Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Zfp110
Z-value: 1.39
Transcription factors associated with Zfp110
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zfp110
|
ENSMUSG00000058638.7 | zinc finger protein 110 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zfp110 | mm10_v2_chr7_+_12834743_12834811 | 0.09 | 6.1e-01 | Click! |
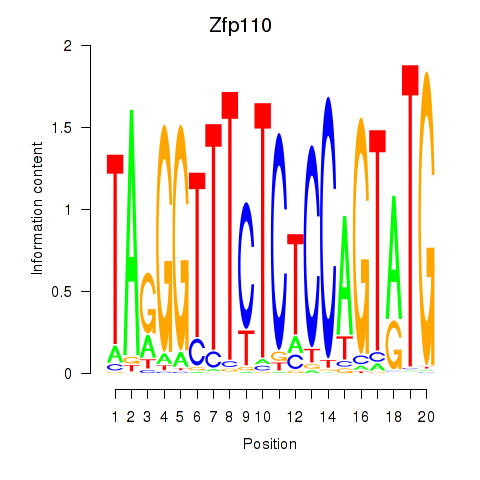
Activity profile of Zfp110 motif
Sorted Z-values of Zfp110 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_+_21691860 | 35.91 |
ENSMUST00000072133.4
|
Gm10226
|
predicted gene 10226 |
| chr17_+_21690766 | 32.91 |
ENSMUST00000097384.1
|
Gm10509
|
predicted gene 10509 |
| chr9_-_124493793 | 32.54 |
ENSMUST00000178787.1
|
Gm21836
|
predicted gene, 21836 |
| chr19_-_61297069 | 28.25 |
ENSMUST00000179346.1
|
Gm21060
|
predicted gene, 21060 |
| chr2_+_177508570 | 21.58 |
ENSMUST00000108940.2
|
Gm14403
|
predicted gene 14403 |
| chr17_+_16972910 | 19.03 |
ENSMUST00000071374.5
|
BC002059
|
cDNA sequence BC002059 |
| chr13_-_66852017 | 18.04 |
ENSMUST00000059329.6
|
Gm17449
|
predicted gene, 17449 |
| chr13_-_66851513 | 17.48 |
ENSMUST00000169322.1
|
Gm17404
|
predicted gene, 17404 |
| chr8_-_69373383 | 15.40 |
ENSMUST00000072427.4
|
Gm10033
|
predicted gene 10033 |
| chr2_-_175131864 | 13.84 |
ENSMUST00000108929.2
|
Gm14399
|
predicted gene 14399 |
| chr5_-_110046486 | 13.68 |
ENSMUST00000167969.1
|
Gm17655
|
predicted gene, 17655 |
| chr2_+_176236860 | 11.23 |
ENSMUST00000166464.1
|
2210418O10Rik
|
RIKEN cDNA 2210418O10 gene |
| chr2_-_176917518 | 9.91 |
ENSMUST00000108931.2
|
Gm14296
|
predicted gene 14296 |
| chr12_-_20900867 | 9.40 |
ENSMUST00000079237.5
|
Zfp125
|
zinc finger protein 125 |
| chr13_-_66227573 | 9.05 |
ENSMUST00000167981.2
|
Gm10772
|
predicted gene 10772 |
| chr7_-_42578588 | 8.92 |
ENSMUST00000179470.1
|
Gm21028
|
predicted gene, 21028 |
| chr12_-_23780265 | 7.70 |
ENSMUST00000072014.4
|
Gm10330
|
predicted gene 10330 |
| chr13_+_65512678 | 6.28 |
ENSMUST00000081471.2
|
Gm10139
|
predicted gene 10139 |
| chr17_-_22007301 | 4.13 |
ENSMUST00000075018.3
|
Gm9772
|
predicted gene 9772 |
| chr8_-_69373914 | 2.81 |
ENSMUST00000095282.1
|
Gm10311
|
predicted gene 10311 |
| chr2_-_150255591 | 2.04 |
ENSMUST00000063463.5
|
Gm21994
|
predicted gene 21994 |
| chr17_+_25370575 | 1.47 |
ENSMUST00000024999.8
|
Tpsg1
|
tryptase gamma 1 |
| chr17_+_25370550 | 1.36 |
ENSMUST00000162021.1
|
Tpsg1
|
tryptase gamma 1 |
| chrX_-_93632113 | 1.30 |
ENSMUST00000006856.2
|
Pola1
|
polymerase (DNA directed), alpha 1 |
| chr2_-_59160644 | 1.28 |
ENSMUST00000077687.5
|
Ccdc148
|
coiled-coil domain containing 148 |
| chr12_-_103675464 | 1.23 |
ENSMUST00000095451.1
|
Gm46
|
predicted gene 46 |
| chr10_+_69925484 | 1.19 |
ENSMUST00000182692.1
ENSMUST00000092433.5 |
Ank3
|
ankyrin 3, epithelial |
| chr9_+_4309719 | 1.16 |
ENSMUST00000049648.7
|
Kbtbd3
|
kelch repeat and BTB (POZ) domain containing 3 |
| chr2_-_32775330 | 1.12 |
ENSMUST00000161089.1
ENSMUST00000066478.2 ENSMUST00000161950.1 |
Ttc16
|
tetratricopeptide repeat domain 16 |
| chr15_-_103215285 | 0.83 |
ENSMUST00000122182.1
ENSMUST00000108813.3 ENSMUST00000127191.1 |
Cbx5
|
chromobox 5 |
| chr4_-_94979063 | 0.80 |
ENSMUST00000075872.3
|
Mysm1
|
myb-like, SWIRM and MPN domains 1 |
| chr6_+_17306415 | 0.80 |
ENSMUST00000150901.1
|
Cav1
|
caveolin 1, caveolae protein |
| chrY_+_897782 | 0.78 |
ENSMUST00000055032.7
|
Kdm5d
|
lysine (K)-specific demethylase 5D |
| chr4_-_43669141 | 0.75 |
ENSMUST00000056474.6
|
Fam221b
|
family with sequence similarity 221, member B |
| chr7_-_24333959 | 0.59 |
ENSMUST00000069562.4
|
Tescl
|
tescalcin-like |
| chr2_-_150485091 | 0.59 |
ENSMUST00000109914.1
|
Zfp345
|
zinc finger protein 345 |
| chr12_-_16800674 | 0.56 |
ENSMUST00000162112.1
|
Greb1
|
gene regulated by estrogen in breast cancer protein |
| chr14_-_55116935 | 0.53 |
ENSMUST00000022819.5
|
Jph4
|
junctophilin 4 |
| chr7_-_104288094 | 0.37 |
ENSMUST00000098179.2
|
Trim5
|
tripartite motif-containing 5 |
| chr7_+_92062392 | 0.33 |
ENSMUST00000098308.2
|
Dlg2
|
discs, large homolog 2 (Drosophila) |
| chr6_+_22288221 | 0.32 |
ENSMUST00000128245.1
ENSMUST00000031681.3 ENSMUST00000148639.1 |
Wnt16
|
wingless-related MMTV integration site 16 |
| chr4_+_90223625 | 0.30 |
ENSMUST00000080541.4
|
Zfp352
|
zinc finger protein 352 |
| chr7_-_122021143 | 0.21 |
ENSMUST00000033160.8
|
Gga2
|
golgi associated, gamma adaptin ear containing, ARF binding protein 2 |
| chr2_-_119541513 | 0.18 |
ENSMUST00000171024.1
|
Exd1
|
exonuclease 3'-5' domain containing 1 |
| chr2_+_59160838 | 0.17 |
ENSMUST00000102754.4
ENSMUST00000168631.1 ENSMUST00000123908.1 |
Pkp4
|
plakophilin 4 |
| chr4_+_134864536 | 0.16 |
ENSMUST00000030627.7
|
Rhd
|
Rh blood group, D antigen |
| chr6_-_53978662 | 0.16 |
ENSMUST00000166545.1
|
Cpvl
|
carboxypeptidase, vitellogenic-like |
| chr4_+_43669266 | 0.13 |
ENSMUST00000107864.1
|
Tmem8b
|
transmembrane protein 8B |
| chr5_-_66173051 | 0.11 |
ENSMUST00000113726.1
|
Rbm47
|
RNA binding motif protein 47 |
| chr9_-_4309432 | 0.10 |
ENSMUST00000051589.7
|
Aasdhppt
|
aminoadipate-semialdehyde dehydrogenase-phosphopantetheinyl transferase |
Network of associatons between targets according to the STRING database.
First level regulatory network of Zfp110
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) leading strand elongation(GO:0006272) |
| 0.2 | 0.8 | GO:0035522 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.1 | 0.7 | GO:1900085 | negative regulation of peptidyl-tyrosine autophosphorylation(GO:1900085) negative regulation of inward rectifier potassium channel activity(GO:1903609) |
| 0.1 | 0.8 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 0.1 | 1.2 | GO:0010650 | positive regulation of cell communication by electrical coupling(GO:0010650) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.1 | 0.3 | GO:0090403 | oxidative stress-induced premature senescence(GO:0090403) |
| 0.1 | 0.2 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.0 | 0.5 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.0 | 0.1 | GO:0006553 | lysine metabolic process(GO:0006553) |
| 0.0 | 0.3 | GO:0045161 | neuronal ion channel clustering(GO:0045161) |
| 0.0 | 0.1 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 50.9 | GO:0006355 | regulation of transcription, DNA-templated(GO:0006355) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.1 | 0.5 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.1 | 0.7 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.1 | 0.4 | GO:1990462 | omegasome(GO:1990462) |
| 0.1 | 1.2 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.2 | GO:1990923 | PET complex(GO:1990923) |
| 0.0 | 0.8 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 0.3 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 1.2 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0070320 | inward rectifier potassium channel inhibitor activity(GO:0070320) |
| 0.1 | 0.8 | GO:0032453 | histone demethylase activity (H3-K4 specific)(GO:0032453) |
| 0.1 | 1.3 | GO:0019103 | pyrimidine nucleotide binding(GO:0019103) |
| 0.0 | 53.7 | GO:0001071 | nucleic acid binding transcription factor activity(GO:0001071) transcription factor activity, sequence-specific DNA binding(GO:0003700) |
| 0.0 | 0.8 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 0.4 | GO:0008329 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.0 | 0.2 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 1.2 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 2.8 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.2 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) serine-type exopeptidase activity(GO:0070008) |
| 0.0 | 0.3 | GO:0004385 | guanylate kinase activity(GO:0004385) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 2.1 | PID E2F PATHWAY | E2F transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.3 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 0.7 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 1.2 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |