Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
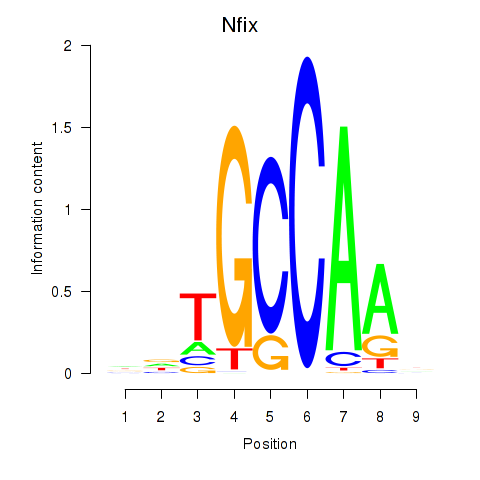
Results for Nfix
Z-value: 1.24
Transcription factors associated with Nfix
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nfix
|
ENSMUSG00000001911.10 | nuclear factor I/X |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nfix | mm10_v2_chr8_-_84773381_84773427 | 0.44 | 7.1e-03 | Click! |
Activity profile of Nfix motif
Sorted Z-values of Nfix motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_97417730 | 9.89 |
ENSMUST00000043077.7
|
Thrsp
|
thyroid hormone responsive |
| chr8_-_93131271 | 4.68 |
ENSMUST00000034189.8
|
Ces1c
|
carboxylesterase 1C |
| chr7_+_140763739 | 4.38 |
ENSMUST00000026552.7
|
Cyp2e1
|
cytochrome P450, family 2, subfamily e, polypeptide 1 |
| chr7_+_140845562 | 4.19 |
ENSMUST00000035300.5
|
Scgb1c1
|
secretoglobin, family 1C, member 1 |
| chr5_-_77115145 | 3.69 |
ENSMUST00000081964.5
|
Hopx
|
HOP homeobox |
| chr6_-_141856171 | 3.20 |
ENSMUST00000165990.1
ENSMUST00000163678.1 |
Slco1a4
|
solute carrier organic anion transporter family, member 1a4 |
| chr10_-_109010955 | 2.95 |
ENSMUST00000105276.1
ENSMUST00000064054.7 |
Syt1
|
synaptotagmin I |
| chr19_-_46672883 | 2.89 |
ENSMUST00000026012.7
|
Cyp17a1
|
cytochrome P450, family 17, subfamily a, polypeptide 1 |
| chr3_-_129332713 | 2.86 |
ENSMUST00000029658.7
|
Enpep
|
glutamyl aminopeptidase |
| chr5_-_87337165 | 2.66 |
ENSMUST00000031195.2
|
Ugt2a3
|
UDP glucuronosyltransferase 2 family, polypeptide A3 |
| chr19_+_44333092 | 2.59 |
ENSMUST00000058856.8
|
Scd4
|
stearoyl-coenzyme A desaturase 4 |
| chr7_-_48848023 | 2.55 |
ENSMUST00000032658.6
|
Csrp3
|
cysteine and glycine-rich protein 3 |
| chr11_-_78422217 | 2.33 |
ENSMUST00000001122.5
|
Slc13a2
|
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 2 |
| chr14_+_65666394 | 2.29 |
ENSMUST00000022610.8
|
Scara5
|
scavenger receptor class A, member 5 (putative) |
| chr10_+_57784859 | 2.18 |
ENSMUST00000020024.5
|
Fabp7
|
fatty acid binding protein 7, brain |
| chr10_+_57784914 | 2.01 |
ENSMUST00000165013.1
|
Fabp7
|
fatty acid binding protein 7, brain |
| chr1_+_88070765 | 1.89 |
ENSMUST00000073772.4
|
Ugt1a9
|
UDP glucuronosyltransferase 1 family, polypeptide A9 |
| chr19_-_43524462 | 1.88 |
ENSMUST00000026196.7
|
Got1
|
glutamate oxaloacetate transaminase 1, soluble |
| chr5_-_87092546 | 1.72 |
ENSMUST00000132667.1
ENSMUST00000145617.1 ENSMUST00000094649.4 |
Ugt2b36
|
UDP glucuronosyltransferase 2 family, polypeptide B36 |
| chr16_+_42907563 | 1.61 |
ENSMUST00000151244.1
ENSMUST00000114694.2 |
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr10_-_24101951 | 1.61 |
ENSMUST00000170267.1
|
Taar8c
|
trace amine-associated receptor 8C |
| chr11_-_89302545 | 1.60 |
ENSMUST00000061728.3
|
Nog
|
noggin |
| chr2_+_122147680 | 1.58 |
ENSMUST00000102476.4
|
B2m
|
beta-2 microglobulin |
| chr4_-_141623799 | 1.55 |
ENSMUST00000038661.7
|
Slc25a34
|
solute carrier family 25, member 34 |
| chr11_+_101367542 | 1.52 |
ENSMUST00000019469.2
|
G6pc
|
glucose-6-phosphatase, catalytic |
| chr14_+_65666430 | 1.46 |
ENSMUST00000069226.6
|
Scara5
|
scavenger receptor class A, member 5 (putative) |
| chrX_-_143933089 | 1.41 |
ENSMUST00000087313.3
|
Dcx
|
doublecortin |
| chr10_-_89506631 | 1.40 |
ENSMUST00000058126.8
ENSMUST00000105296.2 |
Nr1h4
|
nuclear receptor subfamily 1, group H, member 4 |
| chr5_+_90460889 | 1.40 |
ENSMUST00000031314.8
|
Alb
|
albumin |
| chr11_+_78290841 | 1.39 |
ENSMUST00000046361.4
|
BC030499
|
cDNA sequence BC030499 |
| chr17_-_73950172 | 1.36 |
ENSMUST00000024866.4
|
Xdh
|
xanthine dehydrogenase |
| chr15_-_101924725 | 1.34 |
ENSMUST00000023797.6
|
Krt4
|
keratin 4 |
| chr1_+_133365160 | 1.32 |
ENSMUST00000129213.1
|
Etnk2
|
ethanolamine kinase 2 |
| chrX_+_38316177 | 1.30 |
ENSMUST00000016471.2
ENSMUST00000115134.1 |
Atp1b4
|
ATPase, (Na+)/K+ transporting, beta 4 polypeptide |
| chr3_-_145032765 | 1.30 |
ENSMUST00000029919.5
|
Clca3
|
chloride channel calcium activated 3 |
| chr11_-_100121558 | 1.28 |
ENSMUST00000007275.2
|
Krt13
|
keratin 13 |
| chr1_-_162898484 | 1.25 |
ENSMUST00000143123.1
|
Fmo2
|
flavin containing monooxygenase 2 |
| chr5_+_92387846 | 1.24 |
ENSMUST00000138687.1
ENSMUST00000124509.1 |
Art3
|
ADP-ribosyltransferase 3 |
| chr17_-_24209377 | 1.24 |
ENSMUST00000024931.4
|
Ntn3
|
netrin 3 |
| chrX_-_8193387 | 1.24 |
ENSMUST00000143223.1
ENSMUST00000033509.8 |
Ebp
|
phenylalkylamine Ca2+ antagonist (emopamil) binding protein |
| chr4_-_46991842 | 1.23 |
ENSMUST00000107749.2
|
Gabbr2
|
gamma-aminobutyric acid (GABA) B receptor, 2 |
| chr7_+_131032061 | 1.23 |
ENSMUST00000084509.3
|
Dmbt1
|
deleted in malignant brain tumors 1 |
| chr5_+_92387673 | 1.21 |
ENSMUST00000145072.1
|
Art3
|
ADP-ribosyltransferase 3 |
| chr9_-_103222063 | 1.19 |
ENSMUST00000170904.1
|
Trf
|
transferrin |
| chrX_-_100412587 | 1.18 |
ENSMUST00000033567.8
|
Awat2
|
acyl-CoA wax alcohol acyltransferase 2 |
| chr11_-_116654245 | 1.14 |
ENSMUST00000021166.5
|
Cygb
|
cytoglobin |
| chr6_+_78380700 | 1.09 |
ENSMUST00000101272.1
|
Reg3a
|
regenerating islet-derived 3 alpha |
| chr15_+_10952332 | 1.08 |
ENSMUST00000022853.8
ENSMUST00000110523.1 |
C1qtnf3
|
C1q and tumor necrosis factor related protein 3 |
| chr7_-_119479249 | 1.08 |
ENSMUST00000033263.4
|
Umod
|
uromodulin |
| chr7_-_90129339 | 1.07 |
ENSMUST00000181189.1
|
2310010J17Rik
|
RIKEN cDNA 2310010J17 gene |
| chr6_-_136875794 | 1.07 |
ENSMUST00000032342.1
|
Mgp
|
matrix Gla protein |
| chr11_+_99864476 | 1.04 |
ENSMUST00000092694.3
|
Gm11559
|
predicted gene 11559 |
| chr18_+_43963234 | 1.03 |
ENSMUST00000069245.7
|
Spink5
|
serine peptidase inhibitor, Kazal type 5 |
| chr13_-_41847626 | 1.03 |
ENSMUST00000121404.1
|
Adtrp
|
androgen dependent TFPI regulating protein |
| chr6_+_141524379 | 1.01 |
ENSMUST00000032362.9
|
Slco1c1
|
solute carrier organic anion transporter family, member 1c1 |
| chr2_+_32646586 | 1.00 |
ENSMUST00000009705.7
ENSMUST00000167841.1 |
Eng
|
endoglin |
| chr16_+_41532851 | 0.96 |
ENSMUST00000078873.4
|
Lsamp
|
limbic system-associated membrane protein |
| chr16_+_43235856 | 0.95 |
ENSMUST00000146708.1
|
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr3_+_92288566 | 0.95 |
ENSMUST00000090872.4
|
Sprr2a3
|
small proline-rich protein 2A3 |
| chr1_+_87594545 | 0.93 |
ENSMUST00000165109.1
ENSMUST00000070898.5 |
Neu2
|
neuraminidase 2 |
| chr13_-_41847599 | 0.91 |
ENSMUST00000179758.1
|
Adtrp
|
androgen dependent TFPI regulating protein |
| chr12_-_75735729 | 0.91 |
ENSMUST00000021450.4
|
Sgpp1
|
sphingosine-1-phosphate phosphatase 1 |
| chr4_+_82065924 | 0.91 |
ENSMUST00000161588.1
|
Gm5860
|
predicted gene 5860 |
| chr7_+_27195781 | 0.90 |
ENSMUST00000108379.1
ENSMUST00000179391.1 |
BC024978
|
cDNA sequence BC024978 |
| chr5_+_24413406 | 0.90 |
ENSMUST00000049346.5
|
Asic3
|
acid-sensing (proton-gated) ion channel 3 |
| chr7_+_28180226 | 0.90 |
ENSMUST00000172467.1
|
Dyrk1b
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1b |
| chr2_+_133552159 | 0.88 |
ENSMUST00000028836.6
|
Bmp2
|
bone morphogenetic protein 2 |
| chr3_-_39359128 | 0.88 |
ENSMUST00000056409.2
|
Gm9845
|
predicted pseudogene 9845 |
| chr7_-_105399991 | 0.88 |
ENSMUST00000118726.1
ENSMUST00000074686.7 ENSMUST00000122327.1 ENSMUST00000179474.1 ENSMUST00000048079.6 |
Fam160a2
|
family with sequence similarity 160, member A2 |
| chr7_+_28180272 | 0.86 |
ENSMUST00000173223.1
|
Dyrk1b
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1b |
| chr15_-_101712891 | 0.85 |
ENSMUST00000023709.5
|
Krt5
|
keratin 5 |
| chr6_+_78370877 | 0.85 |
ENSMUST00000096904.3
|
Reg3b
|
regenerating islet-derived 3 beta |
| chr3_+_123267445 | 0.85 |
ENSMUST00000047923.7
|
Sec24d
|
Sec24 related gene family, member D (S. cerevisiae) |
| chr11_-_43836243 | 0.84 |
ENSMUST00000167574.1
|
Adra1b
|
adrenergic receptor, alpha 1b |
| chr4_+_102430047 | 0.84 |
ENSMUST00000172616.1
|
Pde4b
|
phosphodiesterase 4B, cAMP specific |
| chr3_-_89279633 | 0.83 |
ENSMUST00000118860.1
ENSMUST00000029566.2 |
Efna1
|
ephrin A1 |
| chr4_-_129227883 | 0.82 |
ENSMUST00000106051.1
|
C77080
|
expressed sequence C77080 |
| chr12_-_56535047 | 0.80 |
ENSMUST00000178477.2
|
Nkx2-1
|
NK2 homeobox 1 |
| chr8_+_31091593 | 0.78 |
ENSMUST00000161713.1
|
Dusp26
|
dual specificity phosphatase 26 (putative) |
| chr1_-_163725123 | 0.78 |
ENSMUST00000159679.1
|
Mettl11b
|
methyltransferase like 11B |
| chr17_-_34862122 | 0.78 |
ENSMUST00000154526.1
|
Cfb
|
complement factor B |
| chr2_-_71750083 | 0.77 |
ENSMUST00000180494.1
|
Gm17250
|
predicted gene, 17250 |
| chr8_-_119635553 | 0.75 |
ENSMUST00000061828.3
|
Kcng4
|
potassium voltage-gated channel, subfamily G, member 4 |
| chr12_-_76795489 | 0.75 |
ENSMUST00000082431.3
|
Gpx2
|
glutathione peroxidase 2 |
| chr4_-_119415494 | 0.74 |
ENSMUST00000063642.2
|
Ccdc30
|
coiled-coil domain containing 30 |
| chr17_-_34862473 | 0.74 |
ENSMUST00000025229.4
ENSMUST00000176203.2 ENSMUST00000128767.1 |
Cfb
|
complement factor B |
| chr18_-_39490649 | 0.73 |
ENSMUST00000115567.1
|
Nr3c1
|
nuclear receptor subfamily 3, group C, member 1 |
| chr4_+_82065855 | 0.73 |
ENSMUST00000151038.1
|
Gm5860
|
predicted gene 5860 |
| chr15_-_64922290 | 0.72 |
ENSMUST00000023007.5
|
Adcy8
|
adenylate cyclase 8 |
| chr6_+_139843648 | 0.72 |
ENSMUST00000087657.6
|
Pik3c2g
|
phosphatidylinositol 3-kinase, C2 domain containing, gamma polypeptide |
| chr10_-_95415484 | 0.72 |
ENSMUST00000172070.1
ENSMUST00000150432.1 |
Socs2
|
suppressor of cytokine signaling 2 |
| chr17_-_35626634 | 0.70 |
ENSMUST00000164502.2
|
Gm9573
|
predicted gene 9573 |
| chrX_+_101377267 | 0.70 |
ENSMUST00000052130.7
|
Gjb1
|
gap junction protein, beta 1 |
| chr17_+_45506825 | 0.70 |
ENSMUST00000024733.7
|
Aars2
|
alanyl-tRNA synthetase 2, mitochondrial (putative) |
| chr10_-_127370535 | 0.68 |
ENSMUST00000026472.8
|
Inhbc
|
inhibin beta-C |
| chr2_-_26092149 | 0.68 |
ENSMUST00000114159.2
|
Nacc2
|
nucleus accumbens associated 2, BEN and BTB (POZ) domain containing |
| chr8_-_64733534 | 0.68 |
ENSMUST00000141021.1
|
Sc4mol
|
sterol-C4-methyl oxidase-like |
| chr4_+_102570065 | 0.68 |
ENSMUST00000097950.2
|
Pde4b
|
phosphodiesterase 4B, cAMP specific |
| chr6_-_126166726 | 0.68 |
ENSMUST00000112244.2
ENSMUST00000050484.7 |
Ntf3
|
neurotrophin 3 |
| chr4_-_129261394 | 0.67 |
ENSMUST00000145261.1
|
C77080
|
expressed sequence C77080 |
| chr16_-_3908639 | 0.67 |
ENSMUST00000115859.1
|
1700037C18Rik
|
RIKEN cDNA 1700037C18 gene |
| chr7_+_130936172 | 0.66 |
ENSMUST00000006367.7
|
Htra1
|
HtrA serine peptidase 1 |
| chr1_+_169929929 | 0.66 |
ENSMUST00000175731.1
|
1700084C01Rik
|
RIKEN cDNA 1700084C01 gene |
| chr8_+_94472763 | 0.64 |
ENSMUST00000053085.5
|
Nlrc5
|
NLR family, CARD domain containing 5 |
| chr11_+_77462605 | 0.64 |
ENSMUST00000130255.1
|
Coro6
|
coronin 6 |
| chr18_-_23038656 | 0.64 |
ENSMUST00000081423.6
|
Nol4
|
nucleolar protein 4 |
| chr9_+_89199319 | 0.64 |
ENSMUST00000138109.1
|
Mthfs
|
5, 10-methenyltetrahydrofolate synthetase |
| chr19_-_34879452 | 0.63 |
ENSMUST00000036584.5
|
Pank1
|
pantothenate kinase 1 |
| chr11_+_115462464 | 0.63 |
ENSMUST00000106532.3
ENSMUST00000092445.5 ENSMUST00000153466.1 |
Slc16a5
|
solute carrier family 16 (monocarboxylic acid transporters), member 5 |
| chr10_-_40142247 | 0.63 |
ENSMUST00000092566.6
|
Slc16a10
|
solute carrier family 16 (monocarboxylic acid transporters), member 10 |
| chr5_-_84417359 | 0.62 |
ENSMUST00000113401.1
|
Epha5
|
Eph receptor A5 |
| chr11_+_67798269 | 0.62 |
ENSMUST00000168612.1
ENSMUST00000040574.4 |
Dhrs7c
|
dehydrogenase/reductase (SDR family) member 7C |
| chr7_-_15879844 | 0.61 |
ENSMUST00000172758.1
ENSMUST00000044434.6 |
Crx
|
cone-rod homeobox containing gene |
| chr12_-_55492587 | 0.60 |
ENSMUST00000021413.7
|
Nfkbia
|
nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor, alpha |
| chr15_-_78206391 | 0.60 |
ENSMUST00000120592.1
|
Pvalb
|
parvalbumin |
| chr6_-_125313844 | 0.60 |
ENSMUST00000032489.7
|
Ltbr
|
lymphotoxin B receptor |
| chr7_+_28179469 | 0.59 |
ENSMUST00000085901.6
ENSMUST00000172761.1 |
Dyrk1b
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1b |
| chr5_+_120511213 | 0.59 |
ENSMUST00000111890.2
ENSMUST00000076051.5 ENSMUST00000147496.1 |
Slc8b1
|
solute carrier family 8 (sodium/lithium/calcium exchanger), member B1 |
| chr7_+_114745685 | 0.59 |
ENSMUST00000136645.1
ENSMUST00000169913.1 |
Insc
|
inscuteable homolog (Drosophila) |
| chrX_-_48208566 | 0.58 |
ENSMUST00000037960.4
|
Zdhhc9
|
zinc finger, DHHC domain containing 9 |
| chr5_-_86906937 | 0.58 |
ENSMUST00000031181.9
ENSMUST00000113333.1 |
Ugt2b34
|
UDP glucuronosyltransferase 2 family, polypeptide B34 |
| chr6_-_33060256 | 0.58 |
ENSMUST00000066379.4
|
Chchd3
|
coiled-coil-helix-coiled-coil-helix domain containing 3 |
| chr5_-_124352233 | 0.57 |
ENSMUST00000111472.1
|
Cdk2ap1
|
CDK2 (cyclin-dependent kinase 2)-associated protein 1 |
| chr1_+_88055467 | 0.56 |
ENSMUST00000173325.1
|
Ugt1a10
|
UDP glycosyltransferase 1 family, polypeptide A10 |
| chr13_-_26770157 | 0.56 |
ENSMUST00000055915.3
|
Hdgfl1
|
hepatoma derived growth factor-like 1 |
| chr2_-_160327494 | 0.55 |
ENSMUST00000099127.2
|
Gm826
|
predicted gene 826 |
| chr6_-_5256226 | 0.55 |
ENSMUST00000125686.1
ENSMUST00000031773.2 |
Pon3
|
paraoxonase 3 |
| chr5_-_103977326 | 0.55 |
ENSMUST00000120320.1
|
Hsd17b13
|
hydroxysteroid (17-beta) dehydrogenase 13 |
| chr7_-_132599637 | 0.54 |
ENSMUST00000054562.3
|
Nkx1-2
|
NK1 transcription factor related, locus 2 (Drosophila) |
| chrX_-_48208870 | 0.54 |
ENSMUST00000088935.3
|
Zdhhc9
|
zinc finger, DHHC domain containing 9 |
| chr19_-_11829024 | 0.54 |
ENSMUST00000061235.2
|
Olfr1417
|
olfactory receptor 1417 |
| chr6_+_34746368 | 0.54 |
ENSMUST00000142716.1
|
Cald1
|
caldesmon 1 |
| chr14_+_65837302 | 0.53 |
ENSMUST00000022614.5
|
Ccdc25
|
coiled-coil domain containing 25 |
| chr13_+_41655697 | 0.52 |
ENSMUST00000067176.8
|
Gm5082
|
predicted gene 5082 |
| chr4_+_139653538 | 0.52 |
ENSMUST00000030510.7
ENSMUST00000166773.1 |
Tas1r2
|
taste receptor, type 1, member 2 |
| chr2_-_147186389 | 0.51 |
ENSMUST00000109970.3
ENSMUST00000067075.5 |
Nkx2-2
|
NK2 homeobox 2 |
| chr10_+_4611971 | 0.51 |
ENSMUST00000105590.1
ENSMUST00000067086.7 |
Esr1
|
estrogen receptor 1 (alpha) |
| chr19_+_46623387 | 0.51 |
ENSMUST00000111855.4
|
Wbp1l
|
WW domain binding protein 1 like |
| chr6_-_33060172 | 0.50 |
ENSMUST00000115091.1
ENSMUST00000127666.1 |
Chchd3
|
coiled-coil-helix-coiled-coil-helix domain containing 3 |
| chr6_+_127015542 | 0.50 |
ENSMUST00000000187.5
|
Fgf6
|
fibroblast growth factor 6 |
| chr3_+_90080442 | 0.50 |
ENSMUST00000127955.1
|
Tpm3
|
tropomyosin 3, gamma |
| chr9_-_87731248 | 0.49 |
ENSMUST00000034991.7
|
Tbx18
|
T-box18 |
| chr11_+_98383811 | 0.49 |
ENSMUST00000008021.2
|
Tcap
|
titin-cap |
| chr11_-_84068357 | 0.49 |
ENSMUST00000100705.4
|
Dusp14
|
dual specificity phosphatase 14 |
| chr5_+_65934922 | 0.49 |
ENSMUST00000153624.1
|
Chrna9
|
cholinergic receptor, nicotinic, alpha polypeptide 9 |
| chr5_-_103977404 | 0.48 |
ENSMUST00000112803.2
|
Hsd17b13
|
hydroxysteroid (17-beta) dehydrogenase 13 |
| chr1_-_56969864 | 0.47 |
ENSMUST00000177424.1
|
Satb2
|
special AT-rich sequence binding protein 2 |
| chr8_-_83955205 | 0.47 |
ENSMUST00000098595.2
|
Gm10644
|
predicted gene 10644 |
| chr5_-_103977360 | 0.47 |
ENSMUST00000048118.8
|
Hsd17b13
|
hydroxysteroid (17-beta) dehydrogenase 13 |
| chr11_+_67238017 | 0.46 |
ENSMUST00000018632.4
|
Myh4
|
myosin, heavy polypeptide 4, skeletal muscle |
| chr12_-_81532840 | 0.46 |
ENSMUST00000169158.1
ENSMUST00000164431.1 ENSMUST00000163402.1 ENSMUST00000166664.1 ENSMUST00000164386.1 |
Synj2bp
Gm20498
|
synaptojanin 2 binding protein predicted gene 20498 |
| chr8_-_36732897 | 0.45 |
ENSMUST00000098826.3
|
Dlc1
|
deleted in liver cancer 1 |
| chr9_-_44965519 | 0.45 |
ENSMUST00000125642.1
ENSMUST00000117506.1 ENSMUST00000117549.1 |
Ube4a
|
ubiquitination factor E4A, UFD2 homolog (S. cerevisiae) |
| chr4_-_82065889 | 0.45 |
ENSMUST00000143755.1
|
Gm11264
|
predicted gene 11264 |
| chr2_+_59484645 | 0.45 |
ENSMUST00000028369.5
|
Dapl1
|
death associated protein-like 1 |
| chr6_-_115251839 | 0.45 |
ENSMUST00000032462.6
|
Timp4
|
tissue inhibitor of metalloproteinase 4 |
| chr7_+_62476306 | 0.45 |
ENSMUST00000097132.3
|
Atp5l-ps1
|
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit g, pseudogene 1 |
| chr9_+_32372409 | 0.44 |
ENSMUST00000047334.8
|
Kcnj1
|
potassium inwardly-rectifying channel, subfamily J, member 1 |
| chr7_+_26757153 | 0.42 |
ENSMUST00000077855.6
|
Cyp2b19
|
cytochrome P450, family 2, subfamily b, polypeptide 19 |
| chr4_+_116720920 | 0.42 |
ENSMUST00000045542.6
ENSMUST00000106459.1 |
Tesk2
|
testis-specific kinase 2 |
| chr5_-_38480131 | 0.41 |
ENSMUST00000143758.1
ENSMUST00000067886.5 |
Slc2a9
|
solute carrier family 2 (facilitated glucose transporter), member 9 |
| chr10_+_116986314 | 0.41 |
ENSMUST00000020378.4
|
Best3
|
bestrophin 3 |
| chr2_+_20737306 | 0.41 |
ENSMUST00000114606.1
ENSMUST00000114608.1 |
Etl4
|
enhancer trap locus 4 |
| chr4_+_95579463 | 0.40 |
ENSMUST00000150830.1
ENSMUST00000134012.2 |
Fggy
|
FGGY carbohydrate kinase domain containing |
| chr6_-_112288837 | 0.40 |
ENSMUST00000053559.4
|
5031434C07Rik
|
RIKEN cDNA 5031434C07 gene |
| chr12_-_40445754 | 0.40 |
ENSMUST00000069692.8
ENSMUST00000069637.7 |
Zfp277
|
zinc finger protein 277 |
| chr3_+_129199960 | 0.40 |
ENSMUST00000173645.2
|
Pitx2
|
paired-like homeodomain transcription factor 2 |
| chr7_+_27653906 | 0.39 |
ENSMUST00000008088.7
|
Ttc9b
|
tetratricopeptide repeat domain 9B |
| chr11_-_84068554 | 0.39 |
ENSMUST00000164891.1
|
Dusp14
|
dual specificity phosphatase 14 |
| chr10_-_95415283 | 0.39 |
ENSMUST00000119917.1
|
Socs2
|
suppressor of cytokine signaling 2 |
| chr6_-_34910869 | 0.39 |
ENSMUST00000081214.5
|
Wdr91
|
WD repeat domain 91 |
| chr7_+_125829653 | 0.38 |
ENSMUST00000124223.1
|
D430042O09Rik
|
RIKEN cDNA D430042O09 gene |
| chr11_-_120624973 | 0.38 |
ENSMUST00000106183.2
ENSMUST00000080202.5 |
Sirt7
|
sirtuin 7 |
| chr5_+_102768771 | 0.38 |
ENSMUST00000112852.1
|
Arhgap24
|
Rho GTPase activating protein 24 |
| chr2_+_33216051 | 0.38 |
ENSMUST00000004208.5
|
Angptl2
|
angiopoietin-like 2 |
| chr6_-_129451906 | 0.38 |
ENSMUST00000037481.7
|
Clec1a
|
C-type lectin domain family 1, member a |
| chr6_+_90122643 | 0.37 |
ENSMUST00000174204.1
|
Vmn1r51
|
vomeronasal 1 receptor 51 |
| chr10_-_58675631 | 0.37 |
ENSMUST00000003312.4
|
Edar
|
ectodysplasin-A receptor |
| chr16_-_59555752 | 0.37 |
ENSMUST00000179383.1
ENSMUST00000044604.8 |
Crybg3
|
beta-gamma crystallin domain containing 3 |
| chr18_-_20114767 | 0.36 |
ENSMUST00000038710.5
|
Dsc1
|
desmocollin 1 |
| chrX_-_141874870 | 0.36 |
ENSMUST00000182079.1
|
Gm15294
|
predicted gene 15294 |
| chr10_+_24076500 | 0.36 |
ENSMUST00000051133.5
|
Taar8a
|
trace amine-associated receptor 8A |
| chr4_-_14621669 | 0.35 |
ENSMUST00000143105.1
|
Slc26a7
|
solute carrier family 26, member 7 |
| chr1_-_52500679 | 0.35 |
ENSMUST00000069792.7
|
Nab1
|
Ngfi-A binding protein 1 |
| chr5_+_24428208 | 0.35 |
ENSMUST00000115049.2
|
Slc4a2
|
solute carrier family 4 (anion exchanger), member 2 |
| chr7_+_104329471 | 0.35 |
ENSMUST00000180136.1
ENSMUST00000178316.1 |
Trim34b
|
tripartite motif-containing 34B |
| chr19_+_56287911 | 0.35 |
ENSMUST00000095948.4
|
Habp2
|
hyaluronic acid binding protein 2 |
| chr11_+_67200137 | 0.34 |
ENSMUST00000129018.1
|
Myh1
|
myosin, heavy polypeptide 1, skeletal muscle, adult |
| chr1_-_86111970 | 0.34 |
ENSMUST00000027431.6
|
Htr2b
|
5-hydroxytryptamine (serotonin) receptor 2B |
| chr11_+_115912001 | 0.33 |
ENSMUST00000132961.1
|
Smim6
|
small integral membrane protein 6 |
| chr9_+_44101722 | 0.33 |
ENSMUST00000161703.1
ENSMUST00000161381.1 ENSMUST00000034654.7 |
Mfrp
|
membrane-type frizzled-related protein |
| chr2_-_37443096 | 0.33 |
ENSMUST00000102789.2
ENSMUST00000067043.4 ENSMUST00000112932.1 |
Zbtb26
Zbtb6
|
zinc finger and BTB domain containing 26 zinc finger and BTB domain containing 6 |
| chr6_-_78378851 | 0.33 |
ENSMUST00000089667.1
ENSMUST00000167492.1 |
Reg3d
|
regenerating islet-derived 3 delta |
| chr8_+_58911755 | 0.33 |
ENSMUST00000062978.6
|
BC030500
|
cDNA sequence BC030500 |
| chr10_+_59616063 | 0.33 |
ENSMUST00000095646.1
|
Gm10322
|
predicted gene 10322 |
| chr19_+_56287943 | 0.32 |
ENSMUST00000166049.1
|
Habp2
|
hyaluronic acid binding protein 2 |
| chrX_-_60147643 | 0.32 |
ENSMUST00000033478.4
ENSMUST00000101531.3 |
Mcf2
|
mcf.2 transforming sequence |
| chrX_-_155623325 | 0.31 |
ENSMUST00000038665.5
|
Ptchd1
|
patched domain containing 1 |
| chr5_+_104775911 | 0.31 |
ENSMUST00000055593.7
|
Gm8258
|
predicted gene 8258 |
| chr13_+_23695791 | 0.31 |
ENSMUST00000041052.2
|
Hist1h1t
|
histone cluster 1, H1t |
Network of associatons between targets according to the STRING database.
First level regulatory network of Nfix
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.6 | GO:1903919 | regulation of actin filament severing(GO:1903918) negative regulation of actin filament severing(GO:1903919) |
| 0.6 | 2.6 | GO:1903966 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.6 | 1.9 | GO:0006533 | fumarate metabolic process(GO:0006106) aspartate biosynthetic process(GO:0006532) aspartate catabolic process(GO:0006533) |
| 0.5 | 1.6 | GO:0060300 | regulation of cytokine activity(GO:0060300) |
| 0.5 | 1.4 | GO:0034971 | histone H3-R17 methylation(GO:0034971) |
| 0.5 | 1.4 | GO:0051659 | maintenance of mitochondrion location(GO:0051659) |
| 0.5 | 3.7 | GO:0043415 | positive regulation of skeletal muscle tissue regeneration(GO:0043415) |
| 0.4 | 0.9 | GO:0003134 | BMP signaling pathway involved in heart induction(GO:0003130) endodermal-mesodermal cell signaling(GO:0003133) endodermal-mesodermal cell signaling involved in heart induction(GO:0003134) |
| 0.4 | 2.9 | GO:0002003 | angiotensin maturation(GO:0002003) |
| 0.3 | 2.8 | GO:0097460 | ferrous iron import into cell(GO:0097460) ferrous iron import across plasma membrane(GO:0098707) |
| 0.3 | 1.0 | GO:0003032 | detection of oxygen(GO:0003032) cardiac jelly development(GO:1905072) |
| 0.3 | 5.1 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.3 | 0.8 | GO:0001982 | baroreceptor response to decreased systemic arterial blood pressure(GO:0001982) |
| 0.3 | 0.6 | GO:0046271 | coumarin metabolic process(GO:0009804) phenylpropanoid catabolic process(GO:0046271) |
| 0.3 | 1.4 | GO:0009115 | xanthine catabolic process(GO:0009115) |
| 0.3 | 1.1 | GO:0071638 | negative regulation of monocyte chemotactic protein-1 production(GO:0071638) |
| 0.3 | 2.9 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.3 | 0.8 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.3 | 0.8 | GO:1902310 | positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 0.3 | 1.5 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.2 | 0.7 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.2 | 0.9 | GO:0006668 | sphinganine-1-phosphate metabolic process(GO:0006668) |
| 0.2 | 0.7 | GO:1900477 | negative regulation of G1/S transition of mitotic cell cycle by negative regulation of transcription from RNA polymerase II promoter(GO:1900477) |
| 0.2 | 5.8 | GO:0017144 | drug metabolic process(GO:0017144) |
| 0.2 | 8.6 | GO:0010866 | regulation of triglyceride biosynthetic process(GO:0010866) |
| 0.2 | 1.1 | GO:0072236 | metanephric loop of Henle development(GO:0072236) |
| 0.2 | 4.2 | GO:0060134 | prepulse inhibition(GO:0060134) |
| 0.2 | 3.8 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.2 | 1.0 | GO:1900425 | negative regulation of defense response to bacterium(GO:1900425) |
| 0.2 | 0.8 | GO:0014028 | notochord formation(GO:0014028) positive regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902961) positive regulation of aspartic-type peptidase activity(GO:1905247) |
| 0.2 | 1.2 | GO:0010025 | wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 0.2 | 3.6 | GO:0006704 | glucocorticoid biosynthetic process(GO:0006704) |
| 0.2 | 0.6 | GO:0050973 | detection of mechanical stimulus involved in equilibrioception(GO:0050973) |
| 0.2 | 0.9 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.2 | 0.5 | GO:0021530 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.2 | 0.5 | GO:0072198 | mesenchymal cell proliferation involved in ureter development(GO:0072198) regulation of mesenchymal cell proliferation involved in ureter development(GO:0072199) positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 0.1 | 1.3 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.1 | 1.1 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.1 | 1.1 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.1 | 0.7 | GO:0007403 | glial cell fate determination(GO:0007403) |
| 0.1 | 1.6 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.1 | 0.3 | GO:0007208 | phospholipase C-activating serotonin receptor signaling pathway(GO:0007208) |
| 0.1 | 1.5 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.1 | 4.2 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.1 | 0.5 | GO:0060523 | prostate epithelial cord elongation(GO:0060523) |
| 0.1 | 1.1 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.1 | 0.4 | GO:0045897 | regulation of transcription during mitosis(GO:0045896) positive regulation of transcription during mitosis(GO:0045897) |
| 0.1 | 1.1 | GO:0010838 | positive regulation of keratinocyte proliferation(GO:0010838) negative regulation of keratinocyte differentiation(GO:0045617) |
| 0.1 | 0.5 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.1 | 0.6 | GO:0070427 | nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070427) |
| 0.1 | 0.3 | GO:0021998 | neural plate mediolateral regionalization(GO:0021998) mesoderm structural organization(GO:0048338) paraxial mesoderm structural organization(GO:0048352) positive regulation of cardiac ventricle development(GO:1904414) |
| 0.1 | 0.5 | GO:0035995 | skeletal muscle myosin thick filament assembly(GO:0030241) detection of muscle stretch(GO:0035995) |
| 0.1 | 0.2 | GO:0021558 | trochlear nerve development(GO:0021558) |
| 0.1 | 0.2 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 0.1 | 0.6 | GO:0045345 | MHC class I biosynthetic process(GO:0045341) regulation of MHC class I biosynthetic process(GO:0045343) positive regulation of MHC class I biosynthetic process(GO:0045345) |
| 0.1 | 0.7 | GO:0060718 | chorionic trophoblast cell differentiation(GO:0060718) |
| 0.1 | 1.1 | GO:0042407 | cristae formation(GO:0042407) |
| 0.1 | 0.6 | GO:0009396 | folic acid-containing compound biosynthetic process(GO:0009396) |
| 0.1 | 1.4 | GO:0021860 | pyramidal neuron development(GO:0021860) |
| 0.1 | 0.7 | GO:0015868 | purine ribonucleotide transport(GO:0015868) |
| 0.1 | 0.6 | GO:1903207 | neuron death in response to hydrogen peroxide(GO:0036476) regulation of hydrogen peroxide-induced neuron death(GO:1903207) negative regulation of hydrogen peroxide-induced neuron death(GO:1903208) |
| 0.1 | 0.6 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.1 | 1.2 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.1 | 0.8 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.1 | 0.2 | GO:1901475 | pyruvate transmembrane transport(GO:1901475) |
| 0.1 | 0.7 | GO:0002862 | negative regulation of inflammatory response to antigenic stimulus(GO:0002862) |
| 0.1 | 3.4 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.1 | 2.5 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.1 | 0.5 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.1 | 0.2 | GO:2000211 | regulation of glutamate metabolic process(GO:2000211) |
| 0.1 | 2.6 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.1 | 0.4 | GO:0070294 | renal sodium ion absorption(GO:0070294) regulation of inward rectifier potassium channel activity(GO:1901979) |
| 0.1 | 0.2 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.1 | 0.3 | GO:0046618 | drug export(GO:0046618) |
| 0.0 | 0.1 | GO:0046166 | alditol catabolic process(GO:0019405) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.0 | 0.4 | GO:0060662 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.0 | 0.2 | GO:0061589 | calcium activated phosphatidylserine scrambling(GO:0061589) |
| 0.0 | 1.1 | GO:0071378 | growth hormone receptor signaling pathway(GO:0060396) cellular response to growth hormone stimulus(GO:0071378) |
| 0.0 | 1.0 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.0 | 0.4 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.0 | 0.6 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.0 | 0.7 | GO:0039694 | viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) |
| 0.0 | 0.6 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.4 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.0 | 0.5 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.0 | 0.1 | GO:0002587 | negative regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002587) |
| 0.0 | 0.2 | GO:0061739 | protein lipidation involved in autophagosome assembly(GO:0061739) |
| 0.0 | 1.2 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.7 | GO:2000010 | positive regulation of protein localization to cell surface(GO:2000010) |
| 0.0 | 0.2 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.0 | 0.6 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.0 | 0.7 | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
| 0.0 | 0.6 | GO:0043011 | myeloid dendritic cell differentiation(GO:0043011) |
| 0.0 | 0.1 | GO:0045006 | DNA deamination(GO:0045006) |
| 0.0 | 0.1 | GO:1903972 | negative regulation of interleukin-6-mediated signaling pathway(GO:0070104) regulation of macrophage colony-stimulating factor signaling pathway(GO:1902226) regulation of response to macrophage colony-stimulating factor(GO:1903969) regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903972) |
| 0.0 | 0.1 | GO:0014719 | skeletal muscle satellite cell activation(GO:0014719) |
| 0.0 | 0.1 | GO:0045829 | negative regulation of isotype switching(GO:0045829) regulation of isotype switching to IgA isotypes(GO:0048296) |
| 0.0 | 0.4 | GO:0019321 | pentose metabolic process(GO:0019321) |
| 0.0 | 0.2 | GO:0019896 | axonal transport of mitochondrion(GO:0019896) |
| 0.0 | 0.1 | GO:0070350 | white fat cell proliferation(GO:0070343) regulation of white fat cell proliferation(GO:0070350) |
| 0.0 | 0.2 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.0 | 0.1 | GO:0071211 | protein targeting to vacuole involved in autophagy(GO:0071211) lysosomal membrane organization(GO:0097212) positive regulation of protein folding(GO:1903334) |
| 0.0 | 0.3 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.0 | 0.9 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 0.5 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.0 | 0.3 | GO:0033280 | response to vitamin D(GO:0033280) |
| 0.0 | 0.1 | GO:1903564 | regulation of protein localization to cilium(GO:1903564) |
| 0.0 | 0.5 | GO:0001502 | cartilage condensation(GO:0001502) |
| 0.0 | 0.3 | GO:0014823 | response to activity(GO:0014823) |
| 0.0 | 0.2 | GO:0047484 | regulation of response to osmotic stress(GO:0047484) |
| 0.0 | 0.8 | GO:0006821 | chloride transport(GO:0006821) |
| 0.0 | 1.4 | GO:0006695 | cholesterol biosynthetic process(GO:0006695) secondary alcohol biosynthetic process(GO:1902653) |
| 0.0 | 0.2 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.0 | 0.1 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.0 | 0.9 | GO:0008333 | endosome to lysosome transport(GO:0008333) |
| 0.0 | 0.9 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.3 | GO:0021794 | thalamus development(GO:0021794) |
| 0.0 | 0.1 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.0 | 0.2 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.0 | 0.1 | GO:0038044 | transforming growth factor-beta secretion(GO:0038044) |
| 0.0 | 0.7 | GO:0007616 | long-term memory(GO:0007616) |
| 0.0 | 0.3 | GO:0060080 | inhibitory postsynaptic potential(GO:0060080) |
| 0.0 | 0.1 | GO:1904217 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.0 | 2.3 | GO:0098656 | anion transmembrane transport(GO:0098656) |
| 0.0 | 0.1 | GO:1900019 | regulation of protein kinase C activity(GO:1900019) positive regulation of protein kinase C activity(GO:1900020) |
| 0.0 | 0.3 | GO:0030204 | chondroitin sulfate metabolic process(GO:0030204) |
| 0.0 | 0.2 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.0 | 0.4 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.0 | 0.3 | GO:0050912 | detection of chemical stimulus involved in sensory perception of taste(GO:0050912) |
| 0.0 | 0.2 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.0 | 0.1 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 0.6 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.3 | GO:2000772 | regulation of cellular senescence(GO:2000772) |
| 0.0 | 0.8 | GO:0046889 | positive regulation of lipid biosynthetic process(GO:0046889) |
| 0.0 | 0.1 | GO:0060613 | hypothalamus gonadotrophin-releasing hormone neuron differentiation(GO:0021886) hypothalamus gonadotrophin-releasing hormone neuron development(GO:0021888) fat pad development(GO:0060613) positive regulation of intrinsic apoptotic signaling pathway by p53 class mediator(GO:1902255) |
| 0.0 | 0.4 | GO:0050907 | detection of chemical stimulus involved in sensory perception(GO:0050907) |
| 0.0 | 0.2 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.9 | GO:0031983 | vesicle lumen(GO:0031983) |
| 0.3 | 1.0 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.3 | 2.8 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.2 | 1.2 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.2 | 1.2 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.2 | 2.9 | GO:0031045 | dense core granule(GO:0031045) |
| 0.1 | 1.3 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.1 | 0.9 | GO:0070695 | FHF complex(GO:0070695) |
| 0.1 | 1.1 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.1 | 3.6 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 0.5 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.1 | 0.9 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.1 | 1.6 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.1 | 0.5 | GO:0030478 | actin cap(GO:0030478) |
| 0.1 | 0.8 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 0.2 | GO:0098842 | postsynaptic early endosome(GO:0098842) |
| 0.0 | 0.5 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.0 | 0.2 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.0 | 0.3 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.0 | 0.3 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.5 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.1 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) |
| 0.0 | 0.7 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 5.9 | GO:0031227 | intrinsic component of endoplasmic reticulum membrane(GO:0031227) |
| 0.0 | 0.8 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.0 | 0.6 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.2 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.0 | 0.7 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.7 | GO:0090568 | nuclear transcriptional repressor complex(GO:0090568) |
| 0.0 | 0.7 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 1.2 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 0.2 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.0 | 0.9 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.6 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.3 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.2 | GO:0005766 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.0 | 0.1 | GO:0099522 | lysosomal lumen(GO:0043202) region of cytosol(GO:0099522) postsynaptic cytosol(GO:0099524) |
| 0.0 | 1.5 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.2 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.1 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.8 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.2 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.0 | 0.4 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.1 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.7 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.0 | 0.8 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.3 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.2 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 0.3 | GO:0033270 | paranode region of axon(GO:0033270) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.8 | GO:0070287 | ferritin receptor activity(GO:0070287) |
| 0.7 | 2.9 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.6 | 2.6 | GO:0032896 | palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 0.6 | 1.9 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) L-phenylalanine aminotransferase activity(GO:0070546) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.5 | 1.5 | GO:0050309 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.4 | 2.6 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.4 | 1.2 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.3 | 1.1 | GO:0004096 | catalase activity(GO:0004096) |
| 0.3 | 1.4 | GO:0038181 | bile acid receptor activity(GO:0038181) |
| 0.3 | 1.4 | GO:0016726 | xanthine dehydrogenase activity(GO:0004854) oxidoreductase activity, acting on CH or CH2 groups, NAD or NADP as acceptor(GO:0016726) |
| 0.3 | 0.8 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.2 | 0.7 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.2 | 1.1 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.2 | 1.3 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.2 | 4.5 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.2 | 0.8 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.2 | 1.2 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.2 | 7.7 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.2 | 0.9 | GO:0052794 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.2 | 0.6 | GO:0004063 | aryldialkylphosphatase activity(GO:0004063) |
| 0.2 | 2.9 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.2 | 1.8 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.2 | 2.5 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.2 | 0.5 | GO:0031798 | type 1 metabotropic glutamate receptor binding(GO:0031798) RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.2 | 1.2 | GO:0015091 | ferric iron transmembrane transporter activity(GO:0015091) trivalent inorganic cation transmembrane transporter activity(GO:0072510) |
| 0.2 | 0.6 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.2 | 1.2 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.2 | 1.1 | GO:0019864 | IgG binding(GO:0019864) |
| 0.1 | 0.7 | GO:0038049 | glucocorticoid receptor activity(GO:0004883) transcription factor activity, ligand-activated RNA polymerase II transcription factor binding(GO:0038049) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.1 | 0.6 | GO:0086038 | calcium:sodium antiporter activity involved in regulation of cardiac muscle cell membrane potential(GO:0086038) |
| 0.1 | 1.5 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.1 | 0.9 | GO:0019211 | phosphatase activator activity(GO:0019211) |
| 0.1 | 1.0 | GO:0005534 | galactose binding(GO:0005534) |
| 0.1 | 4.8 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.1 | 0.4 | GO:0019150 | D-ribulokinase activity(GO:0019150) |
| 0.1 | 0.6 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.1 | 2.9 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 0.7 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.1 | 0.5 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.1 | 0.7 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.1 | 0.4 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) |
| 0.1 | 5.7 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.1 | 0.4 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.1 | 0.9 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.1 | 0.2 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.1 | 0.6 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.1 | 0.3 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.1 | 0.2 | GO:0001716 | L-amino-acid oxidase activity(GO:0001716) |
| 0.1 | 0.2 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.1 | 0.9 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.1 | 0.2 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.1 | 0.5 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.1 | 0.8 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) |
| 0.0 | 0.7 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.2 | GO:0032029 | myosin tail binding(GO:0032029) |
| 0.0 | 2.8 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.7 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.1 | GO:0097677 | STAT family protein binding(GO:0097677) |
| 0.0 | 0.2 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 1.5 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.6 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.7 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 1.1 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.2 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.5 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.2 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.7 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.5 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.3 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.3 | GO:0051378 | serotonin binding(GO:0051378) |
| 0.0 | 0.6 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.0 | 4.7 | GO:0052689 | carboxylic ester hydrolase activity(GO:0052689) |
| 0.0 | 0.7 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.4 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.5 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.1 | GO:0004947 | bradykinin receptor activity(GO:0004947) |
| 0.0 | 0.5 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.6 | GO:0005542 | folic acid binding(GO:0005542) |
| 0.0 | 0.3 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.0 | 0.1 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.0 | 0.6 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 0.6 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 0.9 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.4 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.6 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 0.6 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.8 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.4 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.2 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.0 | 2.3 | GO:0015293 | symporter activity(GO:0015293) |
| 0.0 | 1.6 | GO:0019955 | cytokine binding(GO:0019955) |
| 0.0 | 0.5 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 0.6 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.0 | 0.2 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.1 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.2 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.2 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.8 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.0 | 1.1 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.0 | 0.2 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 2.2 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.5 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 1.1 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.1 | GO:0015194 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.2 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 0.1 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.0 | 0.2 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.7 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 3.5 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.1 | 1.5 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.1 | 2.9 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.8 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 0.7 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 1.4 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 1.4 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.8 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 1.5 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 1.2 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.7 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 0.8 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 1.1 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 0.6 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 1.1 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.5 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 1.0 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.6 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 4.4 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.5 | 1.6 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.2 | 2.9 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.2 | 2.9 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.2 | 2.4 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.2 | 2.6 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 1.9 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.1 | 1.9 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.1 | 1.5 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 1.4 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.1 | 2.9 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 1.7 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 1.3 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 1.7 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.1 | 1.4 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 0.7 | REACTOME HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Highly calcium permeable postsynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.7 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 1.0 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.6 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.8 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.6 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.0 | 0.4 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 1.6 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.7 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.3 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.7 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 1.2 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 1.5 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.0 | 0.5 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.0 | 1.1 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.0 | 0.9 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.3 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.9 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.6 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.6 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.5 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.2 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.7 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.6 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.1 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 0.3 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 1.5 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.0 | 1.0 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |