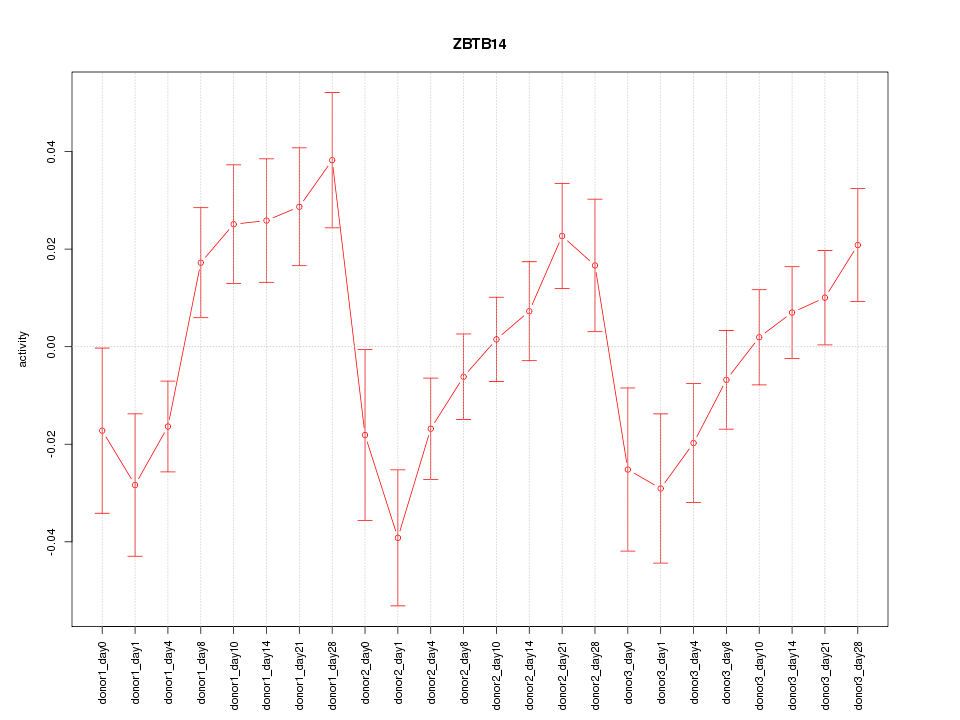
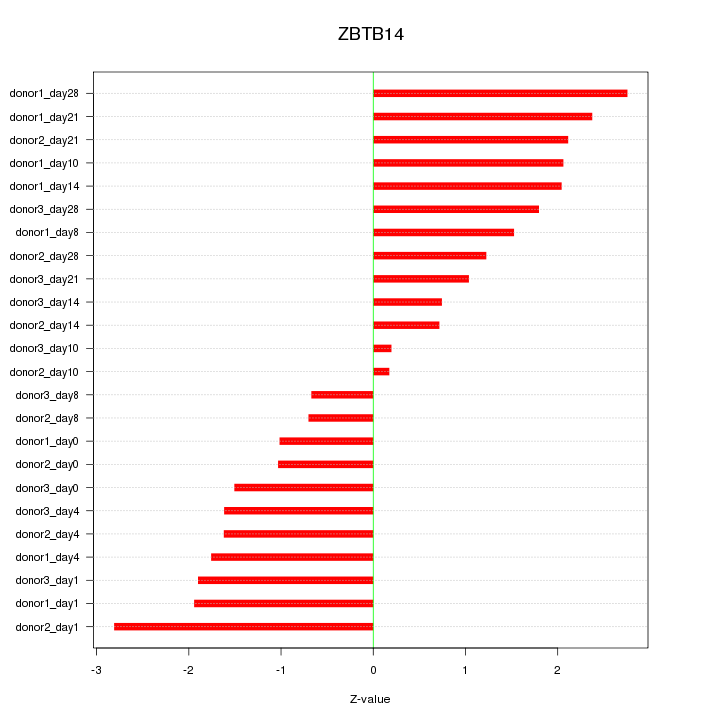
Motif ID: ZBTB14
Z-value: 1.637
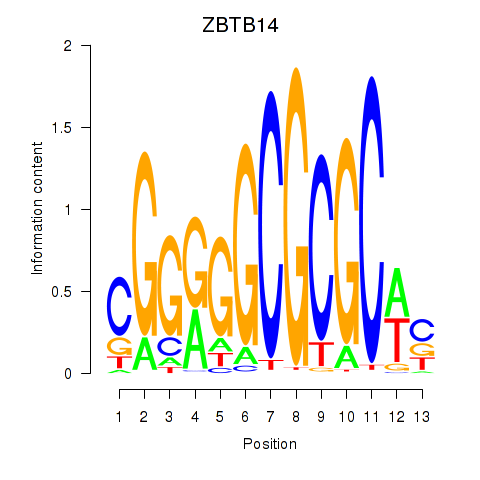
Transcription factors associated with ZBTB14:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| ZBTB14 | ENSG00000198081.6 | ZBTB14 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZBTB14 | hg19_v2_chr18_-_5296001_5296045 | 0.33 | 1.2e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.3 | 16.4 | GO:0019303 | D-ribose catabolic process(GO:0019303) |
| 2.5 | 7.6 | GO:1904899 | regulation of hepatic stellate cell proliferation(GO:1904897) positive regulation of hepatic stellate cell proliferation(GO:1904899) hepatic stellate cell proliferation(GO:1990922) |
| 2.3 | 7.0 | GO:0050760 | negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 1.2 | 3.6 | GO:0035283 | rhombomere 5 development(GO:0021571) central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.8 | 6.5 | GO:0045607 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.8 | 2.4 | GO:1904048 | regulation of spontaneous neurotransmitter secretion(GO:1904048) |
| 0.7 | 2.7 | GO:0043335 | protein unfolding(GO:0043335) |
| 0.6 | 12.8 | GO:0019614 | catechol-containing compound catabolic process(GO:0019614) catecholamine catabolic process(GO:0042424) |
| 0.6 | 3.6 | GO:0071630 | nucleus-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071630) |
| 0.6 | 1.8 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.6 | 5.0 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.5 | 3.1 | GO:0071500 | cellular response to nitrosative stress(GO:0071500) |
| 0.5 | 6.3 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.5 | 6.7 | GO:1903025 | regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903025) |
| 0.5 | 1.4 | GO:0042351 | 'de novo' GDP-L-fucose biosynthetic process(GO:0042351) |
| 0.5 | 1.9 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 0.4 | 2.6 | GO:0032380 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.4 | 1.3 | GO:0021966 | corticospinal neuron axon guidance(GO:0021966) |
| 0.4 | 3.2 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.4 | 1.2 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.4 | 1.6 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.4 | 3.0 | GO:0071233 | cellular response to leucine(GO:0071233) |
| 0.4 | 1.5 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 0.4 | 1.5 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.4 | 4.3 | GO:0030853 | negative regulation of granulocyte differentiation(GO:0030853) |
| 0.4 | 2.2 | GO:0006540 | glutamate decarboxylation to succinate(GO:0006540) |
| 0.4 | 1.8 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 0.4 | 2.1 | GO:0072675 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 0.3 | 1.0 | GO:1990927 | short-term synaptic potentiation(GO:1990926) calcium ion regulated lysosome exocytosis(GO:1990927) |
| 0.3 | 1.0 | GO:0061188 | negative regulation of chromatin silencing at rDNA(GO:0061188) |
| 0.3 | 1.0 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.3 | 0.7 | GO:0061502 | early endosome to recycling endosome transport(GO:0061502) |
| 0.3 | 0.3 | GO:0072053 | renal inner medulla development(GO:0072053) renal outer medulla development(GO:0072054) |
| 0.3 | 4.1 | GO:0070070 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.3 | 2.2 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.3 | 0.3 | GO:0031116 | positive regulation of microtubule polymerization(GO:0031116) |
| 0.3 | 0.9 | GO:2000834 | androgen secretion(GO:0035935) regulation of androgen secretion(GO:2000834) positive regulation of androgen secretion(GO:2000836) |
| 0.3 | 0.9 | GO:0045976 | negative regulation of mitotic cell cycle, embryonic(GO:0045976) |
| 0.3 | 0.9 | GO:0048010 | vascular endothelial growth factor receptor signaling pathway(GO:0048010) |
| 0.3 | 0.8 | GO:0010693 | negative regulation of alkaline phosphatase activity(GO:0010693) |
| 0.3 | 6.0 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.3 | 3.6 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.2 | 0.7 | GO:0019243 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.2 | 0.7 | GO:0016256 | N-glycan processing to lysosome(GO:0016256) |
| 0.2 | 2.5 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.2 | 0.7 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.2 | 1.7 | GO:2000467 | positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.2 | 0.4 | GO:0030860 | regulation of polarized epithelial cell differentiation(GO:0030860) |
| 0.2 | 0.6 | GO:0000294 | nuclear-transcribed mRNA catabolic process, endonucleolytic cleavage-dependent decay(GO:0000294) |
| 0.2 | 2.3 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.2 | 1.2 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.2 | 0.8 | GO:0097026 | dendritic cell dendrite assembly(GO:0097026) |
| 0.2 | 0.6 | GO:0031508 | pericentric heterochromatin assembly(GO:0031508) |
| 0.2 | 0.8 | GO:1900224 | positive regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900224) |
| 0.2 | 7.7 | GO:0000038 | very long-chain fatty acid metabolic process(GO:0000038) |
| 0.2 | 1.1 | GO:0050893 | sensory processing(GO:0050893) |
| 0.2 | 0.6 | GO:0015729 | thiosulfate transport(GO:0015709) oxaloacetate transport(GO:0015729) malate transport(GO:0015743) malate transmembrane transport(GO:0071423) oxaloacetate(2-) transmembrane transport(GO:1902356) |
| 0.2 | 1.1 | GO:0010637 | negative regulation of mitochondrial fusion(GO:0010637) |
| 0.2 | 0.5 | GO:0031587 | positive regulation of inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0031587) |
| 0.2 | 0.5 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 0.2 | 0.5 | GO:0032240 | mRNA export from nucleus in response to heat stress(GO:0031990) negative regulation of nucleobase-containing compound transport(GO:0032240) positive regulation of chromatin assembly or disassembly(GO:0045799) negative regulation of RNA export from nucleus(GO:0046832) |
| 0.2 | 0.5 | GO:1902303 | regulation of heart rate by hormone(GO:0003064) negative regulation of potassium ion export(GO:1902303) |
| 0.2 | 0.2 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.2 | 5.2 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.2 | 0.5 | GO:0035494 | SNARE complex disassembly(GO:0035494) |
| 0.2 | 0.5 | GO:0034553 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.2 | 1.3 | GO:0070445 | oligodendrocyte progenitor proliferation(GO:0070444) regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 0.2 | 0.3 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.2 | 0.5 | GO:0016340 | calcium-dependent cell-matrix adhesion(GO:0016340) |
| 0.2 | 1.1 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) |
| 0.2 | 0.2 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.2 | 0.5 | GO:0031443 | fast-twitch skeletal muscle fiber contraction(GO:0031443) |
| 0.2 | 0.6 | GO:0003172 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) regulation of synaptic activity(GO:0060025) |
| 0.1 | 0.7 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.1 | 0.4 | GO:0006434 | seryl-tRNA aminoacylation(GO:0006434) |
| 0.1 | 0.4 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.1 | 0.4 | GO:0006679 | glucosylceramide biosynthetic process(GO:0006679) |
| 0.1 | 0.4 | GO:0051311 | meiotic metaphase I plate congression(GO:0043060) meiotic spindle midzone assembly(GO:0051257) meiotic metaphase plate congression(GO:0051311) |
| 0.1 | 0.4 | GO:0090427 | activation of meiosis(GO:0090427) |
| 0.1 | 1.1 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.1 | 0.6 | GO:2000729 | olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 0.1 | 0.7 | GO:0003186 | tricuspid valve morphogenesis(GO:0003186) |
| 0.1 | 0.6 | GO:0016095 | polyprenol catabolic process(GO:0016095) |
| 0.1 | 4.4 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.1 | 1.1 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.1 | 0.5 | GO:1902361 | mitochondrial pyruvate transport(GO:0006850) mitochondrial pyruvate transmembrane transport(GO:1902361) |
| 0.1 | 0.5 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.1 | 0.4 | GO:0071139 | resolution of recombination intermediates(GO:0071139) resolution of mitotic recombination intermediates(GO:0071140) |
| 0.1 | 0.4 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) |
| 0.1 | 0.5 | GO:0060120 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.1 | 0.7 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.1 | 1.9 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.1 | 0.4 | GO:0034970 | histone H3-R2 methylation(GO:0034970) |
| 0.1 | 0.6 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.1 | 1.6 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.1 | 2.1 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) |
| 0.1 | 0.5 | GO:1902965 | regulation of protein localization to early endosome(GO:1902965) positive regulation of protein localization to early endosome(GO:1902966) |
| 0.1 | 0.8 | GO:0000710 | meiotic mismatch repair(GO:0000710) |
| 0.1 | 0.3 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 0.1 | 4.0 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.1 | 0.3 | GO:0090310 | negative regulation of methylation-dependent chromatin silencing(GO:0090310) |
| 0.1 | 0.1 | GO:0019336 | phenol-containing compound catabolic process(GO:0019336) |
| 0.1 | 0.2 | GO:0048633 | positive regulation of skeletal muscle tissue growth(GO:0048633) |
| 0.1 | 2.3 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.1 | 7.3 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.1 | 0.6 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.1 | 0.9 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.1 | 1.2 | GO:0043615 | astrocyte cell migration(GO:0043615) |
| 0.1 | 1.1 | GO:0044782 | cilium organization(GO:0044782) |
| 0.1 | 1.6 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.1 | 0.6 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.1 | 1.2 | GO:0002634 | regulation of germinal center formation(GO:0002634) |
| 0.1 | 0.3 | GO:0001544 | initiation of primordial ovarian follicle growth(GO:0001544) |
| 0.1 | 0.6 | GO:0006499 | N-terminal protein lipidation(GO:0006498) N-terminal protein myristoylation(GO:0006499) |
| 0.1 | 0.6 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 0.1 | 2.0 | GO:0086016 | AV node cell action potential(GO:0086016) AV node cell to bundle of His cell signaling(GO:0086027) |
| 0.1 | 1.5 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.1 | 0.5 | GO:0030070 | insulin processing(GO:0030070) |
| 0.1 | 0.9 | GO:1904153 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.1 | 1.6 | GO:0031441 | negative regulation of mRNA 3'-end processing(GO:0031441) |
| 0.1 | 1.0 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.1 | 2.1 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 0.5 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.1 | 0.7 | GO:0030917 | midbrain-hindbrain boundary development(GO:0030917) |
| 0.1 | 1.1 | GO:2000580 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.1 | 0.6 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.1 | 0.4 | GO:2000323 | negative regulation of glucocorticoid receptor signaling pathway(GO:2000323) |
| 0.1 | 0.4 | GO:0045074 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.1 | 0.3 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.1 | 1.6 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.1 | 0.3 | GO:0045719 | negative regulation of glycogen biosynthetic process(GO:0045719) |
| 0.1 | 0.3 | GO:0097056 | selenocysteinyl-tRNA(Sec) biosynthetic process(GO:0097056) |
| 0.1 | 0.2 | GO:0060024 | rhythmic synaptic transmission(GO:0060024) |
| 0.1 | 0.6 | GO:0097068 | response to thyroxine(GO:0097068) response to L-phenylalanine derivative(GO:1904386) |
| 0.1 | 0.4 | GO:0075044 | autophagy of host cells involved in interaction with symbiont(GO:0075044) autophagy involved in symbiotic interaction(GO:0075071) |
| 0.1 | 0.6 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.1 | 2.3 | GO:0045956 | positive regulation of calcium ion-dependent exocytosis(GO:0045956) |
| 0.1 | 0.3 | GO:0002086 | diaphragm contraction(GO:0002086) |
| 0.1 | 4.1 | GO:0051973 | positive regulation of telomerase activity(GO:0051973) |
| 0.1 | 0.4 | GO:0072364 | regulation of cellular ketone metabolic process by regulation of transcription from RNA polymerase II promoter(GO:0072364) |
| 0.1 | 1.0 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.1 | 0.5 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.1 | 0.4 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.1 | 0.4 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) |
| 0.1 | 0.4 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 0.1 | 0.4 | GO:0015692 | lead ion transport(GO:0015692) |
| 0.1 | 0.8 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.1 | 2.8 | GO:0046710 | GDP metabolic process(GO:0046710) |
| 0.1 | 0.8 | GO:0015824 | proline transport(GO:0015824) |
| 0.1 | 0.4 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.1 | 1.1 | GO:0032310 | prostaglandin secretion(GO:0032310) |
| 0.1 | 0.2 | GO:1903697 | negative regulation of microvillus assembly(GO:1903697) |
| 0.1 | 1.1 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.1 | 0.1 | GO:0019046 | release from viral latency(GO:0019046) |
| 0.1 | 6.6 | GO:0043647 | inositol phosphate metabolic process(GO:0043647) |
| 0.1 | 0.7 | GO:0007097 | nuclear migration(GO:0007097) |
| 0.1 | 0.3 | GO:1901563 | response to camptothecin(GO:1901563) |
| 0.1 | 0.1 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 0.1 | 0.5 | GO:1904690 | regulation of cap-independent translational initiation(GO:1903677) positive regulation of cap-independent translational initiation(GO:1903679) regulation of cytoplasmic translational initiation(GO:1904688) positive regulation of cytoplasmic translational initiation(GO:1904690) |
| 0.1 | 10.7 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.1 | 0.3 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.1 | 8.9 | GO:0030449 | regulation of complement activation(GO:0030449) |
| 0.1 | 2.1 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.1 | 0.4 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.1 | 2.2 | GO:0072348 | sulfur compound transport(GO:0072348) |
| 0.1 | 1.0 | GO:0043982 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.1 | 0.2 | GO:0043974 | histone H3-K27 acetylation(GO:0043974) regulation of histone H3-K27 acetylation(GO:1901674) negative regulation of histone H3-K27 acetylation(GO:1901675) |
| 0.1 | 0.4 | GO:0071816 | tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.1 | 1.4 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.1 | 0.2 | GO:0060166 | olfactory pit development(GO:0060166) |
| 0.1 | 0.7 | GO:0034244 | negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.1 | 0.3 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.1 | 0.8 | GO:1990416 | cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.1 | 0.4 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.1 | 1.2 | GO:0044342 | type B pancreatic cell proliferation(GO:0044342) |
| 0.1 | 0.6 | GO:0006098 | pentose-phosphate shunt(GO:0006098) |
| 0.1 | 0.4 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.1 | 2.6 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.1 | 0.2 | GO:0007518 | myoblast fate determination(GO:0007518) |
| 0.1 | 1.4 | GO:0010954 | positive regulation of protein processing(GO:0010954) |
| 0.1 | 0.2 | GO:0071409 | cellular response to cycloheximide(GO:0071409) |
| 0.1 | 0.2 | GO:0009138 | pyrimidine nucleoside diphosphate metabolic process(GO:0009138) |
| 0.1 | 0.5 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.1 | 0.8 | GO:0035360 | positive regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035360) |
| 0.1 | 0.7 | GO:0071880 | adenylate cyclase-activating adrenergic receptor signaling pathway(GO:0071880) |
| 0.1 | 0.9 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.1 | 0.7 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.1 | 1.5 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 1.2 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.0 | 0.1 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.0 | 0.1 | GO:0003221 | right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 0.0 | 0.3 | GO:0032927 | positive regulation of activin receptor signaling pathway(GO:0032927) |
| 0.0 | 0.1 | GO:1903031 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.0 | 0.6 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.0 | 0.2 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.0 | 0.2 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.0 | 0.3 | GO:0090206 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.0 | 0.5 | GO:0009048 | dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.0 | 0.2 | GO:0032849 | positive regulation of cellular pH reduction(GO:0032849) |
| 0.0 | 0.2 | GO:0034770 | histone H4-K20 methylation(GO:0034770) |
| 0.0 | 0.7 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.0 | 0.1 | GO:0045200 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.0 | 0.3 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.0 | 1.0 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 0.3 | GO:0071105 | response to interleukin-11(GO:0071105) |
| 0.0 | 0.4 | GO:0035865 | cellular response to potassium ion(GO:0035865) |
| 0.0 | 0.3 | GO:1902255 | positive regulation of intrinsic apoptotic signaling pathway by p53 class mediator(GO:1902255) positive regulation of protein monoubiquitination(GO:1902527) |
| 0.0 | 0.8 | GO:1904380 | endoplasmic reticulum mannose trimming(GO:1904380) |
| 0.0 | 0.2 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.0 | 0.9 | GO:0072189 | ureter development(GO:0072189) |
| 0.0 | 0.3 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.0 | 0.3 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.0 | 1.4 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.0 | 0.3 | GO:1902915 | negative regulation of histone ubiquitination(GO:0033183) histone H2A K63-linked ubiquitination(GO:0070535) negative regulation of protein K63-linked ubiquitination(GO:1900045) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.0 | 0.7 | GO:1903071 | positive regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903071) |
| 0.0 | 0.1 | GO:1902570 | protein localization to nucleolus(GO:1902570) |
| 0.0 | 0.8 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.0 | 0.1 | GO:0070781 | response to biotin(GO:0070781) |
| 0.0 | 0.7 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.1 | GO:0010716 | negative regulation of extracellular matrix disassembly(GO:0010716) |
| 0.0 | 0.1 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 0.0 | 0.2 | GO:0060965 | negative regulation of gene silencing by miRNA(GO:0060965) |
| 0.0 | 0.5 | GO:0035826 | rubidium ion transport(GO:0035826) regulation of rubidium ion transport(GO:2000680) |
| 0.0 | 0.2 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.0 | 0.4 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.0 | 0.3 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.1 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.0 | 0.2 | GO:0071353 | cellular response to interleukin-4(GO:0071353) |
| 0.0 | 0.8 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.2 | GO:0009411 | response to UV(GO:0009411) |
| 0.0 | 0.6 | GO:0098828 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.0 | 0.4 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 0.2 | GO:0035633 | maintenance of blood-brain barrier(GO:0035633) |
| 0.0 | 1.9 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.5 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.0 | 0.2 | GO:0007042 | lysosomal lumen acidification(GO:0007042) |
| 0.0 | 0.1 | GO:1902187 | regulation of viral release from host cell(GO:1902186) negative regulation of viral release from host cell(GO:1902187) |
| 0.0 | 0.3 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 | 2.6 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 1.1 | GO:0010863 | positive regulation of phospholipase C activity(GO:0010863) |
| 0.0 | 0.3 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.0 | 0.2 | GO:1903998 | regulation of eating behavior(GO:1903998) |
| 0.0 | 0.2 | GO:1904354 | negative regulation of telomere capping(GO:1904354) |
| 0.0 | 0.1 | GO:0045408 | regulation of interleukin-6 biosynthetic process(GO:0045408) |
| 0.0 | 0.7 | GO:0042789 | mRNA transcription from RNA polymerase II promoter(GO:0042789) |
| 0.0 | 0.6 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.0 | 0.6 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.4 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 0.3 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.0 | 0.3 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.0 | 0.2 | GO:1903756 | regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903756) negative regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903758) |
| 0.0 | 3.1 | GO:0006112 | energy reserve metabolic process(GO:0006112) |
| 0.0 | 0.1 | GO:0060355 | positive regulation of cell adhesion molecule production(GO:0060355) |
| 0.0 | 0.8 | GO:0061458 | reproductive system development(GO:0061458) |
| 0.0 | 0.6 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.0 | 0.2 | GO:0046826 | negative regulation of protein export from nucleus(GO:0046826) |
| 0.0 | 0.9 | GO:0007411 | axon guidance(GO:0007411) |
| 0.0 | 1.1 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 | 0.1 | GO:0051138 | positive regulation of NK T cell differentiation(GO:0051138) |
| 0.0 | 0.1 | GO:0003350 | pulmonary myocardium development(GO:0003350) |
| 0.0 | 0.2 | GO:1990034 | calcium ion export(GO:1901660) calcium ion export from cell(GO:1990034) |
| 0.0 | 0.2 | GO:0045682 | regulation of epidermis development(GO:0045682) |
| 0.0 | 0.3 | GO:0045075 | interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 0.0 | 0.5 | GO:0006349 | regulation of gene expression by genetic imprinting(GO:0006349) |
| 0.0 | 0.7 | GO:2000758 | positive regulation of histone acetylation(GO:0035066) positive regulation of peptidyl-lysine acetylation(GO:2000758) |
| 0.0 | 0.2 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 1.4 | GO:0050885 | neuromuscular process controlling balance(GO:0050885) |
| 0.0 | 0.2 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.0 | 0.1 | GO:0045955 | negative regulation of calcium ion-dependent exocytosis(GO:0045955) |
| 0.0 | 0.4 | GO:0010832 | negative regulation of myotube differentiation(GO:0010832) |
| 0.0 | 0.4 | GO:0045786 | negative regulation of cell cycle(GO:0045786) |
| 0.0 | 0.4 | GO:2000696 | regulation of epithelial cell differentiation involved in kidney development(GO:2000696) |
| 0.0 | 0.2 | GO:0055129 | L-proline biosynthetic process(GO:0055129) |
| 0.0 | 0.1 | GO:0072429 | response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 0.0 | 0.9 | GO:0016574 | histone ubiquitination(GO:0016574) |
| 0.0 | 2.2 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.0 | 1.2 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.0 | 0.2 | GO:0051665 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) membrane raft localization(GO:0051665) |
| 0.0 | 0.3 | GO:0016055 | Wnt signaling pathway(GO:0016055) cell-cell signaling by wnt(GO:0198738) |
| 0.0 | 0.1 | GO:0090219 | negative regulation of lipid kinase activity(GO:0090219) |
| 0.0 | 0.9 | GO:0006362 | transcription elongation from RNA polymerase I promoter(GO:0006362) |
| 0.0 | 0.4 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.0 | 0.3 | GO:0036149 | phosphatidylinositol acyl-chain remodeling(GO:0036149) |
| 0.0 | 1.0 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.8 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 0.2 | GO:0042048 | olfactory behavior(GO:0042048) |
| 0.0 | 1.3 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 0.1 | GO:0042472 | inner ear morphogenesis(GO:0042472) |
| 0.0 | 0.7 | GO:0044275 | cellular carbohydrate catabolic process(GO:0044275) |
| 0.0 | 0.3 | GO:1900016 | negative regulation of cytokine production involved in inflammatory response(GO:1900016) |
| 0.0 | 0.5 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.1 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 0.2 | GO:0060736 | prostate gland growth(GO:0060736) |
| 0.0 | 0.2 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 0.5 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 0.2 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 | 0.2 | GO:0010667 | negative regulation of cardiac muscle cell apoptotic process(GO:0010667) |
| 0.0 | 1.2 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 1.0 | GO:0015949 | nucleobase-containing small molecule interconversion(GO:0015949) |
| 0.0 | 2.8 | GO:0008584 | male gonad development(GO:0008584) development of primary male sexual characteristics(GO:0046546) |
| 0.0 | 0.8 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.0 | 0.1 | GO:0002528 | regulation of vascular permeability involved in acute inflammatory response(GO:0002528) negative regulation of mesenchymal cell proliferation(GO:0072201) |
| 0.0 | 0.1 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.0 | 0.1 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) inositol phosphate-mediated signaling(GO:0048016) |
| 0.0 | 0.2 | GO:0060124 | positive regulation of growth hormone secretion(GO:0060124) |
| 0.0 | 0.7 | GO:0071549 | cellular response to dexamethasone stimulus(GO:0071549) |
| 0.0 | 0.3 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.0 | 0.7 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.0 | 0.2 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.2 | GO:0034243 | regulation of transcription elongation from RNA polymerase II promoter(GO:0034243) |
| 0.0 | 0.3 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.0 | 1.1 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.0 | 0.6 | GO:2000144 | positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.0 | 0.3 | GO:0048706 | embryonic skeletal system development(GO:0048706) |
| 0.0 | 0.0 | GO:0009597 | detection of virus(GO:0009597) |
| 0.0 | 0.1 | GO:1901162 | primary amino compound biosynthetic process(GO:1901162) |
| 0.0 | 0.1 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 | 0.1 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 0.0 | 0.1 | GO:0051418 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.0 | 0.4 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.0 | 0.0 | GO:0046726 | positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.0 | 0.1 | GO:0032057 | negative regulation of translational initiation in response to stress(GO:0032057) |
| 0.0 | 0.1 | GO:0015742 | alpha-ketoglutarate transport(GO:0015742) |
| 0.0 | 0.1 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.0 | 0.3 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.4 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.0 | 0.9 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.0 | 0.1 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.9 | GO:0032755 | positive regulation of interleukin-6 production(GO:0032755) |
| 0.0 | 0.3 | GO:0010804 | negative regulation of tumor necrosis factor-mediated signaling pathway(GO:0010804) |
| 0.0 | 0.3 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 0.5 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.0 | 0.2 | GO:0002755 | MyD88-dependent toll-like receptor signaling pathway(GO:0002755) |
| 0.0 | 0.2 | GO:0045648 | positive regulation of erythrocyte differentiation(GO:0045648) |
| 0.0 | 0.1 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.0 | 1.0 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.0 | 0.2 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.0 | 0.1 | GO:0043586 | tongue development(GO:0043586) |
| 0.0 | 0.6 | GO:0030317 | sperm motility(GO:0030317) |
| 0.0 | 0.1 | GO:0019985 | translesion synthesis(GO:0019985) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 4.1 | GO:0043291 | RAVE complex(GO:0043291) |
| 1.0 | 9.3 | GO:1990761 | growth cone lamellipodium(GO:1990761) |
| 0.6 | 12.9 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.5 | 1.6 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.5 | 3.0 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.4 | 1.3 | GO:0072534 | perineuronal net(GO:0072534) |
| 0.3 | 1.7 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.3 | 2.7 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.3 | 4.7 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.3 | 0.9 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.3 | 0.3 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.3 | 0.8 | GO:0031021 | interphase microtubule organizing center(GO:0031021) |
| 0.3 | 3.4 | GO:0098799 | outer mitochondrial membrane protein complex(GO:0098799) |
| 0.2 | 2.5 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.2 | 1.6 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.2 | 0.4 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.2 | 0.8 | GO:0032301 | MutSalpha complex(GO:0032301) |
| 0.2 | 2.6 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.2 | 0.8 | GO:0097362 | MCM8-MCM9 complex(GO:0097362) |
| 0.2 | 1.5 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.2 | 8.0 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.2 | 1.6 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.2 | 2.8 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.2 | 0.5 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.1 | 0.4 | GO:0005715 | late recombination nodule(GO:0005715) |
| 0.1 | 0.4 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.1 | 0.7 | GO:0001740 | Barr body(GO:0001740) |
| 0.1 | 0.8 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.1 | 0.6 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.1 | 0.8 | GO:0000835 | ER ubiquitin ligase complex(GO:0000835) |
| 0.1 | 1.1 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.1 | 0.6 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.1 | 0.2 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.1 | 0.9 | GO:0072487 | MSL complex(GO:0072487) |
| 0.1 | 0.7 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.1 | 0.5 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) |
| 0.1 | 0.5 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.1 | 0.8 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.1 | 1.6 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.1 | 0.7 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.1 | 2.2 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.1 | 0.8 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.1 | 5.5 | GO:0000786 | nucleosome(GO:0000786) |
| 0.1 | 0.4 | GO:0071818 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.1 | 0.4 | GO:0089717 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.1 | 2.0 | GO:0071682 | endocytic vesicle lumen(GO:0071682) |
| 0.1 | 0.1 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.1 | 4.7 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.1 | 0.4 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.1 | 0.8 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.1 | 0.3 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.1 | 2.0 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.1 | 0.2 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.1 | 0.5 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.1 | 0.4 | GO:0070695 | FHF complex(GO:0070695) |
| 0.1 | 0.2 | GO:0031166 | integral component of vacuolar membrane(GO:0031166) |
| 0.1 | 1.6 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.1 | 0.7 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.1 | 0.9 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 0.5 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.1 | 1.0 | GO:0043220 | Schmidt-Lanterman incisure(GO:0043220) |
| 0.1 | 10.1 | GO:0031514 | motile cilium(GO:0031514) |
| 0.1 | 5.4 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 0.1 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.0 | 0.2 | GO:0000333 | telomerase catalytic core complex(GO:0000333) |
| 0.0 | 0.6 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 3.8 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.0 | 1.1 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.3 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.9 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 0.5 | GO:0032009 | early phagosome(GO:0032009) |
| 0.0 | 0.3 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.2 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 2.3 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 3.0 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.1 | GO:0008275 | gamma-tubulin small complex(GO:0008275) |
| 0.0 | 0.5 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 1.1 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.6 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.5 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 0.2 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.0 | 0.4 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 1.6 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.3 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 4.5 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 0.7 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.0 | 1.7 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.2 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.7 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.0 | 3.9 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 0.8 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.6 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.1 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.0 | 0.9 | GO:0090544 | BAF-type complex(GO:0090544) |
| 0.0 | 0.5 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.0 | 0.2 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.0 | 0.7 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 1.3 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.2 | GO:0098651 | basement membrane collagen trimer(GO:0098651) |
| 0.0 | 0.1 | GO:0031213 | RSF complex(GO:0031213) |
| 0.0 | 0.1 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.2 | GO:0061200 | clathrin-sculpted gamma-aminobutyric acid transport vesicle(GO:0061200) clathrin-sculpted gamma-aminobutyric acid transport vesicle membrane(GO:0061202) |
| 0.0 | 0.3 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 2.6 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.2 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.1 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.5 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 0.5 | GO:0031304 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.1 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.0 | 0.3 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.0 | 0.6 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.2 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.4 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 1.2 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.2 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.3 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 0.2 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.8 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.4 | GO:0005798 | Golgi-associated vesicle(GO:0005798) |
| 0.0 | 0.1 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.2 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.2 | GO:0031970 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.0 | 0.3 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.0 | 0.3 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 0.3 | GO:0032040 | small-subunit processome(GO:0032040) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.2 | 12.5 | GO:0052810 | 1-phosphatidylinositol-5-kinase activity(GO:0052810) |
| 1.4 | 12.8 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.8 | 5.0 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.7 | 2.7 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.6 | 8.4 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.6 | 1.7 | GO:0031783 | corticotropin hormone receptor binding(GO:0031780) type 5 melanocortin receptor binding(GO:0031783) |
| 0.5 | 6.5 | GO:0019826 | oxygen sensor activity(GO:0019826) |
| 0.5 | 2.8 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.5 | 1.4 | GO:0016435 | rRNA (guanine) methyltransferase activity(GO:0016435) |
| 0.5 | 5.5 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.4 | 16.4 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.4 | 4.8 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.4 | 1.6 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.4 | 7.0 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.4 | 2.2 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.3 | 1.6 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.3 | 14.5 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.3 | 0.3 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 0.3 | 1.1 | GO:0016404 | 15-hydroxyprostaglandin dehydrogenase (NAD+) activity(GO:0016404) |
| 0.2 | 1.2 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.2 | 0.7 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.2 | 1.7 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.2 | 0.9 | GO:0004803 | transposase activity(GO:0004803) |
| 0.2 | 2.7 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.2 | 0.6 | GO:0008808 | cardiolipin synthase activity(GO:0008808) phosphatidyltransferase activity(GO:0030572) CDP-diacylglycerol-phosphatidylglycerol phosphatidyltransferase activity(GO:0043337) |
| 0.2 | 0.8 | GO:0032143 | single thymine insertion binding(GO:0032143) |
| 0.2 | 0.6 | GO:0102008 | cytosolic dipeptidase activity(GO:0102008) |
| 0.2 | 5.8 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.2 | 0.6 | GO:0017057 | 6-phosphogluconolactonase activity(GO:0017057) |
| 0.2 | 0.6 | GO:0015117 | thiosulfate transmembrane transporter activity(GO:0015117) oxaloacetate transmembrane transporter activity(GO:0015131) |
| 0.2 | 0.4 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.2 | 0.7 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.2 | 6.4 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.2 | 0.8 | GO:0033857 | diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.2 | 1.0 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.2 | 1.1 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.2 | 1.7 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.2 | 0.9 | GO:0004882 | androgen receptor activity(GO:0004882) |
| 0.2 | 1.4 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.2 | 0.8 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.1 | 0.4 | GO:0004828 | serine-tRNA ligase activity(GO:0004828) |
| 0.1 | 1.0 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.1 | 0.8 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.1 | 0.5 | GO:0043398 | HLH domain binding(GO:0043398) |
| 0.1 | 0.3 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 0.1 | 0.4 | GO:0001225 | RNA polymerase II transcription coactivator binding(GO:0001225) |
| 0.1 | 0.9 | GO:0001165 | RNA polymerase I upstream control element sequence-specific DNA binding(GO:0001165) |
| 0.1 | 4.2 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.1 | 1.3 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.1 | 1.7 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.1 | 0.5 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.1 | 0.3 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.1 | 0.3 | GO:0097158 | pre-mRNA intronic pyrimidine-rich binding(GO:0097158) |
| 0.1 | 0.4 | GO:0032137 | guanine/thymine mispair binding(GO:0032137) |
| 0.1 | 0.3 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.1 | 0.5 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.1 | 0.7 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.1 | 1.6 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.1 | 4.4 | GO:0016409 | palmitoyltransferase activity(GO:0016409) |
| 0.1 | 1.5 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.1 | 0.4 | GO:0035241 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) |
| 0.1 | 0.5 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.1 | 2.1 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.1 | 0.3 | GO:0004781 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.1 | 4.2 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.1 | 1.1 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.1 | 0.4 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 0.5 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.1 | 0.4 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.1 | 2.5 | GO:0016505 | peptidase activator activity involved in apoptotic process(GO:0016505) |
| 0.1 | 1.0 | GO:0031386 | protein tag(GO:0031386) |
| 0.1 | 1.6 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.1 | 0.8 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.1 | 2.4 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 0.7 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.1 | 1.2 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.1 | 0.5 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.1 | 0.4 | GO:0015086 | cadmium ion transmembrane transporter activity(GO:0015086) cobalt ion transmembrane transporter activity(GO:0015087) lead ion transmembrane transporter activity(GO:0015094) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.1 | 5.5 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.1 | 0.2 | GO:0047322 | [hydroxymethylglutaryl-CoA reductase (NADPH)] kinase activity(GO:0047322) [acetyl-CoA carboxylase] kinase activity(GO:0050405) |
| 0.1 | 2.9 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.1 | 1.6 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.1 | 2.1 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 0.8 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.1 | 2.0 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.1 | 2.8 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.1 | 0.3 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.1 | 2.6 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.1 | 0.2 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.1 | 1.1 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.1 | 0.5 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.1 | 1.2 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.1 | 1.7 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.1 | 0.9 | GO:0043996 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.1 | 0.4 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.1 | 0.6 | GO:0051430 | corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.1 | 3.5 | GO:0061733 | peptide-lysine-N-acetyltransferase activity(GO:0061733) |
| 0.1 | 6.2 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.1 | 0.3 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.1 | 1.1 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.1 | 0.9 | GO:0016594 | glycine binding(GO:0016594) |
| 0.1 | 1.9 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.1 | 0.5 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.1 | 0.9 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.1 | 0.8 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.1 | 0.5 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.1 | 0.5 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.0 | 0.6 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.6 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.0 | 0.6 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.3 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.0 | 1.0 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.1 | GO:0015140 | malate transmembrane transporter activity(GO:0015140) |
| 0.0 | 1.2 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.2 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.0 | 1.4 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.9 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.4 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.0 | 1.8 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.0 | 0.1 | GO:0005534 | galactose binding(GO:0005534) |
| 0.0 | 0.3 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.3 | GO:0001640 | adenylate cyclase inhibiting G-protein coupled glutamate receptor activity(GO:0001640) G-protein coupled glutamate receptor activity(GO:0098988) |
| 0.0 | 1.1 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.1 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.0 | 0.7 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.1 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) |
| 0.0 | 0.2 | GO:0008525 | phosphatidylcholine transporter activity(GO:0008525) |
| 0.0 | 0.5 | GO:0043175 | RNA polymerase core enzyme binding(GO:0043175) |
| 0.0 | 0.3 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.3 | GO:0047144 | 2-acylglycerol-3-phosphate O-acyltransferase activity(GO:0047144) |
| 0.0 | 7.1 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.2 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.9 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.5 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 1.6 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.4 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 1.6 | GO:0016620 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor(GO:0016620) |
| 0.0 | 0.7 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.1 | GO:0047273 | galactosylgalactosylglucosylceramide beta-D-acetylgalactosaminyltransferase activity(GO:0047273) |
| 0.0 | 1.4 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.2 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.0 | 0.2 | GO:0003696 | satellite DNA binding(GO:0003696) |
| 0.0 | 0.8 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 1.1 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 1.8 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.0 | 1.8 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 10.9 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.0 | 0.2 | GO:0016618 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.0 | 0.1 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.1 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.0 | 0.1 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.0 | 0.2 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.0 | 0.3 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.1 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.0 | 0.6 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.1 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.0 | 2.8 | GO:0005496 | steroid binding(GO:0005496) |
| 0.0 | 2.4 | GO:0005057 | receptor signaling protein activity(GO:0005057) |
| 0.0 | 0.0 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 0.3 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.0 | 0.4 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.0 | 0.4 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 0.2 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.0 | 0.4 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.0 | 0.1 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) |
| 0.0 | 0.4 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.5 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.4 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.1 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.0 | 1.0 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 0.2 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.2 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 1.4 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 0.2 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.4 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.8 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 2.8 | GO:0016853 | isomerase activity(GO:0016853) |
| 0.0 | 0.7 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.2 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.7 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 1.2 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 0.3 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.0 | 0.1 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.1 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.2 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 2.2 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.1 | GO:0039552 | RIG-I binding(GO:0039552) |
| 0.0 | 0.3 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 0.4 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.1 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.8 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.0 | 1.4 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 1.6 | GO:0042626 | ATPase activity, coupled to transmembrane movement of substances(GO:0042626) |
| 0.0 | 0.2 | GO:0016671 | oxidoreductase activity, acting on a sulfur group of donors, disulfide as acceptor(GO:0016671) |
| 0.0 | 0.6 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.0 | 0.4 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.2 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.4 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.2 | GO:0015166 | polyol transmembrane transporter activity(GO:0015166) |
| 0.0 | 0.2 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 0.2 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.3 | GO:0043548 | phosphatidylinositol 3-kinase binding(GO:0043548) |
| 0.0 | 0.5 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.0 | 0.1 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.1 | GO:0009374 | biotin binding(GO:0009374) |
| 0.0 | 0.5 | GO:0035257 | nuclear hormone receptor binding(GO:0035257) |
| 0.0 | 0.2 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 0.3 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.0 | 0.2 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.0 | 0.4 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.0 | 0.2 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.1 | GO:0000146 | microfilament motor activity(GO:0000146) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 13.8 | PID_WNT_CANONICAL_PATHWAY | Canonical Wnt signaling pathway |
| 0.2 | 7.6 | PID_HIF1A_PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.1 | 9.1 | PID_HES_HEY_PATHWAY | Notch-mediated HES/HEY network |
| 0.1 | 0.8 | ST_GA12_PATHWAY | G alpha 12 Pathway |
| 0.1 | 1.6 | PID_FAK_PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.1 | 0.8 | PID_INTEGRIN3_PATHWAY | Beta3 integrin cell surface interactions |
| 0.1 | 0.3 | PID_EPO_PATHWAY | EPO signaling pathway |
| 0.1 | 2.9 | ST_GRANULE_CELL_SURVIVAL_PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.1 | 8.8 | PID_ERA_GENOMIC_PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.1 | 4.7 | ST_G_ALPHA_I_PATHWAY | G alpha i Pathway |
| 0.1 | 8.0 | PID_AR_PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 1.5 | PID_SMAD2_3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.1 | 1.3 | PID_LPA4_PATHWAY | LPA4-mediated signaling events |
| 0.1 | 3.4 | PID_CDC42_REG_PATHWAY | Regulation of CDC42 activity |
| 0.0 | 2.0 | ST_JNK_MAPK_PATHWAY | JNK MAPK Pathway |
| 0.0 | 0.1 | PID_IL2_PI3K_PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.7 | ST_ADRENERGIC | Adrenergic Pathway |
| 0.0 | 0.9 | PID_LYSOPHOSPHOLIPID_PATHWAY | LPA receptor mediated events |
| 0.0 | 0.4 | PID_HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 1.3 | SIG_PIP3_SIGNALING_IN_B_LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 1.6 | PID_P38_ALPHA_BETA_DOWNSTREAM_PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 0.6 | PID_ARF6_DOWNSTREAM_PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.4 | SIG_IL4RECEPTOR_IN_B_LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.0 | 1.5 | PID_ECADHERIN_STABILIZATION_PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 1.2 | PID_ATM_PATHWAY | ATM pathway |
| 0.0 | 0.5 | PID_VEGFR1_PATHWAY | VEGFR1 specific signals |
| 0.0 | 0.6 | PID_HEDGEHOG_2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.8 | PID_THROMBIN_PAR1_PATHWAY | PAR1-mediated thrombin signaling events |
| 0.0 | 0.3 | PID_S1P_META_PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.0 | 0.6 | PID_GLYPICAN_1PATHWAY | Glypican 1 network |
| 0.0 | 1.0 | PID_DELTA_NP63_PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.3 | PID_INTEGRIN5_PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.3 | PID_AP1_PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.7 | PID_HNF3A_PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.3 | PID_PTP1B_PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 0.5 | PID_FAS_PATHWAY | FAS (CD95) signaling pathway |
| 0.0 | 0.2 | SA_CASPASE_CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.3 | PID_ALK1_PATHWAY | ALK1 signaling events |
| 0.0 | 0.2 | PID_HDAC_CLASSIII_PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.3 | PID_HDAC_CLASSI_PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.6 | NABA_PROTEOGLYCANS | Genes encoding proteoglycans |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 12.1 | REACTOME_SYNTHESIS_OF_PIPS_AT_THE_PLASMA_MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.2 | 0.8 | REACTOME_ELEVATION_OF_CYTOSOLIC_CA2_LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.2 | 3.6 | REACTOME_INITIAL_TRIGGERING_OF_COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.1 | 6.2 | REACTOME_OXYGEN_DEPENDENT_PROLINE_HYDROXYLATION_OF_HYPOXIA_INDUCIBLE_FACTOR_ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.1 | 3.3 | REACTOME_ABCA_TRANSPORTERS_IN_LIPID_HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.1 | 0.5 | REACTOME_RECEPTOR_LIGAND_BINDING_INITIATES_THE_SECOND_PROTEOLYTIC_CLEAVAGE_OF_NOTCH_RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.1 | 3.0 | REACTOME_NA_CL_DEPENDENT_NEUROTRANSMITTER_TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 1.8 | REACTOME_SIGNALING_BY_CONSTITUTIVELY_ACTIVE_EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.1 | 3.9 | REACTOME_RECYCLING_PATHWAY_OF_L1 | Genes involved in Recycling pathway of L1 |
| 0.1 | 2.6 | REACTOME_SIGNALING_BY_NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 1.0 | REACTOME_ROLE_OF_SECOND_MESSENGERS_IN_NETRIN1_SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.1 | 1.5 | REACTOME_SIGNAL_ATTENUATION | Genes involved in Signal attenuation |
| 0.1 | 2.1 | REACTOME_GABA_SYNTHESIS_RELEASE_REUPTAKE_AND_DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.1 | 1.4 | REACTOME_DOWNREGULATION_OF_SMAD2_3_SMAD4_TRANSCRIPTIONAL_ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.1 | 1.1 | REACTOME_HIGHLY_CALCIUM_PERMEABLE_POSTSYNAPTIC_NICOTINIC_ACETYLCHOLINE_RECEPTORS | Genes involved in Highly calcium permeable postsynaptic nicotinic acetylcholine receptors |
| 0.1 | 1.7 | REACTOME_INSULIN_SYNTHESIS_AND_PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.1 | 0.3 | REACTOME_SIGNALING_BY_ERBB4 | Genes involved in Signaling by ERBB4 |
| 0.1 | 0.7 | REACTOME_REGULATION_OF_INSULIN_SECRETION_BY_ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.1 | 1.5 | REACTOME_HOMOLOGOUS_RECOMBINATION_REPAIR_OF_REPLICATION_INDEPENDENT_DOUBLE_STRAND_BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 1.1 | REACTOME_ABC_FAMILY_PROTEINS_MEDIATED_TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.5 | REACTOME_ACTIVATION_OF_THE_AP1_FAMILY_OF_TRANSCRIPTION_FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 2.7 | REACTOME_MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.8 | REACTOME_SIGNAL_REGULATORY_PROTEIN_SIRP_FAMILY_INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 3.0 | REACTOME_PLC_BETA_MEDIATED_EVENTS | Genes involved in PLC beta mediated events |
| 0.0 | 1.3 | REACTOME_FORMATION_OF_TUBULIN_FOLDING_INTERMEDIATES_BY_CCT_TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 0.3 | REACTOME_ACYL_CHAIN_REMODELLING_OF_PI | Genes involved in Acyl chain remodelling of PI |
| 0.0 | 6.4 | REACTOME_FACTORS_INVOLVED_IN_MEGAKARYOCYTE_DEVELOPMENT_AND_PLATELET_PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.6 | REACTOME_ACYL_CHAIN_REMODELLING_OF_PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 0.4 | REACTOME_ABACAVIR_TRANSPORT_AND_METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 1.0 | REACTOME_DEGRADATION_OF_THE_EXTRACELLULAR_MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.8 | REACTOME_REGULATION_OF_INSULIN_LIKE_GROWTH_FACTOR_IGF_ACTIVITY_BY_INSULIN_LIKE_GROWTH_FACTOR_BINDING_PROTEINS_IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 1.2 | REACTOME_SULFUR_AMINO_ACID_METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 1.6 | REACTOME_ION_TRANSPORT_BY_P_TYPE_ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.4 | REACTOME_REGULATED_PROTEOLYSIS_OF_P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.0 | 0.1 | REACTOME_TRAF3_DEPENDENT_IRF_ACTIVATION_PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.0 | 0.5 | REACTOME_SIGNALING_BY_BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.8 | REACTOME_SIGNALING_BY_HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.2 | REACTOME_IRAK2_MEDIATED_ACTIVATION_OF_TAK1_COMPLEX_UPON_TLR7_8_OR_9_STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.4 | REACTOME_MEIOTIC_RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.5 | REACTOME_ACTIVATION_OF_RAC | Genes involved in Activation of Rac |
| 0.0 | 0.7 | REACTOME_TRAF6_MEDIATED_NFKB_ACTIVATION | Genes involved in TRAF6 mediated NF-kB activation |
| 0.0 | 0.5 | REACTOME_REGULATION_OF_AMPK_ACTIVITY_VIA_LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.0 | 0.7 | REACTOME_SYNTHESIS_AND_INTERCONVERSION_OF_NUCLEOTIDE_DI_AND_TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 1.2 | REACTOME_MRNA_SPLICING_MINOR_PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 0.5 | REACTOME_SIGNALING_BY_ROBO_RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.2 | REACTOME_ACTIVATION_OF_CHAPERONES_BY_ATF6_ALPHA | Genes involved in Activation of Chaperones by ATF6-alpha |
| 0.0 | 1.8 | REACTOME_MHC_CLASS_II_ANTIGEN_PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 0.5 | REACTOME_NEP_NS2_INTERACTS_WITH_THE_CELLULAR_EXPORT_MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.0 | 0.6 | REACTOME_ACTIVATION_OF_GENES_BY_ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 0.4 | REACTOME_MITOCHONDRIAL_TRNA_AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.2 | REACTOME_REVERSIBLE_HYDRATION_OF_CARBON_DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.1 | REACTOME_REGULATION_OF_THE_FANCONI_ANEMIA_PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 0.6 | REACTOME_G_ALPHA_Z_SIGNALLING_EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.1 | REACTOME_GAP_JUNCTION_DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.6 | REACTOME_RNA_POL_I_PROMOTER_OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.2 | REACTOME_RAP1_SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.6 | REACTOME_GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.3 | REACTOME_OTHER_SEMAPHORIN_INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 1.0 | REACTOME_NUCLEAR_RECEPTOR_TRANSCRIPTION_PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 2.2 | REACTOME_SIGNALING_BY_RHO_GTPASES | Genes involved in Signaling by Rho GTPases |