Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
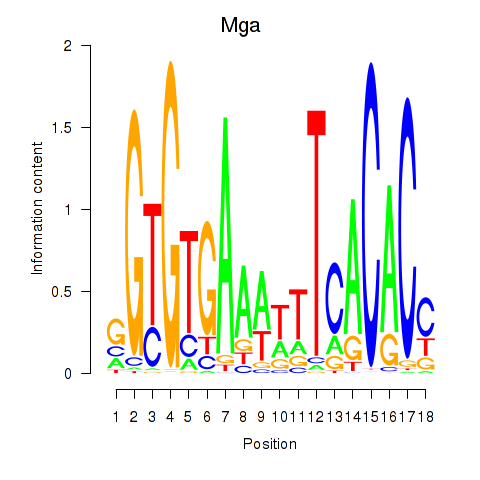
Results for Mga
Z-value: 1.60
Transcription factors associated with Mga
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Mga
|
ENSMUSG00000033943.9 | MAX gene associated |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Mga | mm10_v2_chr2_+_119897212_119897305 | 0.57 | 2.5e-04 | Click! |
Activity profile of Mga motif
Sorted Z-values of Mga motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_-_56102458 | 11.90 |
ENSMUST00000015583.1
|
Ctsg
|
cathepsin G |
| chr16_-_36408349 | 10.81 |
ENSMUST00000023619.6
|
Stfa2
|
stefin A2 |
| chr11_+_116531744 | 5.82 |
ENSMUST00000106387.2
ENSMUST00000100201.3 |
Sphk1
|
sphingosine kinase 1 |
| chr2_+_85136355 | 5.44 |
ENSMUST00000057019.7
|
Aplnr
|
apelin receptor |
| chr11_+_116532441 | 5.08 |
ENSMUST00000106386.1
ENSMUST00000145737.1 ENSMUST00000155102.1 ENSMUST00000063446.6 |
Sphk1
|
sphingosine kinase 1 |
| chr16_+_32186192 | 4.92 |
ENSMUST00000099990.3
|
Bex6
|
brain expressed gene 6 |
| chr14_+_65805832 | 4.55 |
ENSMUST00000022612.3
|
Pbk
|
PDZ binding kinase |
| chr19_+_47228804 | 4.16 |
ENSMUST00000111807.3
|
Neurl1a
|
neuralized homolog 1A (Drosophila) |
| chr11_+_116531097 | 3.75 |
ENSMUST00000138840.1
|
Sphk1
|
sphingosine kinase 1 |
| chr15_-_66801577 | 3.72 |
ENSMUST00000168589.1
|
Sla
|
src-like adaptor |
| chr1_+_52008210 | 3.58 |
ENSMUST00000027277.5
|
Stat4
|
signal transducer and activator of transcription 4 |
| chr3_+_124321031 | 3.43 |
ENSMUST00000058994.4
|
Tram1l1
|
translocation associated membrane protein 1-like 1 |
| chr7_+_13278778 | 3.36 |
ENSMUST00000098814.4
ENSMUST00000146998.1 ENSMUST00000185145.1 |
Lig1
|
ligase I, DNA, ATP-dependent |
| chr6_+_5390387 | 3.17 |
ENSMUST00000183358.1
|
Asb4
|
ankyrin repeat and SOCS box-containing 4 |
| chr3_+_105870858 | 3.15 |
ENSMUST00000164730.1
|
Adora3
|
adenosine A3 receptor |
| chr19_-_40588374 | 3.10 |
ENSMUST00000175932.1
ENSMUST00000176955.1 ENSMUST00000149476.2 |
Aldh18a1
|
aldehyde dehydrogenase 18 family, member A1 |
| chr17_+_35861318 | 2.97 |
ENSMUST00000074259.8
ENSMUST00000174873.1 |
Nrm
|
nurim (nuclear envelope membrane protein) |
| chr12_+_35992900 | 2.90 |
ENSMUST00000020898.5
|
Agr2
|
anterior gradient 2 |
| chrX_+_35888808 | 2.77 |
ENSMUST00000033419.6
|
Dock11
|
dedicator of cytokinesis 11 |
| chr17_-_32403551 | 2.67 |
ENSMUST00000135618.1
ENSMUST00000063824.7 |
Rasal3
|
RAS protein activator like 3 |
| chr4_-_117125618 | 2.64 |
ENSMUST00000183310.1
|
Btbd19
|
BTB (POZ) domain containing 19 |
| chr17_-_32403526 | 2.50 |
ENSMUST00000137458.1
|
Rasal3
|
RAS protein activator like 3 |
| chr11_-_121039400 | 2.45 |
ENSMUST00000026159.5
|
Cd7
|
CD7 antigen |
| chr7_-_3720382 | 2.37 |
ENSMUST00000078451.6
|
Pirb
|
paired Ig-like receptor B |
| chr7_-_104390586 | 2.28 |
ENSMUST00000106828.1
|
Trim30c
|
tripartite motif-containing 30C |
| chr5_+_149265035 | 2.04 |
ENSMUST00000130144.1
ENSMUST00000071130.3 |
Alox5ap
|
arachidonate 5-lipoxygenase activating protein |
| chr3_+_105870898 | 2.02 |
ENSMUST00000010279.5
|
Adora3
|
adenosine A3 receptor |
| chr2_+_3336159 | 1.99 |
ENSMUST00000115089.1
|
Acbd7
|
acyl-Coenzyme A binding domain containing 7 |
| chr4_+_132564051 | 1.96 |
ENSMUST00000070690.7
|
Ptafr
|
platelet-activating factor receptor |
| chr3_+_87376381 | 1.88 |
ENSMUST00000163661.1
ENSMUST00000072480.2 ENSMUST00000167200.1 |
Fcrl1
|
Fc receptor-like 1 |
| chr6_+_123229843 | 1.86 |
ENSMUST00000112554.2
ENSMUST00000024118.4 ENSMUST00000117130.1 |
Clec4n
|
C-type lectin domain family 4, member n |
| chr8_+_12984246 | 1.71 |
ENSMUST00000110873.3
ENSMUST00000173006.1 ENSMUST00000145067.1 |
Mcf2l
|
mcf.2 transforming sequence-like |
| chr11_-_8973266 | 1.66 |
ENSMUST00000154153.1
|
Pkd1l1
|
polycystic kidney disease 1 like 1 |
| chr8_-_13677575 | 1.59 |
ENSMUST00000117551.2
|
Rasa3
|
RAS p21 protein activator 3 |
| chr7_-_30880263 | 1.57 |
ENSMUST00000108125.2
|
Cd22
|
CD22 antigen |
| chr9_-_97111117 | 1.55 |
ENSMUST00000085206.4
|
Slc25a36
|
solute carrier family 25, member 36 |
| chr14_+_32833955 | 1.48 |
ENSMUST00000104926.2
|
Fam170b
|
family with sequence similarity 170, member B |
| chr8_+_83165348 | 1.46 |
ENSMUST00000034145.4
|
Tbc1d9
|
TBC1 domain family, member 9 |
| chr2_-_170131156 | 1.45 |
ENSMUST00000063710.6
|
Zfp217
|
zinc finger protein 217 |
| chr6_-_41636389 | 1.42 |
ENSMUST00000031902.5
|
Trpv6
|
transient receptor potential cation channel, subfamily V, member 6 |
| chr4_-_117156144 | 1.38 |
ENSMUST00000102696.4
|
Rps8
|
ribosomal protein S8 |
| chr10_+_70868633 | 1.36 |
ENSMUST00000058942.5
|
4930533K18Rik
|
RIKEN cDNA 4930533K18 gene |
| chr5_-_120777628 | 1.36 |
ENSMUST00000044833.8
|
Oas3
|
2'-5' oligoadenylate synthetase 3 |
| chr17_-_32350569 | 1.35 |
ENSMUST00000050214.7
|
Akap8l
|
A kinase (PRKA) anchor protein 8-like |
| chr4_+_62525369 | 1.25 |
ENSMUST00000062145.1
|
4933430I17Rik
|
RIKEN cDNA 4933430I17 gene |
| chr5_+_137745730 | 1.08 |
ENSMUST00000100540.3
|
Tsc22d4
|
TSC22 domain family, member 4 |
| chr1_-_130423009 | 1.04 |
ENSMUST00000112488.2
ENSMUST00000119432.1 |
Daf2
|
decay accelerating factor 2 |
| chr17_-_7827289 | 0.99 |
ENSMUST00000167580.1
ENSMUST00000169126.1 |
Fndc1
|
fibronectin type III domain containing 1 |
| chr16_-_36131156 | 0.96 |
ENSMUST00000161638.1
ENSMUST00000096090.2 |
Csta
|
cystatin A |
| chr12_-_54695885 | 0.90 |
ENSMUST00000067272.8
|
Eapp
|
E2F-associated phosphoprotein |
| chr6_-_56923927 | 0.90 |
ENSMUST00000031793.5
|
Nt5c3
|
5'-nucleotidase, cytosolic III |
| chr5_-_5663263 | 0.89 |
ENSMUST00000148193.1
|
A330021E22Rik
|
RIKEN cDNA A330021E22 gene |
| chr2_+_151996505 | 0.81 |
ENSMUST00000109859.2
ENSMUST00000073228.5 |
Slc52a3
|
solute carrier protein family 52, member 3 |
| chr9_+_57940104 | 0.80 |
ENSMUST00000043059.7
|
Sema7a
|
sema domain, immunoglobulin domain (Ig), and GPI membrane anchor, (semaphorin) 7A |
| chr4_-_41314877 | 0.77 |
ENSMUST00000030145.8
|
Dcaf12
|
DDB1 and CUL4 associated factor 12 |
| chr17_+_23679363 | 0.71 |
ENSMUST00000024699.2
|
Cldn6
|
claudin 6 |
| chr10_+_75212065 | 0.67 |
ENSMUST00000105421.2
|
Specc1l
|
sperm antigen with calponin homology and coiled-coil domains 1-like |
| chr2_+_151572606 | 0.66 |
ENSMUST00000028950.8
|
Sdcbp2
|
syndecan binding protein (syntenin) 2 |
| chr11_-_78751656 | 0.65 |
ENSMUST00000059468.4
|
Fam58b
|
family with sequence similarity 58, member B |
| chr9_+_59291565 | 0.64 |
ENSMUST00000026266.7
|
Adpgk
|
ADP-dependent glucokinase |
| chrX_+_20617503 | 0.55 |
ENSMUST00000115375.1
ENSMUST00000115374.1 ENSMUST00000084383.3 |
Rbm10
|
RNA binding motif protein 10 |
| chr7_+_27486910 | 0.52 |
ENSMUST00000008528.7
|
Sertad1
|
SERTA domain containing 1 |
| chr5_+_100196611 | 0.51 |
ENSMUST00000066813.1
|
Gm9932
|
predicted gene 9932 |
| chrX_+_163909132 | 0.49 |
ENSMUST00000033734.7
ENSMUST00000112294.2 |
Ap1s2
|
adaptor-related protein complex 1, sigma 2 subunit |
| chr3_-_146521396 | 0.49 |
ENSMUST00000029838.6
|
Rpf1
|
ribosome production factor 1 homolog (S. cerevisiae) |
| chr11_+_49203465 | 0.46 |
ENSMUST00000150284.1
ENSMUST00000109197.1 ENSMUST00000151228.1 |
Zfp62
|
zinc finger protein 62 |
| chr9_-_117252450 | 0.46 |
ENSMUST00000111773.3
ENSMUST00000068962.7 ENSMUST00000044901.7 |
Rbms3
|
RNA binding motif, single stranded interacting protein |
| chr15_+_79075209 | 0.42 |
ENSMUST00000040518.4
|
Eif3l
|
eukaryotic translation initiation factor 3, subunit L |
| chr3_+_76075583 | 0.39 |
ENSMUST00000160261.1
|
Fstl5
|
follistatin-like 5 |
| chr15_+_51877742 | 0.38 |
ENSMUST00000136129.1
|
Utp23
|
UTP23, small subunit (SSU) processome component, homolog (yeast) |
| chr8_-_84104773 | 0.38 |
ENSMUST00000041367.7
|
Dcaf15
|
DDB1 and CUL4 associated factor 15 |
| chr4_-_62525036 | 0.35 |
ENSMUST00000030091.3
|
Pole3
|
polymerase (DNA directed), epsilon 3 (p17 subunit) |
| chr2_-_129371131 | 0.31 |
ENSMUST00000028881.7
|
Il1b
|
interleukin 1 beta |
| chr8_+_11713259 | 0.31 |
ENSMUST00000134409.1
|
1700128E19Rik
|
RIKEN cDNA 1700128E19 gene |
| chr8_+_95534078 | 0.30 |
ENSMUST00000041569.3
|
Ccdc113
|
coiled-coil domain containing 113 |
| chr11_-_4440745 | 0.29 |
ENSMUST00000109948.1
|
Hormad2
|
HORMA domain containing 2 |
| chr9_-_96478596 | 0.27 |
ENSMUST00000071301.4
|
Rnf7
|
ring finger protein 7 |
| chr8_-_105979413 | 0.26 |
ENSMUST00000034371.7
|
Dpep3
|
dipeptidase 3 |
| chr9_-_96478660 | 0.26 |
ENSMUST00000057500.4
|
Rnf7
|
ring finger protein 7 |
| chr9_-_109702700 | 0.26 |
ENSMUST00000098359.3
|
Fbxw18
|
F-box and WD-40 domain protein 18 |
| chr9_-_109746089 | 0.24 |
ENSMUST00000071917.3
|
Fbxw26
|
F-box and WD-40 domain protein 26 |
| chr12_-_54695829 | 0.22 |
ENSMUST00000162106.1
ENSMUST00000160085.1 ENSMUST00000161592.1 ENSMUST00000163433.1 |
Eapp
|
E2F-associated phosphoprotein |
| chr1_+_190928822 | 0.16 |
ENSMUST00000135364.1
|
Angel2
|
angel homolog 2 (Drosophila) |
| chr15_+_51877429 | 0.15 |
ENSMUST00000137116.2
ENSMUST00000161651.1 ENSMUST00000059599.9 |
Utp23
|
UTP23, small subunit (SSU) processome component, homolog (yeast) |
| chr9_-_109449140 | 0.13 |
ENSMUST00000084984.6
|
Fbxw16
|
F-box and WD-40 domain protein 16 |
| chr11_+_49203285 | 0.11 |
ENSMUST00000109198.1
ENSMUST00000137061.2 |
Zfp62
|
zinc finger protein 62 |
| chr19_-_5610038 | 0.11 |
ENSMUST00000113641.2
|
Kat5
|
K(lysine) acetyltransferase 5 |
| chr1_+_60409612 | 0.09 |
ENSMUST00000052332.8
|
Abi2
|
abl-interactor 2 |
| chr9_-_109626059 | 0.07 |
ENSMUST00000073962.6
|
Fbxw24
|
F-box and WD-40 domain protein 24 |
| chr14_-_52279238 | 0.07 |
ENSMUST00000167116.1
ENSMUST00000100631.4 |
Rab2b
|
RAB2B, member RAS oncogene family |
| chr9_-_109568262 | 0.03 |
ENSMUST00000056745.6
|
Fbxw15
|
F-box and WD-40 domain protein 15 |
| chr2_-_132145057 | 0.02 |
ENSMUST00000028815.8
|
Slc23a2
|
solute carrier family 23 (nucleobase transporters), member 2 |
| chr2_+_83644435 | 0.01 |
ENSMUST00000081591.6
|
Zc3h15
|
zinc finger CCCH-type containing 15 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Mga
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 14.7 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 1.4 | 5.4 | GO:0002030 | inhibitory G-protein coupled receptor phosphorylation(GO:0002030) |
| 1.3 | 11.9 | GO:0070944 | neutrophil mediated killing of bacterium(GO:0070944) |
| 1.0 | 3.1 | GO:0006592 | ornithine biosynthetic process(GO:0006592) |
| 1.0 | 5.2 | GO:0051142 | regulation of NK T cell proliferation(GO:0051140) positive regulation of NK T cell proliferation(GO:0051142) |
| 0.7 | 2.9 | GO:1903896 | positive regulation of IRE1-mediated unfolded protein response(GO:1903896) |
| 0.7 | 2.0 | GO:1904306 | positive regulation of gastro-intestinal system smooth muscle contraction(GO:1904306) |
| 0.6 | 1.7 | GO:0003127 | detection of nodal flow(GO:0003127) detection of endogenous stimulus(GO:0009726) |
| 0.5 | 5.2 | GO:0002553 | histamine production involved in inflammatory response(GO:0002349) histamine secretion involved in inflammatory response(GO:0002441) histamine secretion by mast cell(GO:0002553) |
| 0.5 | 2.0 | GO:0002540 | leukotriene production involved in inflammatory response(GO:0002540) |
| 0.5 | 3.4 | GO:0033567 | DNA replication, Okazaki fragment processing(GO:0033567) |
| 0.4 | 4.2 | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 0.3 | 3.2 | GO:2001214 | positive regulation of vasculogenesis(GO:2001214) |
| 0.3 | 1.5 | GO:0072531 | pyrimidine-containing compound transmembrane transport(GO:0072531) |
| 0.3 | 1.5 | GO:0080154 | regulation of fertilization(GO:0080154) |
| 0.2 | 2.8 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.2 | 0.8 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.2 | 1.4 | GO:0060700 | regulation of ribonuclease activity(GO:0060700) |
| 0.1 | 1.3 | GO:0010793 | regulation of mRNA export from nucleus(GO:0010793) |
| 0.1 | 1.4 | GO:0098703 | parathyroid hormone secretion(GO:0035898) calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.1 | 2.4 | GO:0043011 | myeloid dendritic cell differentiation(GO:0043011) |
| 0.1 | 1.1 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.1 | 0.5 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.1 | 0.3 | GO:0060559 | positive regulation of vitamin metabolic process(GO:0046136) positive regulation of vitamin D biosynthetic process(GO:0060557) positive regulation of calcidiol 1-monooxygenase activity(GO:0060559) |
| 0.1 | 0.4 | GO:0075525 | viral translational termination-reinitiation(GO:0075525) |
| 0.1 | 0.6 | GO:0034393 | positive regulation of smooth muscle cell apoptotic process(GO:0034393) |
| 0.1 | 0.8 | GO:0060907 | positive regulation of macrophage cytokine production(GO:0060907) |
| 0.1 | 1.0 | GO:0034244 | negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.1 | 4.6 | GO:0070303 | negative regulation of stress-activated MAPK cascade(GO:0032873) negative regulation of stress-activated protein kinase signaling cascade(GO:0070303) |
| 0.1 | 1.9 | GO:0050832 | defense response to fungus(GO:0050832) |
| 0.1 | 0.7 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.1 | 0.9 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.1 | 10.8 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 1.7 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 | 3.7 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.0 | 0.5 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.0 | 2.4 | GO:0048873 | homeostasis of number of cells within a tissue(GO:0048873) |
| 0.0 | 1.4 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 1.0 | GO:0010661 | positive regulation of muscle cell apoptotic process(GO:0010661) positive regulation of striated muscle cell apoptotic process(GO:0010663) positive regulation of cardiac muscle cell apoptotic process(GO:0010666) |
| 0.0 | 1.6 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 1.5 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.0 | 1.5 | GO:0007569 | cell aging(GO:0007569) |
| 0.0 | 0.7 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.0 | 0.5 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 3.7 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.4 | 6.8 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.2 | 1.5 | GO:0002081 | outer acrosomal membrane(GO:0002081) |
| 0.2 | 3.0 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.2 | 11.9 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.1 | 4.2 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.1 | 0.3 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.0 | 11.6 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 0.7 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 1.4 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.4 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 1.7 | GO:0034704 | calcium channel complex(GO:0034704) |
| 0.0 | 0.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.8 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.7 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 0.5 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 1.5 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 2.2 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 1.3 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 0.7 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 14.7 | GO:0017050 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 1.0 | 3.1 | GO:0017084 | glutamate 5-kinase activity(GO:0004349) glutamate-5-semialdehyde dehydrogenase activity(GO:0004350) delta1-pyrroline-5-carboxylate synthetase activity(GO:0017084) amino acid kinase activity(GO:0019202) |
| 0.7 | 2.0 | GO:0004051 | arachidonate 5-lipoxygenase activity(GO:0004051) |
| 0.5 | 5.2 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.4 | 2.0 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.4 | 3.4 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.2 | 0.8 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.2 | 2.9 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.1 | 1.4 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.1 | 10.8 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.1 | 0.5 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.1 | 1.7 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 4.2 | GO:0045182 | translation regulator activity(GO:0045182) |
| 0.1 | 3.7 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.1 | 1.6 | GO:0099604 | calcium-release channel activity(GO:0015278) ligand-gated calcium channel activity(GO:0099604) |
| 0.1 | 0.5 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.1 | 0.5 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.1 | 1.6 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.1 | 11.9 | GO:0008201 | heparin binding(GO:0008201) |
| 0.1 | 2.0 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.1 | 1.3 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.1 | 0.8 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.1 | 1.5 | GO:0015215 | nucleotide transmembrane transporter activity(GO:0015215) |
| 0.0 | 0.9 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.7 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 0.3 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 2.9 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.0 | 6.6 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 2.2 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.0 | 0.7 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.3 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.3 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 14.7 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.2 | 11.9 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.1 | 3.9 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.1 | 4.5 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 1.6 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
| 0.0 | 3.7 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.0 | 1.4 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 1.7 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 11.9 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.3 | 14.7 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.3 | 5.2 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.2 | 3.4 | REACTOME PROCESSIVE SYNTHESIS ON THE LAGGING STRAND | Genes involved in Processive synthesis on the lagging strand |
| 0.1 | 3.1 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 3.3 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.1 | 0.9 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.5 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 5.4 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.8 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 1.4 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.7 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |