Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
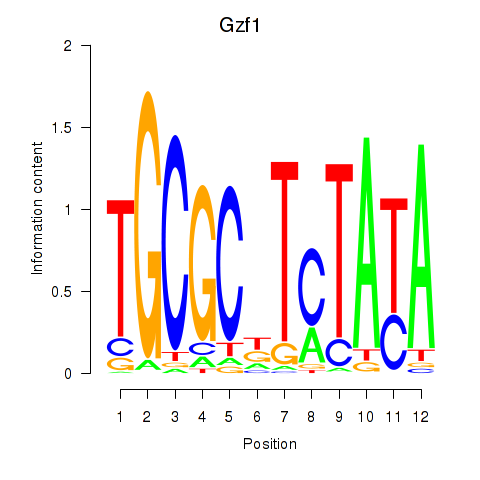
Results for Gzf1
Z-value: 1.10
Transcription factors associated with Gzf1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Gzf1
|
ENSMUSG00000027439.9 | GDNF-inducible zinc finger protein 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Gzf1 | mm10_v2_chr2_+_148681199_148681218 | -0.01 | 9.4e-01 | Click! |
Activity profile of Gzf1 motif
Sorted Z-values of Gzf1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_+_109540979 | 6.89 |
ENSMUST00000129245.1
ENSMUST00000143836.1 ENSMUST00000124106.1 |
Meg3
|
maternally expressed 3 |
| chr4_-_134128707 | 3.04 |
ENSMUST00000105879.1
ENSMUST00000030651.8 |
Sh3bgrl3
|
SH3 domain binding glutamic acid-rich protein-like 3 |
| chr14_+_11227511 | 2.90 |
ENSMUST00000080237.3
|
Rpl21-ps4
|
ribosomal protein L21, pseudogene 4 |
| chr17_+_47594629 | 2.84 |
ENSMUST00000182846.1
|
Ccnd3
|
cyclin D3 |
| chr17_-_55915870 | 2.35 |
ENSMUST00000074828.4
|
Rpl21-ps6
|
ribosomal protein L21, pseudogene 6 |
| chr7_-_30612731 | 2.34 |
ENSMUST00000006476.4
|
Upk1a
|
uroplakin 1A |
| chr14_+_55854115 | 2.31 |
ENSMUST00000168479.1
|
Nynrin
|
NYN domain and retroviral integrase containing |
| chr17_+_48462355 | 2.10 |
ENSMUST00000162132.1
|
Unc5cl
|
unc-5 homolog C (C. elegans)-like |
| chr11_-_33163072 | 2.00 |
ENSMUST00000093201.6
ENSMUST00000101375.4 ENSMUST00000109354.3 ENSMUST00000075641.3 |
Npm1
|
nucleophosmin 1 |
| chr14_+_55853997 | 2.00 |
ENSMUST00000100529.3
|
Nynrin
|
NYN domain and retroviral integrase containing |
| chr8_+_121127827 | 1.54 |
ENSMUST00000181609.1
|
Foxl1
|
forkhead box L1 |
| chr6_-_52204415 | 1.44 |
ENSMUST00000048794.6
|
Hoxa5
|
homeobox A5 |
| chr4_+_154960915 | 1.41 |
ENSMUST00000049621.6
|
Hes5
|
hairy and enhancer of split 5 (Drosophila) |
| chr2_+_162931520 | 1.20 |
ENSMUST00000130411.1
|
Srsf6
|
serine/arginine-rich splicing factor 6 |
| chr8_-_70792392 | 1.09 |
ENSMUST00000166004.1
|
Mast3
|
microtubule associated serine/threonine kinase 3 |
| chr8_-_78508876 | 1.09 |
ENSMUST00000049245.7
|
Rbmxl1
|
RNA binding motif protein, X linked-like-1 |
| chr14_+_19751257 | 0.41 |
ENSMUST00000022340.3
|
Nid2
|
nidogen 2 |
| chr7_-_29168647 | 0.36 |
ENSMUST00000048923.6
|
Spred3
|
sprouty-related, EVH1 domain containing 3 |
| chr10_+_82699007 | 0.33 |
ENSMUST00000020478.7
|
Hcfc2
|
host cell factor C2 |
| chr4_-_129542710 | 0.31 |
ENSMUST00000102597.4
|
Hdac1
|
histone deacetylase 1 |
| chr6_-_52012476 | 0.25 |
ENSMUST00000078214.5
|
Skap2
|
src family associated phosphoprotein 2 |
| chr13_-_55415166 | 0.25 |
ENSMUST00000054146.3
|
Pfn3
|
profilin 3 |
| chr9_-_123978297 | 0.20 |
ENSMUST00000071404.3
|
Ccr1l1
|
chemokine (C-C motif) receptor 1-like 1 |
| chr8_+_105855086 | 0.12 |
ENSMUST00000040445.7
|
Thap11
|
THAP domain containing 11 |
| chr4_-_124862171 | 0.06 |
ENSMUST00000064444.7
|
Maneal
|
mannosidase, endo-alpha-like |
Network of associatons between targets according to the STRING database.
First level regulatory network of Gzf1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.0 | GO:0006407 | cleavage in ITS2 between 5.8S rRNA and LSU-rRNA of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000448) rRNA export from nucleus(GO:0006407) |
| 0.5 | 4.3 | GO:0015074 | DNA integration(GO:0015074) |
| 0.5 | 1.4 | GO:0060435 | bronchiole development(GO:0060435) intestinal epithelial cell maturation(GO:0060574) |
| 0.5 | 1.4 | GO:0042668 | auditory receptor cell fate determination(GO:0042668) negative regulation of forebrain neuron differentiation(GO:2000978) |
| 0.4 | 1.5 | GO:0061146 | Peyer's patch morphogenesis(GO:0061146) |
| 0.2 | 6.9 | GO:0008340 | determination of adult lifespan(GO:0008340) |
| 0.1 | 2.8 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.1 | 1.2 | GO:0045617 | negative regulation of keratinocyte differentiation(GO:0045617) |
| 0.1 | 0.3 | GO:2000676 | fungiform papilla formation(GO:0061198) positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.0 | 3.0 | GO:0030834 | regulation of actin filament depolymerization(GO:0030834) |
| 0.0 | 1.1 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.4 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 2.1 | GO:0046330 | positive regulation of JNK cascade(GO:0046330) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.0 | GO:0001652 | granular component(GO:0001652) |
| 0.2 | 1.1 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.0 | 2.8 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.3 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 3.0 | GO:0030027 | lamellipodium(GO:0030027) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.0 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.2 | 3.0 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 0.3 | GO:0035851 | Krueppel-associated box domain binding(GO:0035851) |
| 0.1 | 0.4 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.1 | 2.8 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 1.2 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.2 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.0 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 2.8 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.3 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 1.4 | PID CDC42 PATHWAY | CDC42 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.8 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 2.0 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 1.2 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.0 | 1.7 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.3 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |