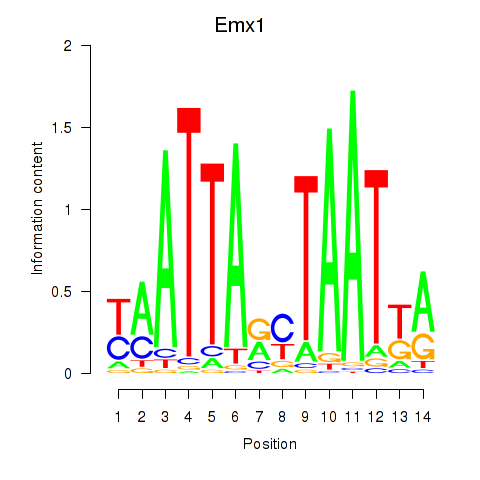
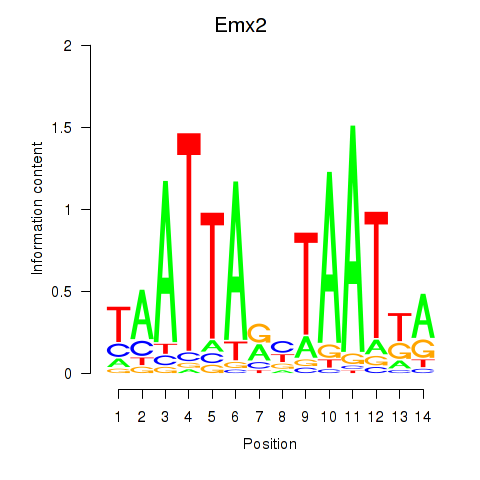
Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Emx1_Emx2
Z-value: 1.20
Transcription factors associated with Emx1_Emx2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Emx1
|
ENSMUSG00000033726.8 | empty spiracles homeobox 1 |
|
Emx2
|
ENSMUSG00000043969.4 | empty spiracles homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Emx2 | mm10_v2_chr19_+_59458372_59458450 | -0.35 | 3.8e-02 | Click! |
| Emx1 | mm10_v2_chr6_+_85187438_85187510 | -0.30 | 7.9e-02 | Click! |
Activity profile of Emx1_Emx2 motif
Sorted Z-values of Emx1_Emx2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_39287074 | 9.82 |
ENSMUST00000003137.8
|
Cyp2c29
|
cytochrome P450, family 2, subfamily c, polypeptide 29 |
| chr3_-_81975742 | 6.66 |
ENSMUST00000029645.8
|
Tdo2
|
tryptophan 2,3-dioxygenase |
| chr7_-_68275098 | 6.21 |
ENSMUST00000135564.1
|
Gm16157
|
predicted gene 16157 |
| chr19_-_39463067 | 5.12 |
ENSMUST00000035488.2
|
Cyp2c38
|
cytochrome P450, family 2, subfamily c, polypeptide 38 |
| chr7_-_48848023 | 5.08 |
ENSMUST00000032658.6
|
Csrp3
|
cysteine and glycine-rich protein 3 |
| chr17_-_36032682 | 4.61 |
ENSMUST00000102678.4
|
H2-T23
|
histocompatibility 2, T region locus 23 |
| chr12_-_98577940 | 4.59 |
ENSMUST00000110113.1
|
Kcnk10
|
potassium channel, subfamily K, member 10 |
| chr18_-_38866702 | 4.28 |
ENSMUST00000115582.1
|
Fgf1
|
fibroblast growth factor 1 |
| chr4_-_61439743 | 4.20 |
ENSMUST00000095049.4
|
Mup15
|
major urinary protein 15 |
| chr5_-_89457763 | 3.90 |
ENSMUST00000049209.8
|
Gc
|
group specific component |
| chr16_+_22918378 | 3.54 |
ENSMUST00000170805.1
|
Fetub
|
fetuin beta |
| chr17_-_31636631 | 3.00 |
ENSMUST00000135425.1
ENSMUST00000151718.1 ENSMUST00000155814.1 |
Cbs
|
cystathionine beta-synthase |
| chr1_+_130826762 | 2.74 |
ENSMUST00000133792.1
|
Pigr
|
polymeric immunoglobulin receptor |
| chr5_-_87337165 | 2.74 |
ENSMUST00000031195.2
|
Ugt2a3
|
UDP glucuronosyltransferase 2 family, polypeptide A3 |
| chr4_-_82505707 | 2.66 |
ENSMUST00000107248.1
ENSMUST00000107247.1 |
Nfib
|
nuclear factor I/B |
| chr7_+_27029074 | 2.61 |
ENSMUST00000075552.5
|
Cyp2a12
|
cytochrome P450, family 2, subfamily a, polypeptide 12 |
| chr1_-_162898665 | 2.58 |
ENSMUST00000111510.1
ENSMUST00000045902.6 |
Fmo2
|
flavin containing monooxygenase 2 |
| chr2_-_67194695 | 2.52 |
ENSMUST00000147939.1
|
Gm13598
|
predicted gene 13598 |
| chr1_-_162898484 | 2.51 |
ENSMUST00000143123.1
|
Fmo2
|
flavin containing monooxygenase 2 |
| chr6_+_42245907 | 2.45 |
ENSMUST00000031897.5
|
Gstk1
|
glutathione S-transferase kappa 1 |
| chr8_-_93363676 | 2.42 |
ENSMUST00000145041.1
|
Ces1h
|
carboxylesterase 1H |
| chr19_-_8218832 | 2.38 |
ENSMUST00000113298.2
|
Slc22a29
|
solute carrier family 22. member 29 |
| chr4_-_82505274 | 2.38 |
ENSMUST00000050872.8
ENSMUST00000064770.2 |
Nfib
|
nuclear factor I/B |
| chr5_-_65428354 | 2.31 |
ENSMUST00000131263.1
|
Ugdh
|
UDP-glucose dehydrogenase |
| chr2_-_157566319 | 2.24 |
ENSMUST00000109528.2
ENSMUST00000088494.2 |
Blcap
|
bladder cancer associated protein homolog (human) |
| chr11_+_109543694 | 2.02 |
ENSMUST00000106696.1
|
Arsg
|
arylsulfatase G |
| chr1_-_174921813 | 2.00 |
ENSMUST00000055294.3
|
Grem2
|
gremlin 2 homolog, cysteine knot superfamily (Xenopus laevis) |
| chr2_-_17460610 | 1.90 |
ENSMUST00000145492.1
|
Nebl
|
nebulette |
| chr1_-_139858684 | 1.87 |
ENSMUST00000094489.3
|
Cfhr2
|
complement factor H-related 2 |
| chr4_-_82505749 | 1.85 |
ENSMUST00000107245.2
ENSMUST00000107246.1 |
Nfib
|
nuclear factor I/B |
| chr5_+_45493374 | 1.85 |
ENSMUST00000046122.6
|
Lap3
|
leucine aminopeptidase 3 |
| chr1_+_58113136 | 1.83 |
ENSMUST00000040999.7
|
Aox3
|
aldehyde oxidase 3 |
| chr5_-_87535113 | 1.74 |
ENSMUST00000120150.1
|
Sult1b1
|
sulfotransferase family 1B, member 1 |
| chr11_-_54249640 | 1.72 |
ENSMUST00000019060.5
|
Csf2
|
colony stimulating factor 2 (granulocyte-macrophage) |
| chr17_+_45734506 | 1.68 |
ENSMUST00000180558.1
|
F630040K05Rik
|
RIKEN cDNA F630040K05 gene |
| chr7_-_26939377 | 1.67 |
ENSMUST00000170227.1
|
Cyp2a22
|
cytochrome P450, family 2, subfamily a, polypeptide 22 |
| chrX_-_8193387 | 1.55 |
ENSMUST00000143223.1
ENSMUST00000033509.8 |
Ebp
|
phenylalkylamine Ca2+ antagonist (emopamil) binding protein |
| chr7_+_132610620 | 1.53 |
ENSMUST00000033241.5
|
Lhpp
|
phospholysine phosphohistidine inorganic pyrophosphate phosphatase |
| chr19_-_39649046 | 1.51 |
ENSMUST00000067328.6
|
Cyp2c67
|
cytochrome P450, family 2, subfamily c, polypeptide 67 |
| chr7_+_44676965 | 1.49 |
ENSMUST00000094460.1
|
2310016G11Rik
|
RIKEN cDNA 2310016G11 gene |
| chr7_-_101845300 | 1.49 |
ENSMUST00000094141.5
|
Folr2
|
folate receptor 2 (fetal) |
| chr19_-_39812744 | 1.46 |
ENSMUST00000162507.1
ENSMUST00000160476.1 |
Cyp2c40
|
cytochrome P450, family 2, subfamily c, polypeptide 40 |
| chr9_-_15301555 | 1.45 |
ENSMUST00000034414.8
|
4931406C07Rik
|
RIKEN cDNA 4931406C07 gene |
| chr1_-_187215454 | 1.45 |
ENSMUST00000183819.1
|
Spata17
|
spermatogenesis associated 17 |
| chr6_+_124304646 | 1.44 |
ENSMUST00000112541.2
ENSMUST00000032234.2 |
Cd163
|
CD163 antigen |
| chr11_+_99873389 | 1.39 |
ENSMUST00000093936.3
|
Krtap9-1
|
keratin associated protein 9-1 |
| chr4_+_138879360 | 1.39 |
ENSMUST00000105804.1
|
Pla2g2e
|
phospholipase A2, group IIE |
| chr6_-_57535422 | 1.29 |
ENSMUST00000042766.3
|
Ppm1k
|
protein phosphatase 1K (PP2C domain containing) |
| chr5_-_3647806 | 1.29 |
ENSMUST00000119783.1
ENSMUST00000007559.8 |
Gatad1
|
GATA zinc finger domain containing 1 |
| chr3_-_96452306 | 1.28 |
ENSMUST00000093126.4
ENSMUST00000098841.3 |
BC107364
|
cDNA sequence BC107364 |
| chr2_+_30171486 | 1.27 |
ENSMUST00000015481.5
|
Endog
|
endonuclease G |
| chr13_-_120215213 | 1.25 |
ENSMUST00000177561.1
|
Gm21762
|
predicted gene, 21762 |
| chr18_-_73754457 | 1.24 |
ENSMUST00000041138.2
|
Elac1
|
elaC homolog 1 (E. coli) |
| chr6_+_129533183 | 1.21 |
ENSMUST00000032264.6
|
Gabarapl1
|
gamma-aminobutyric acid (GABA) A receptor-associated protein-like 1 |
| chr11_-_49113757 | 1.20 |
ENSMUST00000060398.1
|
Olfr1396
|
olfactory receptor 1396 |
| chr3_+_57425314 | 1.20 |
ENSMUST00000029377.7
|
Tm4sf4
|
transmembrane 4 superfamily member 4 |
| chr4_-_150914401 | 1.14 |
ENSMUST00000105675.1
|
Park7
|
Parkinson disease (autosomal recessive, early onset) 7 |
| chr6_+_79818031 | 1.13 |
ENSMUST00000179797.1
|
Gm20594
|
predicted gene, 20594 |
| chr9_+_78191966 | 1.12 |
ENSMUST00000034903.5
|
Gsta4
|
glutathione S-transferase, alpha 4 |
| chr14_+_55560010 | 1.12 |
ENSMUST00000147981.1
ENSMUST00000133256.1 |
Dcaf11
|
DDB1 and CUL4 associated factor 11 |
| chr3_+_127633134 | 1.12 |
ENSMUST00000029587.7
|
Neurog2
|
neurogenin 2 |
| chr14_+_118137101 | 1.11 |
ENSMUST00000022728.2
|
Gpr180
|
G protein-coupled receptor 180 |
| chr8_+_45507768 | 1.07 |
ENSMUST00000067065.7
ENSMUST00000098788.3 ENSMUST00000067107.7 ENSMUST00000171337.2 ENSMUST00000138049.1 ENSMUST00000141039.1 |
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr2_-_5676046 | 1.06 |
ENSMUST00000114987.3
|
Camk1d
|
calcium/calmodulin-dependent protein kinase ID |
| chr1_+_93685574 | 1.05 |
ENSMUST00000027499.6
|
Bok
|
BCL2-related ovarian killer protein |
| chr14_+_69347587 | 1.03 |
ENSMUST00000064831.5
|
Entpd4
|
ectonucleoside triphosphate diphosphohydrolase 4 |
| chr9_-_50746501 | 1.02 |
ENSMUST00000034564.1
|
2310030G06Rik
|
RIKEN cDNA 2310030G06 gene |
| chr16_-_44016387 | 1.02 |
ENSMUST00000036174.3
|
Gramd1c
|
GRAM domain containing 1C |
| chr13_-_4609122 | 1.01 |
ENSMUST00000110691.3
ENSMUST00000091848.5 |
Akr1e1
|
aldo-keto reductase family 1, member E1 |
| chr15_-_33687840 | 0.95 |
ENSMUST00000042021.3
|
Tspyl5
|
testis-specific protein, Y-encoded-like 5 |
| chr18_-_66002612 | 0.93 |
ENSMUST00000120461.1
ENSMUST00000048260.7 |
Lman1
|
lectin, mannose-binding, 1 |
| chr3_+_121953213 | 0.90 |
ENSMUST00000037958.7
ENSMUST00000128366.1 |
Arhgap29
|
Rho GTPase activating protein 29 |
| chr11_+_97029925 | 0.89 |
ENSMUST00000021249.4
|
Scrn2
|
secernin 2 |
| chr6_-_144209471 | 0.88 |
ENSMUST00000038815.7
|
Sox5
|
SRY-box containing gene 5 |
| chr16_+_11406618 | 0.88 |
ENSMUST00000122168.1
|
Snx29
|
sorting nexin 29 |
| chr2_-_175703646 | 0.88 |
ENSMUST00000109027.2
ENSMUST00000179061.1 ENSMUST00000131041.1 |
Gm4245
|
predicted gene 4245 |
| chr12_+_84285232 | 0.88 |
ENSMUST00000123614.1
ENSMUST00000147363.1 ENSMUST00000135001.1 ENSMUST00000146377.1 |
Ptgr2
|
prostaglandin reductase 2 |
| chr6_-_144209448 | 0.87 |
ENSMUST00000077160.5
|
Sox5
|
SRY-box containing gene 5 |
| chr14_-_8258800 | 0.87 |
ENSMUST00000022271.7
|
Acox2
|
acyl-Coenzyme A oxidase 2, branched chain |
| chr9_-_20728219 | 0.84 |
ENSMUST00000034692.7
|
Olfm2
|
olfactomedin 2 |
| chr10_-_86011833 | 0.83 |
ENSMUST00000105304.1
ENSMUST00000061699.5 |
Bpifc
|
BPI fold containing family C |
| chr7_-_5413145 | 0.81 |
ENSMUST00000108569.2
|
Vmn1r58
|
vomeronasal 1 receptor 58 |
| chr2_+_175372436 | 0.81 |
ENSMUST00000131676.1
ENSMUST00000109048.2 ENSMUST00000109047.2 |
Gm4723
|
predicted gene 4723 |
| chr6_-_144209558 | 0.78 |
ENSMUST00000111749.1
ENSMUST00000170367.2 |
Sox5
|
SRY-box containing gene 5 |
| chr4_+_102589687 | 0.77 |
ENSMUST00000097949.4
ENSMUST00000106901.1 |
Pde4b
|
phosphodiesterase 4B, cAMP specific |
| chr9_+_72958785 | 0.77 |
ENSMUST00000098567.2
ENSMUST00000034734.8 |
Dyx1c1
|
dyslexia susceptibility 1 candidate 1 homolog (human) |
| chr5_+_104202609 | 0.76 |
ENSMUST00000066708.5
|
Dmp1
|
dentin matrix protein 1 |
| chr1_-_187215421 | 0.76 |
ENSMUST00000110945.3
ENSMUST00000183931.1 ENSMUST00000027908.6 |
Spata17
|
spermatogenesis associated 17 |
| chr17_+_14943184 | 0.76 |
ENSMUST00000052691.8
ENSMUST00000164837.2 ENSMUST00000174004.1 |
1600012H06Rik
|
RIKEN cDNA 1600012H06 gene |
| chr6_+_37870786 | 0.76 |
ENSMUST00000120428.1
ENSMUST00000031859.7 |
Trim24
|
tripartite motif-containing 24 |
| chr16_-_59553970 | 0.75 |
ENSMUST00000139989.1
|
Crybg3
|
beta-gamma crystallin domain containing 3 |
| chr2_+_93452796 | 0.74 |
ENSMUST00000099693.2
ENSMUST00000162565.1 ENSMUST00000163052.1 |
Gm10804
|
predicted gene 10804 |
| chr2_+_144594054 | 0.73 |
ENSMUST00000136628.1
|
Gm561
|
predicted gene 561 |
| chr19_-_39886730 | 0.73 |
ENSMUST00000168838.1
|
Cyp2c69
|
cytochrome P450, family 2, subfamily c, polypeptide 69 |
| chr18_-_15403680 | 0.73 |
ENSMUST00000079081.6
|
Aqp4
|
aquaporin 4 |
| chr14_-_96519067 | 0.69 |
ENSMUST00000022666.7
|
Klhl1
|
kelch-like 1 |
| chrX_-_160994665 | 0.69 |
ENSMUST00000087104.4
|
Cdkl5
|
cyclin-dependent kinase-like 5 |
| chr7_-_97738222 | 0.69 |
ENSMUST00000084986.6
|
Aqp11
|
aquaporin 11 |
| chr14_-_18893376 | 0.69 |
ENSMUST00000151926.1
|
Ube2e2
|
ubiquitin-conjugating enzyme E2E 2 |
| chr5_+_135009152 | 0.68 |
ENSMUST00000111216.1
ENSMUST00000046999.8 |
Abhd11
|
abhydrolase domain containing 11 |
| chr14_-_51195923 | 0.68 |
ENSMUST00000051274.1
|
Ang2
|
angiogenin, ribonuclease A family, member 2 |
| chr4_+_126609818 | 0.67 |
ENSMUST00000097886.3
ENSMUST00000164362.1 |
5730409E04Rik
|
RIKEN cDNA 5730409E04Rik gene |
| chrX_+_57212110 | 0.67 |
ENSMUST00000033466.1
|
Cd40lg
|
CD40 ligand |
| chr13_-_23934156 | 0.67 |
ENSMUST00000052776.2
|
Hist1h2ba
|
histone cluster 1, H2ba |
| chr8_-_125898291 | 0.66 |
ENSMUST00000047239.6
|
Pcnxl2
|
pecanex-like 2 (Drosophila) |
| chr15_-_36496722 | 0.65 |
ENSMUST00000057486.7
|
Ankrd46
|
ankyrin repeat domain 46 |
| chr11_+_17257558 | 0.64 |
ENSMUST00000000594.2
ENSMUST00000156784.1 |
C1d
|
C1D nuclear receptor co-repressor |
| chr6_-_123853190 | 0.62 |
ENSMUST00000162046.1
|
Vmn2r25
|
vomeronasal 2, receptor 25 |
| chr6_-_117214048 | 0.62 |
ENSMUST00000170447.1
|
Rpl28-ps4
|
ribosomal protein L28, pseudogene 4 |
| chr17_-_45686899 | 0.62 |
ENSMUST00000156254.1
|
Tmem63b
|
transmembrane protein 63b |
| chr7_-_122101735 | 0.61 |
ENSMUST00000139456.1
ENSMUST00000106471.2 ENSMUST00000123296.1 ENSMUST00000033157.3 |
Ndufab1
|
NADH dehydrogenase (ubiquinone) 1, alpha/beta subcomplex, 1 |
| chr7_+_80246375 | 0.61 |
ENSMUST00000058266.6
|
Ttll13
|
tubulin tyrosine ligase-like family, member 13 |
| chr3_+_108571699 | 0.60 |
ENSMUST00000143054.1
|
Taf13
|
TAF13 RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr19_+_29925107 | 0.60 |
ENSMUST00000120388.2
ENSMUST00000144528.1 ENSMUST00000177518.1 |
Il33
|
interleukin 33 |
| chr2_+_69789647 | 0.59 |
ENSMUST00000112266.1
|
Phospho2
|
phosphatase, orphan 2 |
| chr4_+_118961578 | 0.57 |
ENSMUST00000058651.4
|
Lao1
|
L-amino acid oxidase 1 |
| chr10_+_128322443 | 0.57 |
ENSMUST00000026446.2
|
Cnpy2
|
canopy 2 homolog (zebrafish) |
| chr12_-_90969768 | 0.57 |
ENSMUST00000181184.1
|
4930544I03Rik
|
RIKEN cDNA 4930544I03 gene |
| chr3_+_121531603 | 0.57 |
ENSMUST00000180804.1
|
A530020G20Rik
|
RIKEN cDNA A530020G20 gene |
| chr17_+_29274078 | 0.56 |
ENSMUST00000149405.2
|
BC004004
|
cDNA sequence BC004004 |
| chr3_-_151749877 | 0.56 |
ENSMUST00000029671.7
|
Ifi44
|
interferon-induced protein 44 |
| chr17_-_45599603 | 0.56 |
ENSMUST00000171847.1
ENSMUST00000166633.1 ENSMUST00000169729.1 |
Slc29a1
|
solute carrier family 29 (nucleoside transporters), member 1 |
| chr2_-_12419456 | 0.56 |
ENSMUST00000154899.1
ENSMUST00000028105.6 |
Fam188a
|
family with sequence similarity 188, member A |
| chr3_-_75451818 | 0.56 |
ENSMUST00000178270.1
|
Wdr49
|
WD repeat domain 49 |
| chr7_-_14254870 | 0.55 |
ENSMUST00000184731.1
ENSMUST00000076576.6 |
Sult2a6
|
sulfotransferase family 2A, dehydroepiandrosterone (DHEA)-preferring, member 6 |
| chr10_+_81183000 | 0.54 |
ENSMUST00000178422.1
|
Dapk3
|
death-associated protein kinase 3 |
| chr3_-_117077760 | 0.54 |
ENSMUST00000029642.5
|
1700061I17Rik
|
RIKEN cDNA 1700061I17 gene |
| chr14_+_46832127 | 0.54 |
ENSMUST00000068532.8
|
Cgrrf1
|
cell growth regulator with ring finger domain 1 |
| chr7_+_129591859 | 0.54 |
ENSMUST00000084519.5
|
Wdr11
|
WD repeat domain 11 |
| chr10_-_39163794 | 0.53 |
ENSMUST00000076713.4
|
Wisp3
|
WNT1 inducible signaling pathway protein 3 |
| chr6_+_124024758 | 0.53 |
ENSMUST00000032238.3
|
Vmn2r26
|
vomeronasal 2, receptor 26 |
| chr1_-_4785671 | 0.52 |
ENSMUST00000130201.1
ENSMUST00000156816.1 |
Mrpl15
|
mitochondrial ribosomal protein L15 |
| chr3_-_146495115 | 0.52 |
ENSMUST00000093951.2
|
Spata1
|
spermatogenesis associated 1 |
| chrM_+_2743 | 0.52 |
ENSMUST00000082392.1
|
mt-Nd1
|
mitochondrially encoded NADH dehydrogenase 1 |
| chr10_-_107486077 | 0.52 |
ENSMUST00000000445.1
|
Myf5
|
myogenic factor 5 |
| chr8_+_72219726 | 0.51 |
ENSMUST00000003123.8
|
Fam32a
|
family with sequence similarity 32, member A |
| chr10_+_116966274 | 0.51 |
ENSMUST00000033651.3
|
D630029K05Rik
|
RIKEN cDNA D630029K05 gene |
| chr2_+_175469985 | 0.51 |
ENSMUST00000109042.3
ENSMUST00000109002.2 ENSMUST00000109043.2 ENSMUST00000143490.1 |
Gm8923
|
predicted gene 8923 |
| chr9_-_124304718 | 0.50 |
ENSMUST00000071300.6
|
2010315B03Rik
|
RIKEN cDNA 2010315B03 gene |
| chr1_-_141257158 | 0.50 |
ENSMUST00000179202.1
|
Gm4845
|
predicted gene 4845 |
| chrX_+_93156154 | 0.50 |
ENSMUST00000088134.3
|
1700003E24Rik
|
RIKEN cDNA 1700003E24 gene |
| chr9_+_108648720 | 0.49 |
ENSMUST00000098384.2
|
Gm10621
|
predicted gene 10621 |
| chr7_+_26757153 | 0.49 |
ENSMUST00000077855.6
|
Cyp2b19
|
cytochrome P450, family 2, subfamily b, polypeptide 19 |
| chr16_-_26371828 | 0.49 |
ENSMUST00000023154.2
|
Cldn1
|
claudin 1 |
| chr9_-_77399308 | 0.48 |
ENSMUST00000183878.1
|
RP23-264N13.2
|
RP23-264N13.2 |
| chr2_-_155930018 | 0.48 |
ENSMUST00000152766.1
ENSMUST00000139232.1 ENSMUST00000109632.1 ENSMUST00000006036.6 ENSMUST00000142655.1 ENSMUST00000159238.1 |
Uqcc1
|
ubiquinol-cytochrome c reductase complex assembly factor 1 |
| chr15_-_103565069 | 0.48 |
ENSMUST00000023134.3
|
Glycam1
|
glycosylation dependent cell adhesion molecule 1 |
| chr12_-_84617326 | 0.47 |
ENSMUST00000021666.4
|
Abcd4
|
ATP-binding cassette, sub-family D (ALD), member 4 |
| chr17_-_40319205 | 0.47 |
ENSMUST00000026498.4
|
Crisp1
|
cysteine-rich secretory protein 1 |
| chr10_-_42583628 | 0.47 |
ENSMUST00000019938.4
|
Nr2e1
|
nuclear receptor subfamily 2, group E, member 1 |
| chr4_-_149099802 | 0.47 |
ENSMUST00000103217.4
|
Pex14
|
peroxisomal biogenesis factor 14 |
| chr2_-_155930190 | 0.46 |
ENSMUST00000109636.4
ENSMUST00000109631.1 |
Uqcc1
|
ubiquinol-cytochrome c reductase complex assembly factor 1 |
| chr6_-_101377342 | 0.46 |
ENSMUST00000151175.1
|
Pdzrn3
|
PDZ domain containing RING finger 3 |
| chr17_+_46772635 | 0.45 |
ENSMUST00000071430.5
|
2310039H08Rik
|
RIKEN cDNA 2310039H08 gene |
| chr4_-_86669492 | 0.45 |
ENSMUST00000149700.1
|
Plin2
|
perilipin 2 |
| chrY_+_80135210 | 0.44 |
ENSMUST00000179811.1
|
Gm21760
|
predicted gene, 21760 |
| chr11_-_79962374 | 0.43 |
ENSMUST00000108241.1
ENSMUST00000043152.5 |
Utp6
|
UTP6, small subunit (SSU) processome component, homolog (yeast) |
| chr12_+_80463095 | 0.42 |
ENSMUST00000038185.8
|
Exd2
|
exonuclease 3'-5' domain containing 2 |
| chr11_+_58421103 | 0.41 |
ENSMUST00000013797.2
|
1810065E05Rik
|
RIKEN cDNA 1810065E05 gene |
| chr8_+_21839926 | 0.40 |
ENSMUST00000006745.3
|
Defb2
|
defensin beta 2 |
| chr9_+_34904913 | 0.40 |
ENSMUST00000045091.6
|
Kirrel3
|
kin of IRRE like 3 (Drosophila) |
| chr4_+_145510759 | 0.40 |
ENSMUST00000105742.1
ENSMUST00000136309.1 |
Gm13225
|
predicted gene 13225 |
| chr6_+_66896480 | 0.40 |
ENSMUST00000114222.1
|
Gng12
|
guanine nucleotide binding protein (G protein), gamma 12 |
| chr5_-_139345149 | 0.39 |
ENSMUST00000049630.6
|
Cox19
|
cytochrome c oxidase assembly protein 19 |
| chr18_+_57142782 | 0.39 |
ENSMUST00000139892.1
|
Megf10
|
multiple EGF-like-domains 10 |
| chr8_+_85432686 | 0.38 |
ENSMUST00000180883.1
|
1700051O22Rik
|
RIKEN cDNA 1700051O22 Gene |
| chr13_+_55784558 | 0.38 |
ENSMUST00000021961.5
|
Catsper3
|
cation channel, sperm associated 3 |
| chr15_+_99295087 | 0.37 |
ENSMUST00000128352.1
ENSMUST00000145482.1 |
Prpf40b
|
PRP40 pre-mRNA processing factor 40 homolog B (yeast) |
| chr2_+_110597298 | 0.37 |
ENSMUST00000045972.6
ENSMUST00000111026.2 |
Slc5a12
|
solute carrier family 5 (sodium/glucose cotransporter), member 12 |
| chr1_-_158356258 | 0.36 |
ENSMUST00000004133.8
|
Brinp2
|
bone morphogenic protein/retinoic acid inducible neural-specific 2 |
| chr3_+_137341103 | 0.36 |
ENSMUST00000119475.1
|
Emcn
|
endomucin |
| chr16_+_88728828 | 0.34 |
ENSMUST00000060494.6
|
Krtap13-1
|
keratin associated protein 13-1 |
| chr8_-_21906412 | 0.34 |
ENSMUST00000051965.4
|
Defb11
|
defensin beta 11 |
| chr2_+_74721978 | 0.33 |
ENSMUST00000047904.3
|
Hoxd3
|
homeobox D3 |
| chr6_+_66896397 | 0.33 |
ENSMUST00000043148.6
ENSMUST00000114228.1 ENSMUST00000114227.1 ENSMUST00000114226.1 ENSMUST00000114225.1 ENSMUST00000114224.1 |
Gng12
|
guanine nucleotide binding protein (G protein), gamma 12 |
| chrY_-_35920097 | 0.32 |
ENSMUST00000180332.1
|
Gm20896
|
predicted gene, 20896 |
| chr10_+_102158858 | 0.32 |
ENSMUST00000138522.1
ENSMUST00000163753.1 ENSMUST00000138016.1 |
Mgat4c
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) |
| chr11_-_99851608 | 0.31 |
ENSMUST00000107437.1
|
Krtap4-16
|
keratin associated protein 4-16 |
| chr2_-_119662756 | 0.31 |
ENSMUST00000028768.1
ENSMUST00000110801.1 ENSMUST00000110802.1 |
Ndufaf1
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, assembly factor 1 |
| chr1_+_88306731 | 0.30 |
ENSMUST00000040210.7
|
Trpm8
|
transient receptor potential cation channel, subfamily M, member 8 |
| chr11_+_73371246 | 0.30 |
ENSMUST00000120401.1
ENSMUST00000078952.2 ENSMUST00000170592.1 ENSMUST00000127789.1 |
Olfr376
|
olfactory receptor 376 |
| chrX_-_160138375 | 0.30 |
ENSMUST00000033662.8
|
Pdha1
|
pyruvate dehydrogenase E1 alpha 1 |
| chr5_+_121749196 | 0.28 |
ENSMUST00000161064.1
|
Atxn2
|
ataxin 2 |
| chr3_-_37125943 | 0.26 |
ENSMUST00000029275.5
|
Il2
|
interleukin 2 |
| chr3_+_90541146 | 0.26 |
ENSMUST00000107333.1
ENSMUST00000107331.1 ENSMUST00000098910.2 |
S100a16
|
S100 calcium binding protein A16 |
| chr3_+_92957386 | 0.25 |
ENSMUST00000059053.9
|
Lce3d
|
late cornified envelope 3D |
| chrY_-_35130404 | 0.25 |
ENSMUST00000180170.1
|
Gm20855
|
predicted gene, 20855 |
| chr13_+_23162643 | 0.24 |
ENSMUST00000076180.1
|
Vmn1r219
|
vomeronasal 1 receptor 219 |
| chr6_+_17749170 | 0.24 |
ENSMUST00000053148.7
ENSMUST00000115417.3 |
St7
|
suppression of tumorigenicity 7 |
| chr2_+_151543877 | 0.24 |
ENSMUST00000142271.1
|
Fkbp1a
|
FK506 binding protein 1a |
| chr11_+_94967622 | 0.24 |
ENSMUST00000038928.5
|
Hils1
|
histone H1-like protein in spermatids 1 |
| chr12_+_11438191 | 0.23 |
ENSMUST00000181268.1
|
4930511A02Rik
|
RIKEN cDNA 4930511A02 gene |
| chr10_-_53647080 | 0.23 |
ENSMUST00000169866.1
|
Fam184a
|
family with sequence similarity 184, member A |
| chr11_-_99285260 | 0.23 |
ENSMUST00000017255.3
|
Krt24
|
keratin 24 |
| chr8_-_85432841 | 0.22 |
ENSMUST00000047749.5
|
4921524J17Rik
|
RIKEN cDNA 4921524J17 gene |
| chr7_+_78783119 | 0.22 |
ENSMUST00000032840.4
|
Mrps11
|
mitochondrial ribosomal protein S11 |
| chr14_+_4709238 | 0.21 |
ENSMUST00000169465.1
ENSMUST00000171305.1 |
Gm7876
|
predicted gene 7876 |
| chr7_+_126976338 | 0.21 |
ENSMUST00000032920.3
|
Cdipt
|
CDP-diacylglycerol--inositol 3-phosphatidyltransferase (phosphatidylinositol synthase) |
| chr5_-_28210022 | 0.20 |
ENSMUST00000118882.1
|
Cnpy1
|
canopy 1 homolog (zebrafish) |
| chr10_+_102159000 | 0.20 |
ENSMUST00000020039.6
|
Mgat4c
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) |
Network of associatons between targets according to the STRING database.
First level regulatory network of Emx1_Emx2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 6.7 | GO:0019442 | tryptophan catabolic process to acetyl-CoA(GO:0019442) |
| 1.7 | 6.9 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 1.7 | 5.1 | GO:1903919 | regulation of actin filament severing(GO:1903918) negative regulation of actin filament severing(GO:1903919) |
| 1.2 | 4.6 | GO:0002489 | antigen processing and presentation of endogenous peptide antigen via MHC class Ib via ER pathway(GO:0002488) antigen processing and presentation of endogenous peptide antigen via MHC class Ib via ER pathway, TAP-dependent(GO:0002489) positive regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000566) |
| 1.0 | 3.0 | GO:0019343 | cysteine biosynthetic process via cystathionine(GO:0019343) |
| 0.9 | 5.3 | GO:0072592 | oxygen metabolic process(GO:0072592) |
| 0.9 | 4.3 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.7 | 23.4 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.7 | 2.7 | GO:0002415 | immunoglobulin transcytosis in epithelial cells mediated by polymeric immunoglobulin receptor(GO:0002415) |
| 0.7 | 2.0 | GO:0060300 | regulation of cytokine activity(GO:0060300) |
| 0.4 | 1.1 | GO:1903122 | enzyme active site formation via L-cysteine sulfinic acid(GO:0018323) primary alcohol biosynthetic process(GO:0034309) cellular response to glyoxal(GO:0036471) glycolate biosynthetic process(GO:0046295) negative regulation of TRAIL-activated apoptotic signaling pathway(GO:1903122) regulation of pyrroline-5-carboxylate reductase activity(GO:1903167) positive regulation of pyrroline-5-carboxylate reductase activity(GO:1903168) regulation of tyrosine 3-monooxygenase activity(GO:1903176) positive regulation of tyrosine 3-monooxygenase activity(GO:1903178) L-dopa metabolic process(GO:1903184) L-dopa biosynthetic process(GO:1903185) glyoxal metabolic process(GO:1903189) regulation of L-dopa biosynthetic process(GO:1903195) positive regulation of L-dopa biosynthetic process(GO:1903197) regulation of L-dopa decarboxylase activity(GO:1903198) positive regulation of L-dopa decarboxylase activity(GO:1903200) positive regulation of cellular amino acid biosynthetic process(GO:2000284) |
| 0.4 | 1.8 | GO:0009115 | xanthine catabolic process(GO:0009115) |
| 0.3 | 1.7 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.3 | 3.0 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.3 | 1.3 | GO:1902512 | positive regulation of apoptotic DNA fragmentation(GO:1902512) |
| 0.3 | 4.6 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.3 | 1.7 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.3 | 1.1 | GO:0097360 | chorionic trophoblast cell proliferation(GO:0097360) regulation of chorionic trophoblast cell proliferation(GO:1901382) |
| 0.2 | 1.2 | GO:0042780 | tRNA 3'-end processing(GO:0042780) |
| 0.2 | 0.7 | GO:2001200 | positive regulation of dendritic cell differentiation(GO:2001200) |
| 0.2 | 3.9 | GO:0042359 | vitamin D metabolic process(GO:0042359) |
| 0.2 | 0.4 | GO:0021740 | trigeminal sensory nucleus development(GO:0021730) principal sensory nucleus of trigeminal nerve development(GO:0021740) |
| 0.2 | 0.8 | GO:0070173 | regulation of enamel mineralization(GO:0070173) |
| 0.2 | 0.5 | GO:0043519 | myosin II filament organization(GO:0031038) regulation of myosin II filament organization(GO:0043519) |
| 0.2 | 0.7 | GO:0072014 | proximal tubule development(GO:0072014) |
| 0.2 | 2.4 | GO:0015747 | urate transport(GO:0015747) |
| 0.2 | 0.5 | GO:1903348 | positive regulation of bicellular tight junction assembly(GO:1903348) |
| 0.2 | 2.7 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.2 | 0.5 | GO:0016561 | protein import into peroxisome matrix, translocation(GO:0016561) |
| 0.2 | 0.8 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.1 | 0.7 | GO:0070295 | renal water absorption(GO:0070295) |
| 0.1 | 0.9 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.1 | 0.6 | GO:0002282 | microglial cell activation involved in immune response(GO:0002282) |
| 0.1 | 1.1 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.1 | 0.3 | GO:0044029 | DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.1 | 0.6 | GO:0045716 | positive regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045716) |
| 0.1 | 0.1 | GO:0060974 | cell migration involved in heart formation(GO:0060974) |
| 0.1 | 1.2 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.1 | 0.3 | GO:0001777 | T cell homeostatic proliferation(GO:0001777) regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.1 | 0.7 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.1 | 0.9 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.1 | 0.7 | GO:0035093 | spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.1 | 1.5 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.1 | 3.5 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.1 | 0.6 | GO:0015862 | uridine transport(GO:0015862) |
| 0.1 | 2.2 | GO:0006921 | cellular component disassembly involved in execution phase of apoptosis(GO:0006921) apoptotic nuclear changes(GO:0030262) |
| 0.1 | 1.4 | GO:0034374 | low-density lipoprotein particle remodeling(GO:0034374) |
| 0.1 | 0.6 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.1 | 0.6 | GO:1901727 | positive regulation of histone deacetylase activity(GO:1901727) |
| 0.1 | 0.8 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.1 | 0.2 | GO:0046341 | CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.1 | 0.5 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.1 | 0.2 | GO:0071727 | response to triacyl bacterial lipopeptide(GO:0071725) cellular response to triacyl bacterial lipopeptide(GO:0071727) |
| 0.1 | 0.5 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.1 | 0.8 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.1 | 1.8 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.1 | 2.3 | GO:0001702 | gastrulation with mouth forming second(GO:0001702) |
| 0.1 | 0.8 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.0 | 0.4 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) recognition of apoptotic cell(GO:0043654) |
| 0.0 | 0.3 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.3 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.0 | 0.2 | GO:0060279 | positive regulation of ovulation(GO:0060279) |
| 0.0 | 2.4 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 1.4 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.0 | 0.3 | GO:0050955 | thermoception(GO:0050955) |
| 0.0 | 0.4 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.1 | GO:1905035 | regulation of antifungal innate immune response(GO:1905034) negative regulation of antifungal innate immune response(GO:1905035) |
| 0.0 | 1.0 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.0 | 0.4 | GO:0000729 | DNA double-strand break processing(GO:0000729) |
| 0.0 | 0.5 | GO:0033194 | response to hydroperoxide(GO:0033194) |
| 0.0 | 1.5 | GO:0006695 | cholesterol biosynthetic process(GO:0006695) secondary alcohol biosynthetic process(GO:1902653) |
| 0.0 | 0.2 | GO:0032324 | molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.0 | 0.6 | GO:0000460 | maturation of 5.8S rRNA(GO:0000460) |
| 0.0 | 0.2 | GO:1990000 | amyloid fibril formation(GO:1990000) |
| 0.0 | 0.1 | GO:0060060 | post-embryonic retina morphogenesis in camera-type eye(GO:0060060) |
| 0.0 | 1.1 | GO:0061049 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.7 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.1 | GO:0000960 | mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 0.0 | 0.2 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.0 | 0.5 | GO:0000002 | mitochondrial genome maintenance(GO:0000002) |
| 0.0 | 0.9 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.0 | 0.2 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.0 | 0.1 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) |
| 0.0 | 0.3 | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 0.0 | 0.0 | GO:0071922 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.0 | 0.4 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 0.7 | GO:0007628 | adult walking behavior(GO:0007628) |
| 0.0 | 0.1 | GO:0045590 | negative regulation of regulatory T cell differentiation(GO:0045590) |
| 0.0 | 0.0 | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 0.0 | 0.1 | GO:1902416 | positive regulation of mRNA binding(GO:1902416) |
| 0.0 | 0.1 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 | 0.6 | GO:0007528 | neuromuscular junction development(GO:0007528) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 4.6 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.3 | 1.0 | GO:0097636 | intrinsic component of autophagosome membrane(GO:0097636) integral component of autophagosome membrane(GO:0097637) |
| 0.3 | 6.9 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.1 | 0.5 | GO:1990415 | Pex17p-Pex14p docking complex(GO:1990415) peroxisomal importomer complex(GO:1990429) |
| 0.1 | 0.3 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.1 | 0.8 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.1 | 3.8 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.1 | 1.7 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.1 | 0.4 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.1 | 1.2 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.1 | 0.2 | GO:0043512 | inhibin A complex(GO:0043512) |
| 0.1 | 0.2 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.1 | 0.9 | GO:0033655 | host cell cytoplasm(GO:0030430) host cell cytoplasm part(GO:0033655) |
| 0.0 | 2.3 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.7 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.0 | 0.3 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.0 | 0.4 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.0 | 0.2 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.0 | 0.1 | GO:1990730 | VCP-NSFL1C complex(GO:1990730) |
| 0.0 | 0.6 | GO:0000178 | exosome (RNase complex)(GO:0000178) |
| 0.0 | 1.1 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.8 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.2 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.2 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 0.7 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.8 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.4 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.0 | 0.7 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 0.1 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.0 | 3.8 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 2.4 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 2.9 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.6 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 5.6 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 0.2 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 22.9 | GO:0005783 | endoplasmic reticulum(GO:0005783) |
| 0.0 | 2.1 | GO:0044798 | nuclear transcription factor complex(GO:0044798) |
| 0.0 | 0.6 | GO:0000800 | lateral element(GO:0000800) |
| 0.0 | 0.7 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 14.9 | GO:0033695 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 1.7 | 6.7 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 1.3 | 3.9 | GO:1902271 | D3 vitamins binding(GO:1902271) |
| 1.0 | 3.0 | GO:0098809 | nitrite reductase activity(GO:0098809) |
| 0.8 | 5.1 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.8 | 2.3 | GO:0003979 | UDP-glucose 6-dehydrogenase activity(GO:0003979) |
| 0.5 | 5.1 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.4 | 1.5 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.4 | 1.1 | GO:0036478 | tyrosine 3-monooxygenase activator activity(GO:0036470) L-dopa decarboxylase activator activity(GO:0036478) |
| 0.4 | 1.8 | GO:0016726 | aldehyde oxidase activity(GO:0004031) xanthine dehydrogenase activity(GO:0004854) oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) oxidoreductase activity, acting on CH or CH2 groups, NAD or NADP as acceptor(GO:0016726) |
| 0.4 | 4.6 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.3 | 1.7 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.3 | 2.7 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) |
| 0.3 | 0.9 | GO:0047522 | 13-prostaglandin reductase activity(GO:0036132) 15-oxoprostaglandin 13-oxidase activity(GO:0047522) |
| 0.3 | 4.6 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.3 | 4.3 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.3 | 8.5 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 0.2 | 3.5 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.2 | 1.5 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.2 | 0.6 | GO:0001716 | L-amino-acid oxidase activity(GO:0001716) |
| 0.2 | 7.0 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.2 | 2.4 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.1 | 1.0 | GO:0045134 | uridine-diphosphatase activity(GO:0045134) |
| 0.1 | 0.6 | GO:0033883 | pyridoxal phosphatase activity(GO:0033883) |
| 0.1 | 1.5 | GO:0051870 | methotrexate binding(GO:0051870) |
| 0.1 | 2.0 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.1 | 0.9 | GO:0016725 | oxidoreductase activity, acting on CH or CH2 groups(GO:0016725) |
| 0.1 | 0.7 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.1 | 0.8 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.1 | 0.7 | GO:0015288 | porin activity(GO:0015288) |
| 0.1 | 2.4 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.1 | 0.6 | GO:0050656 | 3'-phosphoadenosine 5'-phosphosulfate binding(GO:0050656) |
| 0.1 | 1.0 | GO:0036122 | BMP binding(GO:0036122) |
| 0.1 | 1.2 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.1 | 2.7 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 0.5 | GO:0043199 | sulfate binding(GO:0043199) |
| 0.1 | 0.3 | GO:0034604 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.1 | 0.2 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.1 | 0.4 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.1 | 1.1 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.1 | 0.4 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.7 | GO:0015250 | water channel activity(GO:0015250) |
| 0.0 | 0.4 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.7 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.0 | 0.9 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 0.3 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 1.2 | GO:0016891 | endoribonuclease activity, producing 5'-phosphomonoesters(GO:0016891) |
| 0.0 | 0.2 | GO:0071723 | lipopeptide binding(GO:0071723) |
| 0.0 | 0.1 | GO:0047936 | glucose 1-dehydrogenase [NAD(P)] activity(GO:0047936) |
| 0.0 | 0.9 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 1.1 | GO:1900750 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.0 | 1.9 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.0 | 0.5 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 1.4 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.0 | 0.2 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.0 | 0.3 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.0 | 0.9 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 1.4 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.2 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.0 | 0.2 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.0 | 0.3 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 0.2 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 1.1 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.1 | GO:0001565 | phorbol ester receptor activity(GO:0001565) non-kinase phorbol ester receptor activity(GO:0001566) |
| 0.0 | 0.5 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 0.8 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 1.0 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.2 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 1.1 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 2.4 | GO:0052689 | carboxylic ester hydrolase activity(GO:0052689) |
| 0.0 | 1.3 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.6 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.0 | 0.1 | GO:0051381 | histamine binding(GO:0051381) |
| 0.0 | 0.5 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.7 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.6 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 0.2 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.4 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 0.2 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.0 | 0.1 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 1.5 | GO:0004519 | endonuclease activity(GO:0004519) |
| 0.0 | 2.3 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.0 | 0.1 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 0.0 | 0.1 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 7.0 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 2.9 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 4.6 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 2.7 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 1.1 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.7 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.5 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 0.8 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.8 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.8 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 4.6 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.4 | 6.7 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.3 | 6.9 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.3 | 4.3 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.2 | 2.0 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.2 | 2.3 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.1 | 1.7 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.1 | 3.0 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.1 | 4.5 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.1 | 1.4 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.1 | 1.7 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.1 | 2.0 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 1.5 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 0.7 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.1 | 2.3 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.9 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.0 | 0.5 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 1.1 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.8 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.2 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.3 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.7 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.0 | 0.7 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.8 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.4 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.2 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.6 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.2 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 0.7 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.6 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.5 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.5 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.2 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.4 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |