron development(GO:0021860) |
| 0.0 | 0.3 | GO:2000811 | negative regulation of anoikis(GO:2000811) |
| 0.0 | 0.1 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.1 | GO:0097298 | regulation of nucleus size(GO:0097298) |
| 0.0 | 0.1 | GO:0030578 | PML body organization(GO:0030578) |
| 0.0 | 0.2 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.6 | GO:0031116 | positive regulation of microtubule polymerization(GO:0031116) |
| 0.0 | 0.2 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.0 | 0.4 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
| 0.0 | 0.1 | GO:0032962 | positive regulation of inositol trisphosphate biosynthetic process(GO:0032962) |
| 0.0 | 2.5 | GO:0007051 | spindle organization(GO:0007051) |
| 0.0 | 0.3 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.0 | 0.2 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 | 0.5 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.2 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.3 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 | 0.1 | GO:0070561 | vitamin D receptor signaling pathway(GO:0070561) regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.0 | 0.5 | GO:0072348 | sulfur compound transport(GO:0072348) |
| 0.0 | 0.4 | GO:0036075 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 0.7 | GO:0001662 | behavioral fear response(GO:0001662) |
| 0.0 | 0.1 | GO:0021819 | layer formation in cerebral cortex(GO:0021819) |
| 0.0 | 0.2 | GO:0009128 | purine nucleoside monophosphate catabolic process(GO:0009128) adenosine metabolic process(GO:0046085) |
| 0.0 | 0.1 | GO:0006701 | progesterone biosynthetic process(GO:0006701) |
| 0.0 | 0.4 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 | 0.2 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.0 | 0.1 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.0 | 0.0 | GO:1904016 | response to Thyroglobulin triiodothyronine(GO:1904016) cellular response to Thyroglobulin triiodothyronine(GO:1904017) |
| 0.0 | 0.7 | GO:1904377 | positive regulation of protein localization to plasma membrane(GO:1903078) positive regulation of protein localization to cell periphery(GO:1904377) |
| 0.0 | 0.2 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.0 | GO:1990859 | cellular response to endothelin(GO:1990859) |
| 0.0 | 0.2 | GO:0015747 | urate transport(GO:0015747) |
| 0.0 | 0.3 | GO:0009649 | entrainment of circadian clock(GO:0009649) |
| 0.0 | 0.2 | GO:0035066 | positive regulation of histone acetylation(GO:0035066) |
| 0.0 | 0.3 | GO:0042501 | serine phosphorylation of STAT protein(GO:0042501) |
| 0.0 | 0.4 | GO:0031648 | protein destabilization(GO:0031648) |
| 0.0 | 0.2 | GO:0097435 | fibril organization(GO:0097435) |
| 0.0 | 0.2 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 3.0 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.3 | 1.3 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.2 | 0.7 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.2 | 1.2 | GO:0005784 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.2 | 1.7 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.2 | 0.8 | GO:0097059 | CNTFR-CLCF1 complex(GO:0097059) |
| 0.2 | 2.7 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.2 | 1.0 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.2 | 0.7 | GO:1990590 | ATF1-ATF4 transcription factor complex(GO:1990590) |
| 0.2 | 0.9 | GO:0072487 | MSL complex(GO:0072487) |
| 0.2 | 4.1 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.2 | 0.5 | GO:0031417 | NatC complex(GO:0031417) |
| 0.1 | 0.7 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.1 | 1.6 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.1 | 2.3 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.1 | 1.0 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.1 | 1.7 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.1 | 1.6 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.1 | 0.6 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.1 | 0.3 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.1 | 0.7 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 0.1 | 0.5 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.1 | 0.3 | GO:1990421 | subtelomeric heterochromatin(GO:1990421) nuclear subtelomeric heterochromatin(GO:1990707) |
| 0.1 | 0.2 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.1 | 0.2 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.1 | 0.4 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.1 | 1.8 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.1 | 1.3 | GO:0010369 | chromocenter(GO:0010369) |
| 0.1 | 0.7 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 1.3 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 0.6 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.0 | 0.5 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.0 | 0.3 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.8 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 0.4 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.0 | 0.2 | GO:0000235 | astral microtubule(GO:0000235) |
| 0.0 | 0.3 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 0.8 | GO:0043189 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 0.9 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.2 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.0 | 0.2 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 1.3 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 1.4 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 0.2 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.2 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.0 | 0.2 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.0 | 1.1 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.3 | GO:0090568 | nuclear transcriptional repressor complex(GO:0090568) |
| 0.0 | 0.6 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.4 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 1.6 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.7 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.3 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.0 | 4.4 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 0.3 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 0.4 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.8 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.0 | 0.8 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.3 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.0 | 0.2 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.0 | 0.2 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 0.0 | 4.3 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 0.6 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.1 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.0 | 2.7 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.2 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 0.9 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.8 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.6 | GO:0031228 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 0.3 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.0 | 0.2 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.3 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 1.0 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 2.7 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 0.1 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.7 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.2 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 0.0 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.0 | 1.3 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.2 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 0.2 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.7 | GO:0017153 | sodium:dicarboxylate symporter activity(GO:0017153) |
| 0.8 | 2.3 | GO:0016964 | alpha-2 macroglobulin receptor activity(GO:0016964) |
| 0.7 | 2.7 | GO:0008802 | betaine-aldehyde dehydrogenase activity(GO:0008802) |
| 0.3 | 1.3 | GO:0015154 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.3 | 2.2 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.3 | 0.9 | GO:0047661 | racemase and epimerase activity, acting on amino acids and derivatives(GO:0016855) racemase activity, acting on amino acids and derivatives(GO:0036361) amino-acid racemase activity(GO:0047661) |
| 0.3 | 0.9 | GO:0070905 | serine binding(GO:0070905) |
| 0.3 | 1.7 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.3 | 1.3 | GO:0038049 | glucocorticoid receptor activity(GO:0004883) transcription factor activity, ligand-activated RNA polymerase II transcription factor binding(GO:0038049) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.3 | 2.8 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.2 | 0.7 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.2 | 0.9 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.2 | 0.6 | GO:0098918 | structural constituent of synapse(GO:0098918) structural constituent of postsynaptic actin cytoskeleton(GO:0098973) |
| 0.2 | 0.8 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.2 | 1.2 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.2 | 2.3 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.2 | 0.7 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.2 | 0.2 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 0.2 | 3.0 | GO:0070888 | E-box binding(GO:0070888) |
| 0.2 | 1.7 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.2 | 0.5 | GO:0047522 | 13-prostaglandin reductase activity(GO:0036132) 15-oxoprostaglandin 13-oxidase activity(GO:0047522) |
| 0.2 | 1.0 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.2 | 0.5 | GO:0005118 | sevenless binding(GO:0005118) |
| 0.2 | 0.5 | GO:0030977 | taurine binding(GO:0030977) |
| 0.1 | 1.8 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 0.6 | GO:0004096 | catalase activity(GO:0004096) |
| 0.1 | 0.6 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.1 | 0.7 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.1 | 1.1 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.1 | 0.3 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.1 | 1.0 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.1 | 0.8 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.1 | 0.4 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.1 | 0.6 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.1 | 0.3 | GO:0071820 | N-box binding(GO:0071820) |
| 0.1 | 0.7 | GO:0004897 | ciliary neurotrophic factor receptor activity(GO:0004897) |
| 0.1 | 0.5 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.1 | 0.4 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.1 | 2.6 | GO:0004675 | transmembrane receptor protein serine/threonine kinase activity(GO:0004675) |
| 0.1 | 1.0 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.1 | 0.7 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.1 | 0.8 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.1 | 0.4 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.1 | 0.3 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.1 | 0.4 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.1 | 0.2 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.1 | 2.2 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.1 | 0.5 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.1 | 0.7 | GO:0046790 | virion binding(GO:0046790) |
| 0.1 | 0.5 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.1 | 0.6 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.1 | 0.2 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.1 | 0.5 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.1 | 2.6 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.1 | 1.1 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) |
| 0.1 | 1.0 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 0.4 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.1 | 0.4 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.1 | 0.6 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.1 | 0.5 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.1 | 1.3 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.1 | 0.5 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.1 | 0.4 | GO:0051733 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.1 | 0.4 | GO:0001226 | RNA polymerase II transcription corepressor binding(GO:0001226) |
| 0.1 | 0.5 | GO:0099529 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) |
| 0.1 | 0.2 | GO:0031208 | POZ domain binding(GO:0031208) |
| 0.1 | 0.7 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.1 | 1.4 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.1 | 0.2 | GO:0071936 | coreceptor activity involved in Wnt signaling pathway(GO:0071936) |
| 0.1 | 1.7 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.1 | 0.6 | GO:0035014 | phosphatidylinositol 3-kinase regulator activity(GO:0035014) |
| 0.1 | 0.5 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.1 | 0.8 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.1 | 3.2 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.1 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 0.0 | 0.3 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.2 | GO:0070320 | inward rectifier potassium channel inhibitor activity(GO:0070320) |
| 0.0 | 0.7 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 0.6 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.3 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.0 | 0.2 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.0 | 0.3 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.0 | 0.4 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 0.0 | 0.6 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 2.4 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 0.2 | GO:0050220 | prostaglandin-D synthase activity(GO:0004667) prostaglandin-E synthase activity(GO:0050220) |
| 0.0 | 0.3 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 0.5 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.2 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.4 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.3 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.7 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.0 | 0.9 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 1.3 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.0 | 1.1 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 1.1 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.2 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.0 | 0.5 | GO:0055106 | ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.0 | 0.1 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.5 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.2 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.0 | 0.4 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.0 | 0.4 | GO:0019104 | DNA N-glycosylase activity(GO:0019104) |
| 0.0 | 1.8 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.4 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.0 | 0.2 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 2.7 | GO:0101005 | thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 0.2 | GO:0008079 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.1 | GO:0032453 | histone demethylase activity (H3-K4 specific)(GO:0032453) |
| 0.0 | 0.4 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 1.1 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 13.9 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.7 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.1 | GO:0051022 | GDP-dissociation inhibitor binding(GO:0051021) Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.0 | 0.3 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.0 | 0.2 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 1.4 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 1.8 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.1 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.4 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.0 | 0.8 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.3 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 1.2 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.0 | 0.1 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.7 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.0 | 0.2 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.9 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.6 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 1.2 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.1 | GO:0031686 | A1 adenosine receptor binding(GO:0031686) |
| 0.0 | 0.3 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.3 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.2 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.0 | 0.2 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.0 | 0.5 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.0 | 1.3 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 1.4 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 0.2 | GO:0016918 | retinal binding(GO:0016918) |
| 0.0 | 0.2 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.5 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.3 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.5 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 3.2 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 0.3 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.3 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.2 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.3 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.0 | 0.3 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 0.2 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.0 | 0.1 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.2 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.6 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 1.0 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.9 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 8.2 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 0.9 | PID MYC PATHWAY | C-MYC pathway |
| 0.1 | 1.0 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.1 | 1.7 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 0.1 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.1 | 4.2 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.1 | 1.8 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.1 | 3.0 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.1 | 1.8 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.1 | 2.2 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 2.1 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 1.2 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 2.2 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.8 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.9 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 3.6 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 1.4 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.7 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 1.7 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.3 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 1.0 | ST ADRENERGIC | Adrenergic Pathway |
| 0.0 | 0.3 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 0.8 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.6 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 1.4 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.4 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.6 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.5 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 0.6 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.3 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.2 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.0 | 0.6 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 0.7 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.7 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.4 | PID AURORA B PATHWAY | Aurora B signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 4.1 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.2 | 2.7 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.2 | 0.7 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.2 | 2.9 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.1 | 2.3 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.1 | 4.5 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 2.2 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 1.3 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.1 | 0.9 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.1 | 1.4 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.1 | 3.7 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.1 | 2.7 | REACTOME LIPOPROTEIN METABOLISM | Genes involved in Lipoprotein metabolism |
| 0.1 | 0.8 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.1 | 2.0 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 1.1 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.1 | 2.2 | REACTOME FATTY ACYL COA BIOSYNTHESIS | Genes involved in Fatty Acyl-CoA Biosynthesis |
| 0.1 | 0.8 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.1 | 0.7 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.1 | 0.2 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.1 | 0.3 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.1 | 0.7 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.9 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.7 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 1.0 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 1.1 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.9 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.7 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.4 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.0 | 0.5 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.0 | 0.5 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.3 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 0.5 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 2.3 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 1.0 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.5 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.0 | 1.2 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.9 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.3 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.5 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.9 | REACTOME TRANSCRIPTIONAL ACTIVITY OF SMAD2 SMAD3 SMAD4 HETEROTRIMER | Genes involved in Transcriptional activity of SMAD2/SMAD3:SMAD4 heterotrimer |
| 0.0 | 1.1 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 1.1 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.5 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.2 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.2 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.2 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.0 | 0.6 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 3.3 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.2 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 0.2 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.2 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.0 | 0.3 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |