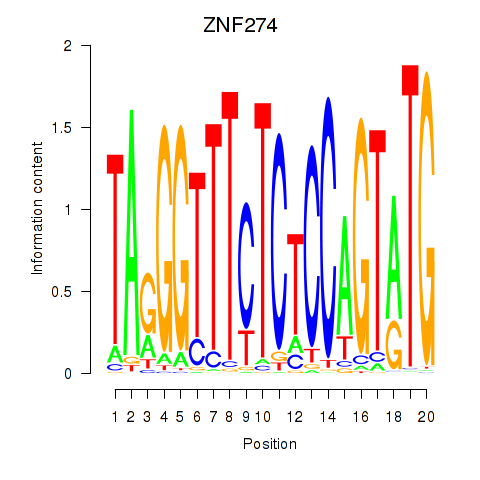
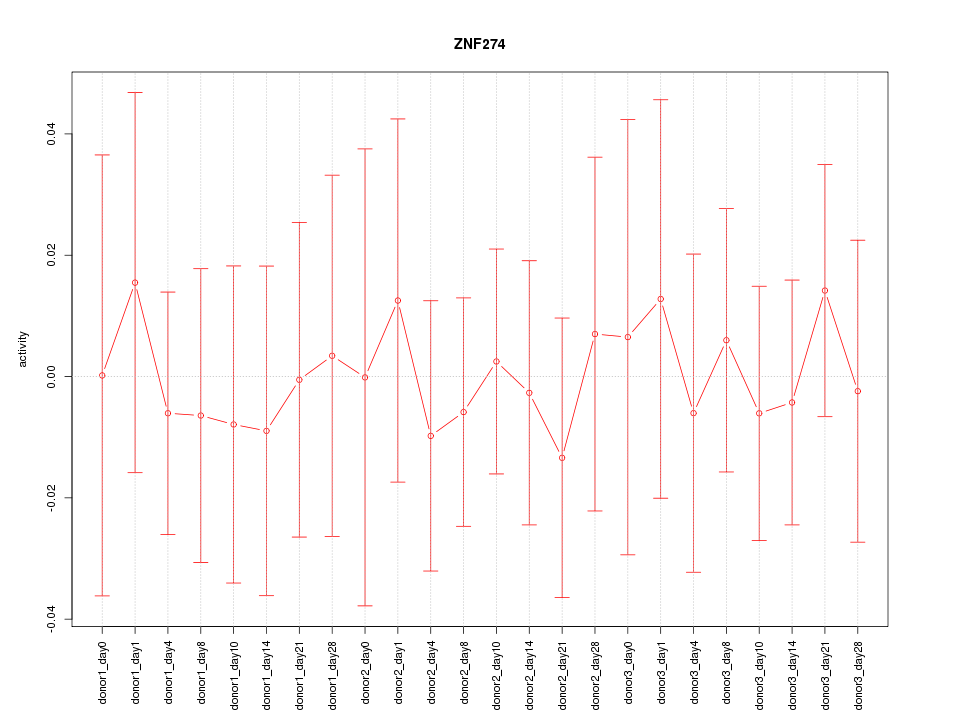
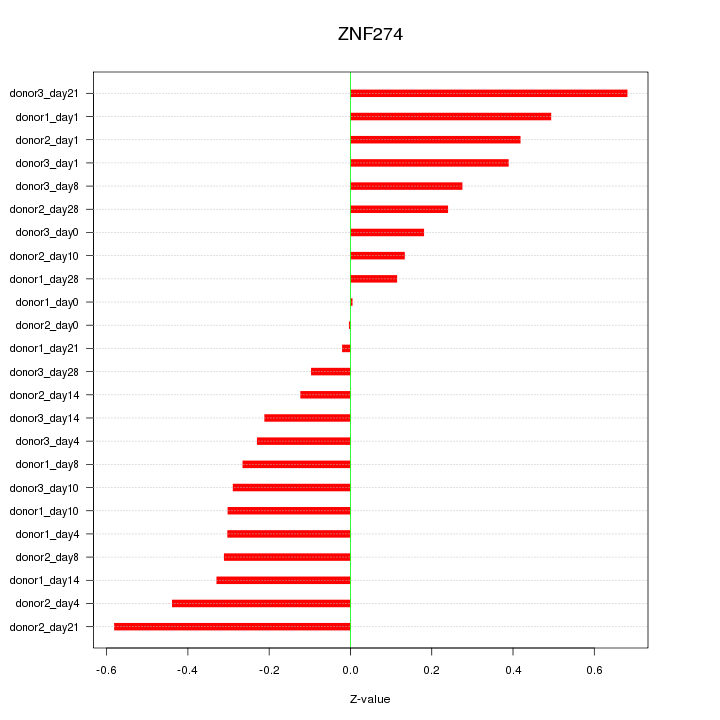
|
chr1_+_45205498
|
0.672
|
ENST00000372218.4
|
KIF2C
|
kinesin family member 2C
|
|
chr1_+_45205478
|
0.667
|
ENST00000452259.1
ENST00000372224.4
|
KIF2C
|
kinesin family member 2C
|
|
chr12_-_54653313
|
0.329
|
ENST00000550411.1
ENST00000439541.2
|
CBX5
|
chromobox homolog 5
|
|
chr2_+_11864458
|
0.304
|
ENST00000396098.1
ENST00000396099.1
ENST00000425416.2
|
LPIN1
|
lipin 1
|
|
chr6_+_31553901
|
0.272
|
ENST00000418507.2
ENST00000438075.2
ENST00000376100.3
ENST00000376111.4
|
LST1
|
leukocyte specific transcript 1
|
|
chr6_+_31553978
|
0.248
|
ENST00000376096.1
ENST00000376099.1
ENST00000376110.3
|
LST1
|
leukocyte specific transcript 1
|
|
chr2_-_42991257
|
0.246
|
ENST00000378661.2
|
OXER1
|
oxoeicosanoid (OXE) receptor 1
|
|
chr16_+_29818857
|
0.242
|
ENST00000567444.1
|
MAZ
|
MYC-associated zinc finger protein (purine-binding transcription factor)
|
|
chr6_+_30687978
|
0.239
|
ENST00000327892.8
ENST00000435534.1
|
TUBB
|
tubulin, beta class I
|
|
chr12_+_113344755
|
0.229
|
ENST00000550883.1
|
OAS1
|
2'-5'-oligoadenylate synthetase 1, 40/46kDa
|
|
chr20_+_60174827
|
0.228
|
ENST00000543233.1
|
CDH4
|
cadherin 4, type 1, R-cadherin (retinal)
|
|
chr22_+_39353527
|
0.227
|
ENST00000249116.2
|
APOBEC3A
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3A
|
|
chr11_+_55653396
|
0.225
|
ENST00000244891.3
|
TRIM51
|
tripartite motif-containing 51
|
|
chr17_-_41132410
|
0.223
|
ENST00000409399.1
ENST00000421990.2
ENST00000409446.3
ENST00000453594.1
|
PTGES3L-AARSD1
PTGES3L
|
PTGES3L-AARSD1 readthrough
prostaglandin E synthase 3 (cytosolic)-like
|
|
chr18_+_9885760
|
0.214
|
ENST00000536353.2
ENST00000584255.1
|
TXNDC2
|
thioredoxin domain containing 2 (spermatozoa)
|
|
chr14_+_22409308
|
0.193
|
ENST00000390441.2
|
TRAV9-2
|
T cell receptor alpha variable 9-2
|
|
chr11_-_10590118
|
0.183
|
ENST00000529598.1
|
LYVE1
|
lymphatic vessel endothelial hyaluronan receptor 1
|
|
chr6_-_32784687
|
0.174
|
ENST00000447394.1
ENST00000438763.2
|
HLA-DOB
|
major histocompatibility complex, class II, DO beta
|
|
chr2_-_89292422
|
0.163
|
ENST00000495489.1
|
IGKV1-8
|
immunoglobulin kappa variable 1-8
|
|
chr12_+_109273806
|
0.163
|
ENST00000228476.3
ENST00000547768.1
|
DAO
|
D-amino-acid oxidase
|
|
chr16_-_80603558
|
0.157
|
ENST00000567317.1
|
RP11-18F14.1
|
RP11-18F14.1
|
|
chr8_-_4852494
|
0.155
|
ENST00000520002.1
ENST00000602557.1
|
CSMD1
|
CUB and Sushi multiple domains 1
|
|
chr3_+_102153859
|
0.153
|
ENST00000306176.1
ENST00000466937.1
|
ZPLD1
|
zona pellucida-like domain containing 1
|
|
chr19_+_6887571
|
0.149
|
ENST00000250572.8
ENST00000381407.5
ENST00000312053.4
ENST00000450315.3
ENST00000381404.4
|
EMR1
|
egf-like module containing, mucin-like, hormone receptor-like 1
|
|
chr8_-_4852218
|
0.144
|
ENST00000400186.3
ENST00000602723.1
|
CSMD1
|
CUB and Sushi multiple domains 1
|
|
chr6_+_36165133
|
0.131
|
ENST00000446974.1
ENST00000454960.1
|
BRPF3
|
bromodomain and PHD finger containing, 3
|
|
chr18_+_9885961
|
0.130
|
ENST00000306084.6
|
TXNDC2
|
thioredoxin domain containing 2 (spermatozoa)
|
|
chr18_+_9885934
|
0.129
|
ENST00000357775.5
|
TXNDC2
|
thioredoxin domain containing 2 (spermatozoa)
|
|
chr14_-_24047965
|
0.127
|
ENST00000397118.3
ENST00000356300.4
|
JPH4
|
junctophilin 4
|
|
chr10_-_29811456
|
0.115
|
ENST00000535393.1
|
SVIL
|
supervillin
|
|
chr5_+_32531893
|
0.105
|
ENST00000512913.1
|
SUB1
|
SUB1 homolog (S. cerevisiae)
|
|
chr10_+_5488564
|
0.097
|
ENST00000449083.1
ENST00000380359.3
|
NET1
|
neuroepithelial cell transforming 1
|
|
chr16_+_29819096
|
0.092
|
ENST00000568411.1
ENST00000563012.1
ENST00000562557.1
|
MAZ
|
MYC-associated zinc finger protein (purine-binding transcription factor)
|
|
chr15_+_89164520
|
0.087
|
ENST00000332810.3
|
AEN
|
apoptosis enhancing nuclease
|
|
chr5_-_74162605
|
0.071
|
ENST00000389156.4
ENST00000510496.1
ENST00000380515.3
|
FAM169A
|
family with sequence similarity 169, member A
|
|
chr16_+_31470179
|
0.070
|
ENST00000538189.1
ENST00000268314.4
|
ARMC5
|
armadillo repeat containing 5
|
|
chr19_+_6135646
|
0.068
|
ENST00000588304.1
ENST00000588485.1
ENST00000588722.1
ENST00000591403.1
ENST00000586696.1
ENST00000589401.1
ENST00000252669.5
|
ACSBG2
|
acyl-CoA synthetase bubblegum family member 2
|
|
chr16_+_29819446
|
0.064
|
ENST00000568282.1
|
MAZ
|
MYC-associated zinc finger protein (purine-binding transcription factor)
|
|
chr16_+_31470143
|
0.062
|
ENST00000457010.2
ENST00000563544.1
|
ARMC5
|
armadillo repeat containing 5
|
|
chrX_+_52920336
|
0.062
|
ENST00000452154.2
|
FAM156B
|
family with sequence similarity 156, member B
|
|
chr16_+_29819372
|
0.061
|
ENST00000568544.1
ENST00000569978.1
|
MAZ
|
MYC-associated zinc finger protein (purine-binding transcription factor)
|
|
chrX_+_119737806
|
0.051
|
ENST00000371317.5
|
MCTS1
|
malignant T cell amplified sequence 1
|
|
chr3_+_50284321
|
0.047
|
ENST00000451956.1
|
GNAI2
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 2
|
|
chr1_+_42928945
|
0.043
|
ENST00000428554.2
|
CCDC30
|
coiled-coil domain containing 30
|
|
chr19_+_40195101
|
0.043
|
ENST00000360675.3
ENST00000601802.1
|
LGALS14
|
lectin, galactoside-binding, soluble, 14
|
|
chr19_+_40194946
|
0.036
|
ENST00000392052.3
|
LGALS14
|
lectin, galactoside-binding, soluble, 14
|
|
chr22_+_24038593
|
0.034
|
ENST00000452208.1
|
RGL4
|
ral guanine nucleotide dissociation stimulator-like 4
|
|
chr7_+_30791743
|
0.032
|
ENST00000013222.5
ENST00000409539.1
|
INMT
|
indolethylamine N-methyltransferase
|
|
chr4_-_121993673
|
0.031
|
ENST00000379692.4
|
NDNF
|
neuron-derived neurotrophic factor
|
|
chr17_-_3461092
|
0.016
|
ENST00000301365.4
ENST00000572519.1
|
TRPV3
|
transient receptor potential cation channel, subfamily V, member 3
|
|
chr2_+_9696028
|
0.011
|
ENST00000607241.1
|
RP11-214N9.1
|
RP11-214N9.1
|
|
chr19_+_41949054
|
0.004
|
ENST00000378187.2
|
C19orf69
|
chromosome 19 open reading frame 69
|