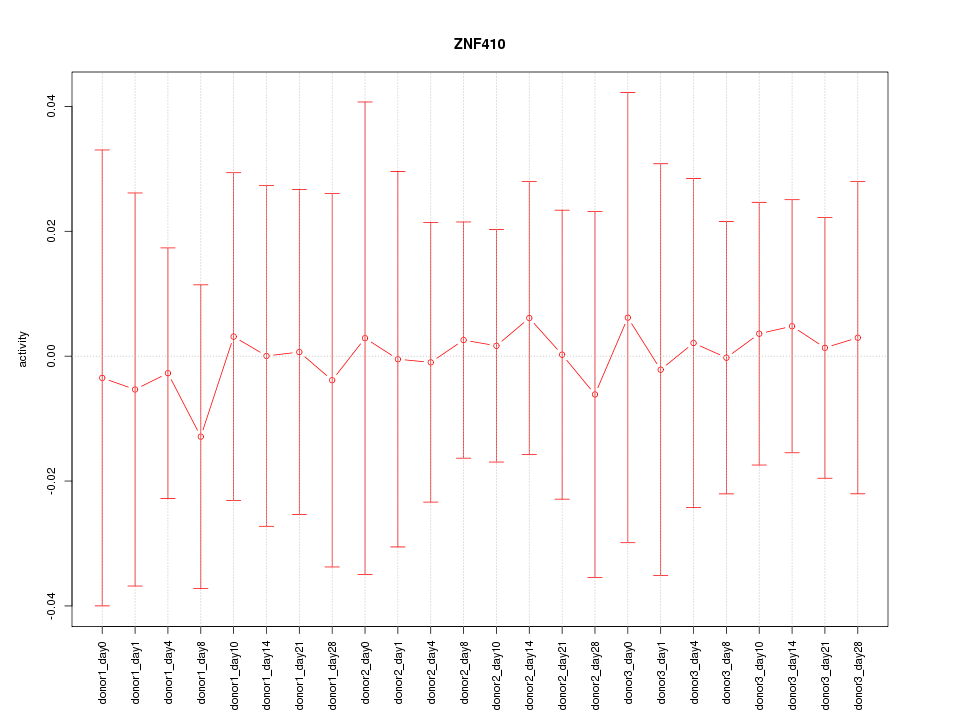
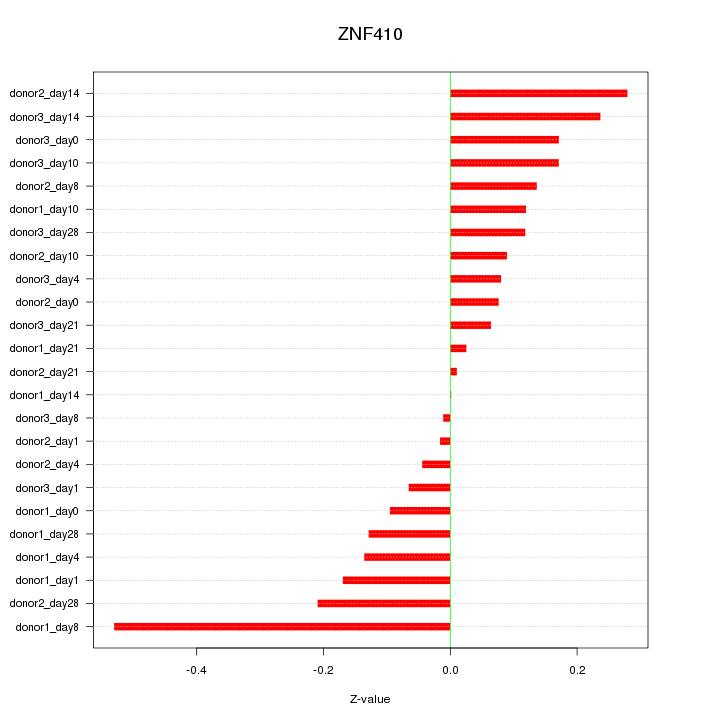
Motif ID: ZNF410
Z-value: 0.167
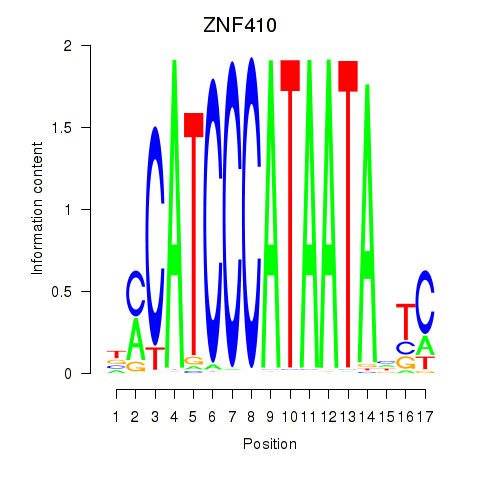
Transcription factors associated with ZNF410:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| ZNF410 | ENSG00000119725.13 | ZNF410 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZNF410 | hg19_v2_chr14_+_74353574_74353589 | -0.07 | 7.3e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | GO:0072553 | terminal button organization(GO:0072553) |
| 0.0 | 0.0 | GO:0090024 | complement component C5a signaling pathway(GO:0038178) negative regulation of granulocyte chemotaxis(GO:0071623) negative regulation of neutrophil chemotaxis(GO:0090024) negative regulation of neutrophil migration(GO:1902623) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 0.0 | GO:0004878 | complement component C5a receptor activity(GO:0004878) |