Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Nr2e3
Z-value: 1.35
Transcription factors associated with Nr2e3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nr2e3
|
ENSMUSG00000032292.2 | nuclear receptor subfamily 2, group E, member 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nr2e3 | mm10_v2_chr9_-_59950068_59950122 | 0.49 | 2.4e-03 | Click! |
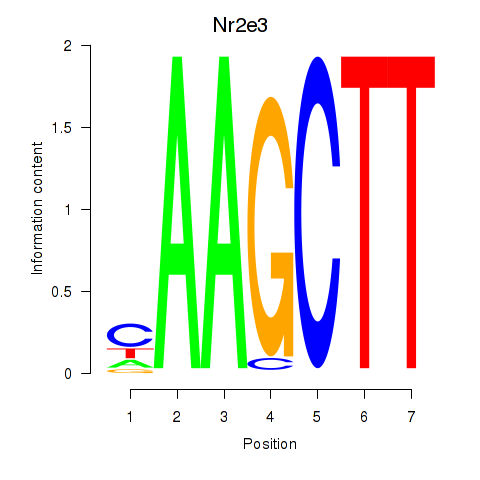
Activity profile of Nr2e3 motif
Sorted Z-values of Nr2e3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_90695706 | 9.16 |
ENSMUST00000069960.5
ENSMUST00000117167.1 |
S100a9
|
S100 calcium binding protein A9 (calgranulin B) |
| chr6_+_142298419 | 6.33 |
ENSMUST00000041993.2
|
Iapp
|
islet amyloid polypeptide |
| chr11_-_55185029 | 6.04 |
ENSMUST00000039305.5
|
Slc36a2
|
solute carrier family 36 (proton/amino acid symporter), member 2 |
| chr10_-_75940633 | 5.86 |
ENSMUST00000059658.4
|
Gm867
|
predicted gene 867 |
| chr19_+_38132767 | 5.49 |
ENSMUST00000025956.5
ENSMUST00000112329.1 |
Pde6c
|
phosphodiesterase 6C, cGMP specific, cone, alpha prime |
| chr3_-_137981523 | 4.12 |
ENSMUST00000136613.1
ENSMUST00000029806.6 |
Dapp1
|
dual adaptor for phosphotyrosine and 3-phosphoinositides 1 |
| chr7_+_100494044 | 3.62 |
ENSMUST00000153287.1
|
Ucp2
|
uncoupling protein 2 (mitochondrial, proton carrier) |
| chr1_-_66935333 | 3.58 |
ENSMUST00000120415.1
ENSMUST00000119429.1 |
Myl1
|
myosin, light polypeptide 1 |
| chr10_-_45470201 | 3.56 |
ENSMUST00000079390.6
|
Lin28b
|
lin-28 homolog B (C. elegans) |
| chr11_+_104608000 | 3.43 |
ENSMUST00000021028.4
|
Itgb3
|
integrin beta 3 |
| chr7_+_100493795 | 3.34 |
ENSMUST00000129324.1
|
Ucp2
|
uncoupling protein 2 (mitochondrial, proton carrier) |
| chr9_+_65587187 | 3.28 |
ENSMUST00000047099.5
ENSMUST00000131483.1 ENSMUST00000141046.1 |
Pif1
|
PIF1 5'-to-3' DNA helicase homolog (S. cerevisiae) |
| chr9_+_65587149 | 3.17 |
ENSMUST00000134538.1
ENSMUST00000136205.1 |
Pif1
|
PIF1 5'-to-3' DNA helicase homolog (S. cerevisiae) |
| chr8_-_57652993 | 3.13 |
ENSMUST00000110316.2
|
Galnt7
|
UDP-N-acetyl-alpha-D-galactosamine: polypeptide N-acetylgalactosaminyltransferase 7 |
| chr13_+_49544443 | 3.12 |
ENSMUST00000177948.1
ENSMUST00000021820.6 |
Aspn
|
asporin |
| chr7_-_67803489 | 3.07 |
ENSMUST00000181235.1
|
4833412C05Rik
|
RIKEN cDNA 4833412C05 gene |
| chr12_+_69963452 | 2.87 |
ENSMUST00000110560.1
|
Gm3086
|
predicted gene 3086 |
| chr7_-_110862944 | 2.69 |
ENSMUST00000033050.3
|
Lyve1
|
lymphatic vessel endothelial hyaluronan receptor 1 |
| chr8_+_88289028 | 2.67 |
ENSMUST00000171456.1
|
Adcy7
|
adenylate cyclase 7 |
| chr8_+_79028587 | 2.55 |
ENSMUST00000119254.1
|
Zfp827
|
zinc finger protein 827 |
| chr3_+_103832562 | 2.51 |
ENSMUST00000062945.5
|
Bcl2l15
|
BCLl2-like 15 |
| chr15_-_77022632 | 2.44 |
ENSMUST00000019037.8
ENSMUST00000169226.1 |
Mb
|
myoglobin |
| chr13_-_92794809 | 2.37 |
ENSMUST00000022213.7
|
Thbs4
|
thrombospondin 4 |
| chr2_-_164356067 | 2.29 |
ENSMUST00000165980.1
|
Slpi
|
secretory leukocyte peptidase inhibitor |
| chr2_-_164356507 | 2.24 |
ENSMUST00000109367.3
|
Slpi
|
secretory leukocyte peptidase inhibitor |
| chr10_+_119992962 | 2.19 |
ENSMUST00000154238.1
|
Grip1
|
glutamate receptor interacting protein 1 |
| chr12_+_95695350 | 2.18 |
ENSMUST00000110117.1
|
Flrt2
|
fibronectin leucine rich transmembrane protein 2 |
| chr8_-_57653023 | 2.17 |
ENSMUST00000034021.5
|
Galnt7
|
UDP-N-acetyl-alpha-D-galactosamine: polypeptide N-acetylgalactosaminyltransferase 7 |
| chr4_-_43040279 | 2.12 |
ENSMUST00000107958.1
ENSMUST00000107959.1 ENSMUST00000152846.1 |
Fam214b
|
family with sequence similarity 214, member B |
| chr18_-_36197232 | 2.07 |
ENSMUST00000115705.1
|
Nrg2
|
neuregulin 2 |
| chr8_+_79028317 | 1.97 |
ENSMUST00000087927.4
ENSMUST00000098614.2 |
Zfp827
|
zinc finger protein 827 |
| chr5_+_76809964 | 1.94 |
ENSMUST00000120818.1
|
C530008M17Rik
|
RIKEN cDNA C530008M17 gene |
| chrX_-_59166080 | 1.93 |
ENSMUST00000119306.1
|
Fgf13
|
fibroblast growth factor 13 |
| chr2_-_45112890 | 1.92 |
ENSMUST00000076836.6
|
Zeb2
|
zinc finger E-box binding homeobox 2 |
| chr13_-_28953690 | 1.90 |
ENSMUST00000067230.5
|
Sox4
|
SRY-box containing gene 4 |
| chr17_+_75178797 | 1.83 |
ENSMUST00000112516.1
ENSMUST00000135447.1 |
Ltbp1
|
latent transforming growth factor beta binding protein 1 |
| chr11_-_109472611 | 1.74 |
ENSMUST00000168740.1
|
Slc16a6
|
solute carrier family 16 (monocarboxylic acid transporters), member 6 |
| chr16_+_17233560 | 1.73 |
ENSMUST00000090190.5
ENSMUST00000115698.2 |
Hic2
|
hypermethylated in cancer 2 |
| chr11_+_67200052 | 1.69 |
ENSMUST00000124516.1
ENSMUST00000018637.8 |
Myh1
|
myosin, heavy polypeptide 1, skeletal muscle, adult |
| chrX_+_163911401 | 1.67 |
ENSMUST00000140845.1
|
Ap1s2
|
adaptor-related protein complex 1, sigma 2 subunit |
| chr17_+_75178911 | 1.65 |
ENSMUST00000112514.1
|
Ltbp1
|
latent transforming growth factor beta binding protein 1 |
| chr13_-_51701041 | 1.50 |
ENSMUST00000110042.1
|
Gm15440
|
predicted gene 15440 |
| chr7_+_141228766 | 1.48 |
ENSMUST00000106027.2
|
Phrf1
|
PHD and ring finger domains 1 |
| chrX_+_166344692 | 1.45 |
ENSMUST00000112223.1
ENSMUST00000112224.1 ENSMUST00000112229.2 ENSMUST00000112228.1 ENSMUST00000112227.2 ENSMUST00000112226.2 |
Gpm6b
|
glycoprotein m6b |
| chr1_+_45981548 | 1.44 |
ENSMUST00000085632.2
|
Rpl23a-ps1
|
ribosomal protein 23A, pseudogene 1 |
| chr13_+_35741313 | 1.41 |
ENSMUST00000163595.2
|
Cdyl
|
chromodomain protein, Y chromosome-like |
| chr1_-_52091066 | 1.40 |
ENSMUST00000105087.1
|
Gm3940
|
predicted gene 3940 |
| chr2_+_105130883 | 1.40 |
ENSMUST00000111098.1
ENSMUST00000111099.1 |
Wt1
|
Wilms tumor 1 homolog |
| chr2_-_140671400 | 1.39 |
ENSMUST00000056760.3
|
Flrt3
|
fibronectin leucine rich transmembrane protein 3 |
| chrX_+_100729917 | 1.38 |
ENSMUST00000019503.7
|
Gdpd2
|
glycerophosphodiester phosphodiesterase domain containing 2 |
| chr3_+_103832741 | 1.36 |
ENSMUST00000106822.1
|
Bcl2l15
|
BCLl2-like 15 |
| chr3_-_87174518 | 1.35 |
ENSMUST00000041732.8
|
Kirrel
|
kin of IRRE like (Drosophila) |
| chr11_-_76571527 | 1.35 |
ENSMUST00000072740.6
|
Abr
|
active BCR-related gene |
| chr3_+_131110350 | 1.35 |
ENSMUST00000066849.6
ENSMUST00000106341.2 ENSMUST00000029611.7 |
Lef1
|
lymphoid enhancer binding factor 1 |
| chr5_-_69542622 | 1.34 |
ENSMUST00000031045.6
|
Yipf7
|
Yip1 domain family, member 7 |
| chr9_-_64951599 | 1.33 |
ENSMUST00000037798.7
|
Slc24a1
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 1 |
| chr19_-_31664356 | 1.32 |
ENSMUST00000073581.5
|
Prkg1
|
protein kinase, cGMP-dependent, type I |
| chr9_-_13245741 | 1.30 |
ENSMUST00000110582.2
|
Jrkl
|
jerky homolog-like (mouse) |
| chr5_-_64960048 | 1.29 |
ENSMUST00000062315.4
|
Tlr6
|
toll-like receptor 6 |
| chr2_-_140671462 | 1.29 |
ENSMUST00000110057.2
|
Flrt3
|
fibronectin leucine rich transmembrane protein 3 |
| chr18_+_67562387 | 1.23 |
ENSMUST00000163749.1
|
Gm17669
|
predicted gene, 17669 |
| chr5_-_65335564 | 1.22 |
ENSMUST00000172780.1
|
Rfc1
|
replication factor C (activator 1) 1 |
| chr4_-_62470868 | 1.20 |
ENSMUST00000135811.1
ENSMUST00000120095.1 ENSMUST00000030087.7 ENSMUST00000107452.1 ENSMUST00000155522.1 |
Wdr31
|
WD repeat domain 31 |
| chr17_-_71475285 | 1.17 |
ENSMUST00000127430.1
|
Smchd1
|
SMC hinge domain containing 1 |
| chr5_+_34999111 | 1.17 |
ENSMUST00000114283.1
|
Rgs12
|
regulator of G-protein signaling 12 |
| chr6_-_13839916 | 1.16 |
ENSMUST00000060442.7
|
Gpr85
|
G protein-coupled receptor 85 |
| chr2_-_73660351 | 1.16 |
ENSMUST00000154258.1
|
Chn1
|
chimerin (chimaerin) 1 |
| chr14_-_77252327 | 1.15 |
ENSMUST00000099431.4
|
Gm10132
|
predicted gene 10132 |
| chr2_-_72986716 | 1.11 |
ENSMUST00000112062.1
|
Gm11084
|
predicted gene 11084 |
| chr10_+_127705170 | 1.11 |
ENSMUST00000079590.5
|
Myo1a
|
myosin IA |
| chr16_-_26105777 | 1.08 |
ENSMUST00000039990.5
|
Leprel1
|
leprecan-like 1 |
| chr7_+_102065713 | 1.08 |
ENSMUST00000094129.2
ENSMUST00000094130.2 ENSMUST00000084843.3 |
Trpc2
|
transient receptor potential cation channel, subfamily C, member 2 |
| chrX_+_6873484 | 1.06 |
ENSMUST00000145302.1
|
Dgkk
|
diacylglycerol kinase kappa |
| chr3_-_88503187 | 1.05 |
ENSMUST00000120377.1
|
Lmna
|
lamin A |
| chr15_+_85116829 | 1.05 |
ENSMUST00000105085.1
|
Gm10923
|
predicted gene 10923 |
| chr9_-_20952838 | 1.02 |
ENSMUST00000004202.9
|
Dnmt1
|
DNA methyltransferase (cytosine-5) 1 |
| chr16_+_97489994 | 1.02 |
ENSMUST00000177820.1
|
Gm9242
|
predicted pseudogene 9242 |
| chr4_-_154636831 | 0.99 |
ENSMUST00000030902.6
ENSMUST00000105637.1 ENSMUST00000070313.7 ENSMUST00000105636.1 ENSMUST00000105638.2 ENSMUST00000097759.2 ENSMUST00000124771.1 |
Prdm16
|
PR domain containing 16 |
| chr9_+_106203108 | 0.99 |
ENSMUST00000024047.5
|
Twf2
|
twinfilin, actin-binding protein, homolog 2 (Drosophila) |
| chr3_-_10208569 | 0.99 |
ENSMUST00000029041.4
|
Fabp4
|
fatty acid binding protein 4, adipocyte |
| chr3_-_88503331 | 0.96 |
ENSMUST00000029699.6
|
Lmna
|
lamin A |
| chr5_-_5266038 | 0.94 |
ENSMUST00000115451.1
ENSMUST00000115452.1 ENSMUST00000131392.1 |
Cdk14
|
cyclin-dependent kinase 14 |
| chr2_+_143546144 | 0.94 |
ENSMUST00000028905.9
|
Pcsk2
|
proprotein convertase subtilisin/kexin type 2 |
| chr8_+_106510853 | 0.93 |
ENSMUST00000080797.6
|
Cdh3
|
cadherin 3 |
| chr7_+_102065485 | 0.92 |
ENSMUST00000106950.1
ENSMUST00000146450.1 |
Trpc2
|
transient receptor potential cation channel, subfamily C, member 2 |
| chr2_-_73660401 | 0.91 |
ENSMUST00000102677.4
|
Chn1
|
chimerin (chimaerin) 1 |
| chr9_-_83146601 | 0.90 |
ENSMUST00000162246.2
ENSMUST00000161796.2 |
Hmgn3
|
high mobility group nucleosomal binding domain 3 |
| chr5_-_22550279 | 0.89 |
ENSMUST00000030872.5
|
Orc5
|
origin recognition complex, subunit 5 |
| chr17_+_35241746 | 0.86 |
ENSMUST00000068056.5
ENSMUST00000174757.1 |
Ddx39b
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 39B |
| chr7_-_19271797 | 0.85 |
ENSMUST00000032561.8
|
Vasp
|
vasodilator-stimulated phosphoprotein |
| chr12_+_88953399 | 0.85 |
ENSMUST00000057634.7
|
Nrxn3
|
neurexin III |
| chr11_-_101095367 | 0.84 |
ENSMUST00000019447.8
|
Psmc3ip
|
proteasome (prosome, macropain) 26S subunit, ATPase 3, interacting protein |
| chr2_+_127909058 | 0.84 |
ENSMUST00000110344.1
|
Acoxl
|
acyl-Coenzyme A oxidase-like |
| chr15_+_6299781 | 0.81 |
ENSMUST00000078019.6
|
Dab2
|
disabled 2, mitogen-responsive phosphoprotein |
| chrX_+_42149288 | 0.80 |
ENSMUST00000115073.2
ENSMUST00000115072.1 |
Stag2
|
stromal antigen 2 |
| chr11_-_98625661 | 0.80 |
ENSMUST00000104933.1
|
Gm12355
|
predicted gene 12355 |
| chr8_+_94667082 | 0.80 |
ENSMUST00000109527.4
|
Arl2bp
|
ADP-ribosylation factor-like 2 binding protein |
| chr19_-_7341433 | 0.78 |
ENSMUST00000165965.1
ENSMUST00000051711.9 ENSMUST00000169541.1 ENSMUST00000165989.1 |
Mark2
|
MAP/microtubule affinity-regulating kinase 2 |
| chr5_+_19907502 | 0.77 |
ENSMUST00000101558.3
|
Magi2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr17_+_35241838 | 0.77 |
ENSMUST00000173731.1
|
Ddx39b
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 39B |
| chr17_+_86963279 | 0.77 |
ENSMUST00000139344.1
|
Rhoq
|
ras homolog gene family, member Q |
| chr13_-_78197815 | 0.76 |
ENSMUST00000127137.2
|
Nr2f1
|
nuclear receptor subfamily 2, group F, member 1 |
| chr3_+_68468162 | 0.76 |
ENSMUST00000182532.1
|
Schip1
|
schwannomin interacting protein 1 |
| chrX_+_144153695 | 0.76 |
ENSMUST00000135687.1
|
A730046J19Rik
|
RIKEN cDNA A730046J19 gene |
| chr2_-_126675538 | 0.73 |
ENSMUST00000103227.1
ENSMUST00000110425.2 ENSMUST00000089745.4 |
Gabpb1
|
GA repeat binding protein, beta 1 |
| chr2_+_124610278 | 0.72 |
ENSMUST00000051419.8
ENSMUST00000077847.5 ENSMUST00000078621.5 ENSMUST00000076335.5 |
Sema6d
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D |
| chrX_+_42149534 | 0.69 |
ENSMUST00000127618.1
|
Stag2
|
stromal antigen 2 |
| chr9_+_113812547 | 0.69 |
ENSMUST00000166734.2
ENSMUST00000111838.2 ENSMUST00000163895.2 |
Clasp2
|
CLIP associating protein 2 |
| chr17_-_17624458 | 0.68 |
ENSMUST00000041047.2
|
Lnpep
|
leucyl/cystinyl aminopeptidase |
| chr11_-_65162904 | 0.67 |
ENSMUST00000093002.5
ENSMUST00000047463.8 |
Arhgap44
|
Rho GTPase activating protein 44 |
| chr14_+_76504478 | 0.66 |
ENSMUST00000022587.9
ENSMUST00000134109.1 |
Tsc22d1
|
TSC22 domain family, member 1 |
| chr15_+_6299797 | 0.66 |
ENSMUST00000159046.1
ENSMUST00000161040.1 |
Dab2
|
disabled 2, mitogen-responsive phosphoprotein |
| chr11_+_67200137 | 0.65 |
ENSMUST00000129018.1
|
Myh1
|
myosin, heavy polypeptide 1, skeletal muscle, adult |
| chr4_-_129640959 | 0.65 |
ENSMUST00000132217.1
ENSMUST00000130017.1 ENSMUST00000154105.1 |
Txlna
|
taxilin alpha |
| chr14_-_26442824 | 0.63 |
ENSMUST00000136635.1
ENSMUST00000125437.1 |
Slmap
|
sarcolemma associated protein |
| chr6_+_122553799 | 0.62 |
ENSMUST00000043301.7
|
Aicda
|
activation-induced cytidine deaminase |
| chr4_-_141053660 | 0.62 |
ENSMUST00000040222.7
|
Crocc
|
ciliary rootlet coiled-coil, rootletin |
| chrX_-_7188713 | 0.61 |
ENSMUST00000004428.7
|
Clcn5
|
chloride channel 5 |
| chr16_-_22439570 | 0.61 |
ENSMUST00000170393.1
|
Etv5
|
ets variant gene 5 |
| chr2_-_126675224 | 0.61 |
ENSMUST00000124972.1
|
Gabpb1
|
GA repeat binding protein, beta 1 |
| chr11_+_111066154 | 0.59 |
ENSMUST00000042970.2
|
Kcnj2
|
potassium inwardly-rectifying channel, subfamily J, member 2 |
| chr15_-_81104999 | 0.59 |
ENSMUST00000109579.2
|
Mkl1
|
MKL (megakaryoblastic leukemia)/myocardin-like 1 |
| chr5_-_65335597 | 0.58 |
ENSMUST00000172660.1
ENSMUST00000172732.1 ENSMUST00000031092.8 |
Rfc1
|
replication factor C (activator 1) 1 |
| chr3_-_110250963 | 0.58 |
ENSMUST00000106567.1
|
Prmt6
|
protein arginine N-methyltransferase 6 |
| chr14_+_20674311 | 0.57 |
ENSMUST00000048657.8
|
Sec24c
|
Sec24 related gene family, member C (S. cerevisiae) |
| chr6_-_122340525 | 0.57 |
ENSMUST00000112600.2
|
Phc1
|
polyhomeotic-like 1 (Drosophila) |
| chr11_+_70018728 | 0.55 |
ENSMUST00000018700.6
ENSMUST00000134376.2 |
Dlg4
|
discs, large homolog 4 (Drosophila) |
| chr1_+_146420434 | 0.54 |
ENSMUST00000163646.1
|
Gm5263
|
predicted gene 5263 |
| chr5_-_123140135 | 0.54 |
ENSMUST00000160099.1
|
AI480526
|
expressed sequence AI480526 |
| chr14_-_46575847 | 0.53 |
ENSMUST00000151828.1
|
Gm15219
|
predicted gene 15219 |
| chr6_+_30568367 | 0.53 |
ENSMUST00000049251.5
|
Cpa4
|
carboxypeptidase A4 |
| chr4_-_129640691 | 0.52 |
ENSMUST00000084264.5
|
Txlna
|
taxilin alpha |
| chr8_-_54718664 | 0.52 |
ENSMUST00000144711.2
ENSMUST00000093510.2 |
Wdr17
|
WD repeat domain 17 |
| chrX_-_134111852 | 0.52 |
ENSMUST00000033610.6
|
Nox1
|
NADPH oxidase 1 |
| chr9_-_77347816 | 0.51 |
ENSMUST00000184138.1
ENSMUST00000184006.1 ENSMUST00000185144.1 ENSMUST00000034910.9 |
Mlip
|
muscular LMNA-interacting protein |
| chr3_-_30509681 | 0.50 |
ENSMUST00000173495.1
|
Mecom
|
MDS1 and EVI1 complex locus |
| chrX_+_169879596 | 0.50 |
ENSMUST00000112105.1
ENSMUST00000078947.5 |
Mid1
|
midline 1 |
| chr2_-_126675461 | 0.50 |
ENSMUST00000103226.3
ENSMUST00000110424.2 |
Gabpb1
|
GA repeat binding protein, beta 1 |
| chr1_-_63214543 | 0.49 |
ENSMUST00000050536.7
|
Gpr1
|
G protein-coupled receptor 1 |
| chr5_+_76183880 | 0.48 |
ENSMUST00000031144.7
|
Tmem165
|
transmembrane protein 165 |
| chr6_-_129622685 | 0.47 |
ENSMUST00000032252.5
|
Klrk1
|
killer cell lectin-like receptor subfamily K, member 1 |
| chrY_-_10643315 | 0.47 |
ENSMUST00000100115.1
|
Gm20775
|
predicted gene, 20775 |
| chr4_-_141053704 | 0.47 |
ENSMUST00000102491.3
|
Crocc
|
ciliary rootlet coiled-coil, rootletin |
| chr2_-_72813665 | 0.47 |
ENSMUST00000136807.1
ENSMUST00000148327.1 |
6430710C18Rik
|
RIKEN cDNA 6430710C18 gene |
| chr17_+_86963077 | 0.47 |
ENSMUST00000024956.8
|
Rhoq
|
ras homolog gene family, member Q |
| chr12_-_34528844 | 0.46 |
ENSMUST00000110819.2
|
Hdac9
|
histone deacetylase 9 |
| chr10_+_111125851 | 0.46 |
ENSMUST00000171120.1
|
Gm5428
|
predicted gene 5428 |
| chr1_-_165160773 | 0.45 |
ENSMUST00000027859.5
|
Tbx19
|
T-box 19 |
| chr9_-_77347787 | 0.45 |
ENSMUST00000184848.1
ENSMUST00000184415.1 |
Mlip
|
muscular LMNA-interacting protein |
| chr14_+_76504185 | 0.43 |
ENSMUST00000177207.1
|
Tsc22d1
|
TSC22 domain family, member 1 |
| chr10_-_120112946 | 0.43 |
ENSMUST00000020449.5
|
Helb
|
helicase (DNA) B |
| chr7_+_83755904 | 0.42 |
ENSMUST00000051522.8
ENSMUST00000042280.7 |
Gm7964
|
predicted gene 7964 |
| chr4_-_120951664 | 0.42 |
ENSMUST00000106280.1
|
Zfp69
|
zinc finger protein 69 |
| chr16_-_52454074 | 0.41 |
ENSMUST00000023312.7
|
Alcam
|
activated leukocyte cell adhesion molecule |
| chr13_-_66852017 | 0.39 |
ENSMUST00000059329.6
|
Gm17449
|
predicted gene, 17449 |
| chr3_-_30509752 | 0.39 |
ENSMUST00000172697.1
|
Mecom
|
MDS1 and EVI1 complex locus |
| chr11_-_102579461 | 0.39 |
ENSMUST00000107081.1
|
Gm11627
|
predicted gene 11627 |
| chr5_-_21645541 | 0.38 |
ENSMUST00000115234.1
ENSMUST00000051358.4 |
Fbxl13
|
F-box and leucine-rich repeat protein 13 |
| chr2_+_18055203 | 0.36 |
ENSMUST00000028076.8
|
Mllt10
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 10 |
| chr1_+_97770158 | 0.36 |
ENSMUST00000112844.3
ENSMUST00000112842.1 ENSMUST00000027571.6 |
Gin1
|
gypsy retrotransposon integrase 1 |
| chr8_-_60954726 | 0.35 |
ENSMUST00000110302.1
|
Clcn3
|
chloride channel 3 |
| chr7_+_27731445 | 0.35 |
ENSMUST00000042641.7
|
Zfp60
|
zinc finger protein 60 |
| chr7_+_27731398 | 0.34 |
ENSMUST00000130997.1
|
Zfp60
|
zinc finger protein 60 |
| chr4_-_11386679 | 0.34 |
ENSMUST00000043781.7
ENSMUST00000108310.1 |
Esrp1
|
epithelial splicing regulatory protein 1 |
| chr11_+_34314757 | 0.34 |
ENSMUST00000165963.1
ENSMUST00000093192.3 |
Fam196b
|
family with sequence similarity 196, member B |
| chr19_+_39113898 | 0.33 |
ENSMUST00000087234.2
|
Cyp2c66
|
cytochrome P450, family 2, subfamily c, polypeptide 66 |
| chr7_+_36698002 | 0.32 |
ENSMUST00000021641.6
|
Tshz3
|
teashirt zinc finger family member 3 |
| chr8_+_104540800 | 0.31 |
ENSMUST00000056051.4
|
Car7
|
carbonic anhydrase 7 |
| chr19_+_23675839 | 0.31 |
ENSMUST00000056396.5
|
Gm6563
|
predicted pseudogene 6563 |
| chrX_+_20617503 | 0.29 |
ENSMUST00000115375.1
ENSMUST00000115374.1 ENSMUST00000084383.3 |
Rbm10
|
RNA binding motif protein 10 |
| chr14_+_62663665 | 0.26 |
ENSMUST00000171692.1
|
Serpine3
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 3 |
| chr1_-_179745185 | 0.26 |
ENSMUST00000131716.1
|
Gm1305
|
predicted gene 1305 |
| chr18_+_61555308 | 0.26 |
ENSMUST00000165721.1
ENSMUST00000115246.2 ENSMUST00000166990.1 ENSMUST00000163205.1 ENSMUST00000170862.1 |
Csnk1a1
|
casein kinase 1, alpha 1 |
| chr1_+_151755339 | 0.25 |
ENSMUST00000059498.5
|
Edem3
|
ER degradation enhancer, mannosidase alpha-like 3 |
| chr4_-_11386757 | 0.25 |
ENSMUST00000108313.1
ENSMUST00000108311.2 |
Esrp1
|
epithelial splicing regulatory protein 1 |
| chr16_+_32271468 | 0.24 |
ENSMUST00000093183.3
|
Smco1
|
single-pass membrane protein with coiled-coil domains 1 |
| chr19_+_4214238 | 0.22 |
ENSMUST00000046506.6
|
Clcf1
|
cardiotrophin-like cytokine factor 1 |
| chr12_-_54795698 | 0.21 |
ENSMUST00000005798.8
|
Snx6
|
sorting nexin 6 |
| chrX_-_94542037 | 0.19 |
ENSMUST00000026142.7
|
Maged1
|
melanoma antigen, family D, 1 |
| chr6_+_82402475 | 0.18 |
ENSMUST00000032122.8
|
Tacr1
|
tachykinin receptor 1 |
| chr4_-_137048695 | 0.18 |
ENSMUST00000049583.7
|
Zbtb40
|
zinc finger and BTB domain containing 40 |
| chrX_-_108834303 | 0.18 |
ENSMUST00000101283.3
ENSMUST00000150434.1 |
Brwd3
|
bromodomain and WD repeat domain containing 3 |
| chr11_-_28584260 | 0.17 |
ENSMUST00000093253.3
ENSMUST00000109502.2 ENSMUST00000042534.8 |
Ccdc85a
|
coiled-coil domain containing 85A |
| chr8_+_125995102 | 0.17 |
ENSMUST00000046765.8
|
Kcnk1
|
potassium channel, subfamily K, member 1 |
| chr11_-_42182924 | 0.15 |
ENSMUST00000020707.5
ENSMUST00000132971.1 |
Gabra1
|
gamma-aminobutyric acid (GABA) A receptor, subunit alpha 1 |
| chr1_+_180111339 | 0.15 |
ENSMUST00000145181.1
|
Cdc42bpa
|
CDC42 binding protein kinase alpha |
| chr4_+_49521176 | 0.14 |
ENSMUST00000042964.6
ENSMUST00000107696.1 |
Zfp189
|
zinc finger protein 189 |
| chr18_-_22850738 | 0.14 |
ENSMUST00000092015.4
ENSMUST00000069215.6 |
Nol4
|
nucleolar protein 4 |
| chr10_+_23949516 | 0.14 |
ENSMUST00000045152.4
|
Taar3
|
trace amine-associated receptor 3 |
| chr7_-_65370908 | 0.11 |
ENSMUST00000032729.6
|
Tjp1
|
tight junction protein 1 |
| chr3_+_69222412 | 0.10 |
ENSMUST00000183126.1
|
Arl14
|
ADP-ribosylation factor-like 14 |
| chr2_+_83724397 | 0.10 |
ENSMUST00000028499.4
ENSMUST00000141725.1 ENSMUST00000111740.2 |
Itgav
|
integrin alpha V |
| chr7_-_87493371 | 0.08 |
ENSMUST00000004770.5
|
Tyr
|
tyrosinase |
| chr2_-_80129458 | 0.08 |
ENSMUST00000102653.1
|
Pde1a
|
phosphodiesterase 1A, calmodulin-dependent |
| chr17_+_24718088 | 0.07 |
ENSMUST00000152407.1
|
Rps2
|
ribosomal protein S2 |
| chrX_+_7790012 | 0.07 |
ENSMUST00000140540.1
|
Gripap1
|
GRIP1 associated protein 1 |
| chr17_-_44814604 | 0.07 |
ENSMUST00000160673.1
|
Runx2
|
runt related transcription factor 2 |
| chrX_-_134111708 | 0.07 |
ENSMUST00000159259.1
ENSMUST00000113275.3 |
Nox1
|
NADPH oxidase 1 |
| chr3_-_140839637 | 0.07 |
ENSMUST00000178845.1
|
Gm6214
|
predicted gene 6214 |
| chr16_+_31422268 | 0.06 |
ENSMUST00000089759.2
|
Bdh1
|
3-hydroxybutyrate dehydrogenase, type 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Nr2e3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.1 | 9.2 | GO:0070488 | neutrophil aggregation(GO:0070488) |
| 1.5 | 6.0 | GO:0035524 | proline transmembrane transport(GO:0035524) glycine import(GO:0036233) |
| 1.2 | 3.5 | GO:0045715 | negative regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045715) |
| 0.9 | 6.4 | GO:0044806 | G-quadruplex DNA unwinding(GO:0044806) |
| 0.9 | 2.7 | GO:0003345 | proepicardium cell migration involved in pericardium morphogenesis(GO:0003345) |
| 0.8 | 6.3 | GO:0097647 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.8 | 2.4 | GO:0071603 | endothelial cell-cell adhesion(GO:0071603) |
| 0.6 | 3.1 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.5 | 1.9 | GO:0035905 | ascending aorta development(GO:0035905) ascending aorta morphogenesis(GO:0035910) |
| 0.5 | 1.4 | GO:2001076 | negative regulation of metanephric glomerulus development(GO:0072299) negative regulation of metanephric glomerular mesangial cell proliferation(GO:0072302) regulation of metanephric ureteric bud development(GO:2001074) positive regulation of metanephric ureteric bud development(GO:2001076) |
| 0.5 | 5.5 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.4 | 1.3 | GO:0070340 | detection of bacterial lipopeptide(GO:0070340) |
| 0.4 | 7.0 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.4 | 2.0 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.4 | 2.0 | GO:0002121 | inter-male aggressive behavior(GO:0002121) |
| 0.4 | 3.6 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.4 | 1.2 | GO:0060821 | inactivation of X chromosome by DNA methylation(GO:0060821) |
| 0.3 | 1.3 | GO:0061153 | negative regulation of interleukin-13 production(GO:0032696) trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) odontoblast differentiation(GO:0071895) regulation of estrogen receptor binding(GO:0071898) negative regulation of estrogen receptor binding(GO:0071899) |
| 0.3 | 1.3 | GO:1902606 | regulation of large conductance calcium-activated potassium channel activity(GO:1902606) positive regulation of large conductance calcium-activated potassium channel activity(GO:1902608) |
| 0.3 | 1.9 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.3 | 0.9 | GO:0051794 | regulation of catagen(GO:0051794) |
| 0.3 | 2.4 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.3 | 1.9 | GO:0045200 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.3 | 1.1 | GO:0032053 | ciliary basal body organization(GO:0032053) |
| 0.3 | 1.4 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) negative regulation of blood vessel remodeling(GO:0060313) |
| 0.3 | 1.0 | GO:0090309 | positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.2 | 1.5 | GO:0032346 | positive regulation of aldosterone metabolic process(GO:0032346) positive regulation of aldosterone biosynthetic process(GO:0032349) leading edge cell differentiation(GO:0035026) |
| 0.2 | 0.9 | GO:0030070 | insulin processing(GO:0030070) |
| 0.2 | 1.2 | GO:0038032 | termination of G-protein coupled receptor signaling pathway(GO:0038032) |
| 0.2 | 1.6 | GO:2000002 | negative regulation of DNA damage checkpoint(GO:2000002) |
| 0.2 | 0.6 | GO:0046087 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 0.2 | 1.6 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.2 | 0.6 | GO:0090076 | relaxation of skeletal muscle(GO:0090076) |
| 0.2 | 0.6 | GO:0045726 | positive regulation of integrin biosynthetic process(GO:0045726) |
| 0.2 | 1.5 | GO:0051581 | negative regulation of neurotransmitter uptake(GO:0051581) negative regulation of serotonin uptake(GO:0051612) |
| 0.2 | 0.5 | GO:0002223 | stimulatory C-type lectin receptor signaling pathway(GO:0002223) |
| 0.2 | 2.7 | GO:0006027 | glycosaminoglycan catabolic process(GO:0006027) |
| 0.1 | 1.5 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.1 | 0.6 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.1 | 1.4 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.1 | 0.7 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 0.1 | 2.7 | GO:0007190 | activation of adenylate cyclase activity(GO:0007190) |
| 0.1 | 0.6 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.1 | 1.9 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.1 | 1.0 | GO:0032532 | regulation of microvillus length(GO:0032532) |
| 0.1 | 4.5 | GO:0019731 | antibacterial humoral response(GO:0019731) |
| 0.1 | 0.3 | GO:0097401 | synaptic vesicle lumen acidification(GO:0097401) |
| 0.1 | 0.4 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.1 | 0.3 | GO:0032849 | positive regulation of cellular pH reduction(GO:0032849) |
| 0.1 | 3.5 | GO:0032967 | positive regulation of collagen biosynthetic process(GO:0032967) |
| 0.1 | 0.6 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.1 | 0.7 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.1 | 0.5 | GO:0032472 | Golgi calcium ion transport(GO:0032472) |
| 0.1 | 4.9 | GO:0006493 | protein O-linked glycosylation(GO:0006493) |
| 0.1 | 0.3 | GO:0050975 | sensory perception of touch(GO:0050975) |
| 0.1 | 0.5 | GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.1 | 0.2 | GO:0051466 | corticotropin-releasing hormone secretion(GO:0043396) regulation of corticotropin-releasing hormone secretion(GO:0043397) positive regulation of corticotropin-releasing hormone secretion(GO:0051466) |
| 0.1 | 1.1 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.1 | 0.3 | GO:0035977 | protein deglycosylation involved in glycoprotein catabolic process(GO:0035977) protein demannosylation(GO:0036507) protein alpha-1,2-demannosylation(GO:0036508) glycoprotein ERAD pathway(GO:0097466) mannose trimming involved in glycoprotein ERAD pathway(GO:1904382) response to glycoprotein(GO:1904587) |
| 0.0 | 0.5 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.0 | 2.1 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 1.0 | GO:0030033 | microvillus assembly(GO:0030033) |
| 0.0 | 0.8 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.0 | 0.5 | GO:0034983 | peptidyl-lysine deacetylation(GO:0034983) |
| 0.0 | 0.5 | GO:0099628 | receptor diffusion trapping(GO:0098953) postsynaptic neurotransmitter receptor diffusion trapping(GO:0098970) neurotransmitter receptor diffusion trapping(GO:0099628) |
| 0.0 | 1.0 | GO:0050872 | white fat cell differentiation(GO:0050872) |
| 0.0 | 0.4 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 1.2 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 0.6 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.8 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.0 | 0.8 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 0.2 | GO:0003051 | angiotensin-mediated drinking behavior(GO:0003051) tachykinin receptor signaling pathway(GO:0007217) operant conditioning(GO:0035106) positive regulation of saliva secretion(GO:0046878) |
| 0.0 | 0.8 | GO:0001553 | luteinization(GO:0001553) |
| 0.0 | 0.8 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.0 | 0.2 | GO:0060075 | regulation of resting membrane potential(GO:0060075) |
| 0.0 | 0.9 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 0.3 | GO:0034393 | positive regulation of smooth muscle cell apoptotic process(GO:0034393) |
| 0.0 | 1.3 | GO:0060292 | long term synaptic depression(GO:0060292) |
| 0.0 | 2.2 | GO:0030838 | positive regulation of actin filament polymerization(GO:0030838) |
| 0.0 | 0.8 | GO:0010614 | negative regulation of cardiac muscle hypertrophy(GO:0010614) |
| 0.0 | 0.8 | GO:0007131 | reciprocal meiotic recombination(GO:0007131) reciprocal DNA recombination(GO:0035825) |
| 0.0 | 3.1 | GO:0060048 | cardiac muscle contraction(GO:0060048) |
| 0.0 | 0.7 | GO:0021591 | ventricular system development(GO:0021591) |
| 0.0 | 0.7 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 0.4 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.6 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.0 | 1.1 | GO:0007173 | epidermal growth factor receptor signaling pathway(GO:0007173) |
| 0.0 | 0.5 | GO:0021983 | pituitary gland development(GO:0021983) |
| 0.0 | 0.2 | GO:0090190 | positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.5 | GO:0038045 | large latent transforming growth factor-beta complex(GO:0038045) |
| 1.1 | 3.4 | GO:0034679 | integrin alpha9-beta1 complex(GO:0034679) |
| 0.4 | 1.5 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.3 | 1.8 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.3 | 2.0 | GO:0005638 | lamin filament(GO:0005638) |
| 0.2 | 0.7 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.2 | 1.2 | GO:0001740 | Barr body(GO:0001740) |
| 0.1 | 1.6 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.1 | 1.3 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.1 | 0.9 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.1 | 7.9 | GO:0005657 | replication fork(GO:0005657) |
| 0.1 | 2.7 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.1 | 0.8 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.1 | 0.5 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.1 | 2.3 | GO:0005859 | muscle myosin complex(GO:0005859) myosin filament(GO:0032982) |
| 0.1 | 0.6 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.1 | 4.7 | GO:0016459 | myosin complex(GO:0016459) |
| 0.1 | 0.6 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.1 | 0.2 | GO:0097059 | CRLF-CLCF1 complex(GO:0097058) CNTFR-CLCF1 complex(GO:0097059) |
| 0.1 | 1.1 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 2.0 | GO:0032590 | dendrite membrane(GO:0032590) |
| 0.0 | 2.7 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.2 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.0 | 0.6 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.1 | GO:0034684 | integrin alphav-beta5 complex(GO:0034684) integrin alphav-beta6 complex(GO:0034685) |
| 0.0 | 0.2 | GO:0097422 | tubular endosome(GO:0097422) |
| 0.0 | 0.6 | GO:0000178 | exosome (RNase complex)(GO:0000178) |
| 0.0 | 2.6 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 17.6 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 0.5 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.3 | GO:0042581 | specific granule(GO:0042581) |
| 0.0 | 1.0 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 1.2 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 1.2 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 6.3 | GO:0098857 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 0.6 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 7.8 | GO:0005774 | vacuolar membrane(GO:0005774) |
| 0.0 | 1.8 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.4 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.7 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 1.4 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.8 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 1.4 | GO:0005884 | actin filament(GO:0005884) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 6.4 | GO:0033680 | ATP-dependent DNA/RNA helicase activity(GO:0033680) |
| 2.0 | 6.0 | GO:0005302 | L-tyrosine transmembrane transporter activity(GO:0005302) |
| 1.2 | 7.0 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 1.1 | 9.2 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) RAGE receptor binding(GO:0050786) |
| 0.9 | 3.5 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.7 | 3.5 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) |
| 0.6 | 1.3 | GO:0035663 | Toll-like receptor 2 binding(GO:0035663) |
| 0.5 | 2.1 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.3 | 1.6 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 0.3 | 2.4 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.3 | 4.9 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.3 | 6.9 | GO:0030553 | cGMP binding(GO:0030553) |
| 0.3 | 5.3 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.2 | 1.3 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.2 | 0.4 | GO:0017116 | single-stranded DNA-dependent ATP-dependent DNA helicase activity(GO:0017116) |
| 0.2 | 1.4 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 0.2 | 1.3 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.2 | 2.0 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.2 | 2.7 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.2 | 1.8 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.2 | 2.1 | GO:0005522 | profilin binding(GO:0005522) |
| 0.2 | 1.4 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.1 | 1.0 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.1 | 1.5 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.1 | 0.6 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.1 | 0.6 | GO:0044020 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) histone methyltransferase activity (H4-R3 specific)(GO:0044020) |
| 0.1 | 4.1 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.1 | 2.7 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 0.6 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.1 | 0.3 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.1 | 3.0 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.1 | 2.4 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.1 | 0.8 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.1 | 1.1 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.1 | 0.5 | GO:0031812 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.1 | 0.3 | GO:0008390 | testosterone 16-alpha-hydroxylase activity(GO:0008390) |
| 0.1 | 0.9 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 0.2 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.1 | 0.6 | GO:0086008 | voltage-gated potassium channel activity involved in cardiac muscle cell action potential repolarization(GO:0086008) |
| 0.1 | 2.0 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 6.3 | GO:0005179 | hormone activity(GO:0005179) |
| 0.1 | 0.8 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 1.9 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.6 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.7 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 3.1 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.8 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.7 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.6 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 4.8 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 1.9 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 1.9 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.3 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.2 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.0 | 1.5 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.0 | 1.0 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.2 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 0.5 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 1.2 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.5 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.0 | 0.5 | GO:0042923 | neuropeptide binding(GO:0042923) |
| 0.0 | 1.0 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 0.9 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.9 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 3.4 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 0.1 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.0 | 1.4 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.3 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 1.4 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 0.5 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.9 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.1 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 1.7 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 0.1 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.0 | 1.0 | GO:0046332 | SMAD binding(GO:0046332) |
| 0.0 | 0.8 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 0.2 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 10.4 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.2 | 5.5 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.1 | 2.7 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.1 | 3.5 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.1 | 1.9 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.1 | 2.1 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 7.0 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 1.3 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.1 | 7.8 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.1 | 3.1 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 4.1 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.1 | 1.8 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.0 | 0.9 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 3.4 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 1.3 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 1.3 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 3.6 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 2.0 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 1.5 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 1.5 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.5 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 0.5 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 1.0 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 4.4 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.6 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 0.8 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 1.3 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.6 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.7 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 2.6 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.6 | PID IL4 2PATHWAY | IL4-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.7 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.2 | 0.7 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.2 | 2.1 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.2 | 3.4 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.2 | 2.7 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.2 | 6.3 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 6.0 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.1 | 1.7 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.1 | 0.9 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.1 | 5.3 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.1 | 1.5 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.1 | 3.1 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 5.4 | REACTOME RESPIRATORY ELECTRON TRANSPORT ATP SYNTHESIS BY CHEMIOSMOTIC COUPLING AND HEAT PRODUCTION BY UNCOUPLING PROTEINS | Genes involved in Respiratory electron transport, ATP synthesis by chemiosmotic coupling, and heat production by uncoupling proteins. |
| 0.1 | 1.0 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 1.3 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 1.1 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.9 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 2.0 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.6 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.9 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.3 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.7 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 5.0 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.2 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 2.1 | REACTOME SIGNALING BY PDGF | Genes involved in Signaling by PDGF |
| 0.0 | 1.3 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 1.5 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 1.3 | REACTOME MYD88 MAL CASCADE INITIATED ON PLASMA MEMBRANE | Genes involved in MyD88:Mal cascade initiated on plasma membrane |
| 0.0 | 0.2 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.7 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.4 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.5 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |